Abstract
Many models of sex-biased dispersal predict that the direction of sex-bias depends upon a species' mating system. In agreement with this, almost all polygynous mammals show male-biased dispersal whereas largely monogamous birds show female-biased dispersal (FBD). The hamadryas baboon (Papio hamadryas hamadryas) is polygynous and so dispersal is predicted to be male biased, as is found in all other baboon subspecies, but there are conflicting field data showing both female and male dispersal. Using 19 autosomal genetic markers genotyped in baboons from four Saudi Arabian populations, we found strong evidence for FBD in post-dispersal adults but not, as expected, in pre-dispersal infants and young juveniles, when we compared male and female: population structure (Fst), inbreeding (Fis), relatedness (r), and the mean assignment index (mAIc). Furthermore, we found evidence for female-biased gene flow as population genetic structure (Fst), was about four times higher for the paternally inherited Y, than for either autosomal markers or for maternally inherited mtDNA. These results contradict the direction of sex-bias predicted by the mating system and show that FBD has evolved recently from an ancestral state of male-biased dispersal. We suggest that the cost–benefit balance of dispersal to males and females is tightly linked to the unique hierarchical social structure of hamadryas baboons and that dispersal and social organization have coevolved.
Keywords: female-biased dispersal, gene flow, sex-specific markers, hamadryas baboons, polygyny
1. Introduction
Dispersal from the natal group in order to breed is a key life history trait that has important consequences for the genetic make-up of populations (Clobert et al. 2001), and is a trait that evolves in response to natural selection (Ferriere et al. 2000). Dispersal is often greater in one sex, and there is considerable debate as to what selective processes lead to sex-biased dispersal (Clobert et al. 2001). The main, non-mutually exclusive, evolutionary models that attempt to explain sex-biased dispersal can be broadly divided into those that are based upon competition among related females for resources (local resource competition, LRC; Greenwood 1980; Clarke et al. 1997), competition between related males for mates (local mate competition, LMC; Hamilton 1967; Dobson 1982; Moore & Ali 1984) and inbreeding avoidance (Bengtsson 1978; Packer 1979; Dobson 1982; Waser et al. 1986; Pusey 1987; Clutton-Brock 1989; Wolff 1994; Perrin & Mazalov 2000). A common prediction of many models is that in polygynous mating systems high LMC should lead to male-biased dispersal, whereas in monogamous species high LRC should lead to female-biased dispersal (FBD). In agreement with these predictions, dispersal is male biased in the majority of polygynous mammal species (Greenwood 1980; Dobson 1982), but typically female biased in monogamous, resource-defending birds (Greenwood 1980; Clarke et al. 1997). The simple mammal/bird comparison is, however, confounded by phylogeny (Pusey 1987; Harvey & Pagel 1991) and sex-bias in dispersal can vary with geographic scale (e.g. Fontanillas et al. 2004; Fraser et al. 2004). Investigations of species with atypical patterns of dispersal and mating, over a range of geographic scales are therefore needed to test the general validity of hypotheses (Pusey & Packer 1987), and to develop a more general framework to study the evolution of sex-biased dispersal (Goudet et al. 2002).
Only a few examples exist of mammalian species that contradict the typical pattern of male dispersal and female philopatry (for reviews see Greenwood 1980 and Dobson 1982). The hamadryas baboon (Papio hamadryas hamadryas) is one species that is often cited as having FBD (e.g. Pusey 1987; Pusey & Packer 1987; Stammbach 1987), however, observations of male dispersal have also been documented (Kummer 1968; Phillips-Conroy et al. 1992), and some authors have suggested that sex-biased dispersal is limited to local scales and so of little significance in hamadryas baboons (Sigg et al. 1982; Abbeglen 1984; Henzi & Barrett 2003; Yamane et al. 2003). Recently, it has been argued that low levels of population genetic differentiation for mitochondrial DNA (mtDNA) are consistent with female dispersal (Hapke et al. 2001; Winney et al. 2004), but without analysis of paternal and/or biparental markers it is impossible to reject the possibility of equal or greater male dispersal. The uncertainty as to whether dispersal is truly female biased is reflected by influential papers on the evolution of dispersal where hamadryas baboons have been categorized as exhibiting either female biased (Marks & Redmond 1987; Pusey 1987) or unbiased dispersal (Greenwood 1980; Dobson 1982; Clutton-Brock 1989). Importantly, all other subspecies of baboon are known to have male-biased dispersal (Packer 1979; Alberts & Altmann 1995), so, if confirmed, FBD in hamadryas baboons would represent a recent and phylogenetically independent example of FBD evolving from the ancestral state. A more thorough investigation of patterns of dispersal is therefore needed in this species.
Estimating sex-biased dispersal by direct observation is not always feasible, especially in animals with long life spans and/or long-distance dispersal (Koenig et al. 1996). Furthermore, direct observations do not allow quantification of how dispersal is translated into gene flow. Genetic methods, using markers of varying modes of inheritance, can give important insights into dispersal at varying geographic scales and also how dispersal relates to historical gene flow (Prugnolle & de Meeus 2002). However, there have been only a few studies investigating FBD in mammals using genetic methods (e.g. Favre et al. 1997; Seielstad et al. 1998; Banks et al. 2002; Bradley et al. 2004).
We tested the hypothesis that dispersal and gene flow in hamadryas baboons is female biased, using genetic data from both sex-specific and bi-parentally inherited markers. First, we tested whether dispersal from natal to breeding group is sex biased by comparing male and female population genetic structure based on 19 autosomal loci in a ‘pre-dispersal’ and a ‘post-dispersal’ sample of baboons from three Saudi Arabian populations (Goudet et al. 2002). This tests dispersal in one generation because in the following generation alleles are randomly assorted in the offspring (Prugnolle & de Meeus 2002). If dispersal is female biased, we expect greater genetic structuring in males than in females in our post-dispersal sample, but no sex differences in the pre-dispersal sample. Second, we investigated whether gene flow is sex biased by comparing the population genetic structure of Y chromosome, mtDNA and autosomal markers. Mitochondrial DNA and the majority of the Y chromosome (the exception being the pseudoautosomal region) do not recombine and are uniparentally inherited, so signatures of historical sex-biased gene flow are maintained in successive generations (Prugnolle & de Meeus 2002). If gene flow is female biased, we expect greater population structure for non-recombining regions of the Y chromosome than for mtDNA and autosomal markers (Laporte & Charlesworth 2002).
2. Material and methods
Hamadryas baboons are found in north-east Africa and western Arabia. In this study we had access to DNA samples from 298 individuals, sampled from four populations (Abha, Baha, Taif and Al-Akhal, figure 1) in the Asir mountain range in the west of Saudi Arabia. Of the 298 individuals 99.0% were of known sex and 87.2% were of known age. Details of how animals were sexed and aged, DNA samples collected and DNA extracted are given in Hammond et al. (submitted) and Winney et al. (2004). Hamadryas social structure is based on the ‘one male unit’ (OMU), which consists of an adult leader male, several adult females and juvenile and infant offspring. In African populations, it is known that several OMUs associate with one another to form clans (Sigg et al. 1982, Abegglen 1984), clans aggregate into bands, and bands congregate at sleeping cliffs to form troops (Henzi & Barrett 2003). In spite of this knowledge about the hierarchical social organization of hamadryas baboons, we had no information on the clan, band or troop structure in any of our four Arabian populations. The only information about social organization available to us was for the Abha population, where we knew the members of seven OMUs which accounted for 78 individuals from the total of 244 (Hammond et al. submitted). Because of this, we have conducted all our analyses at the level of the population.
Figure 1.
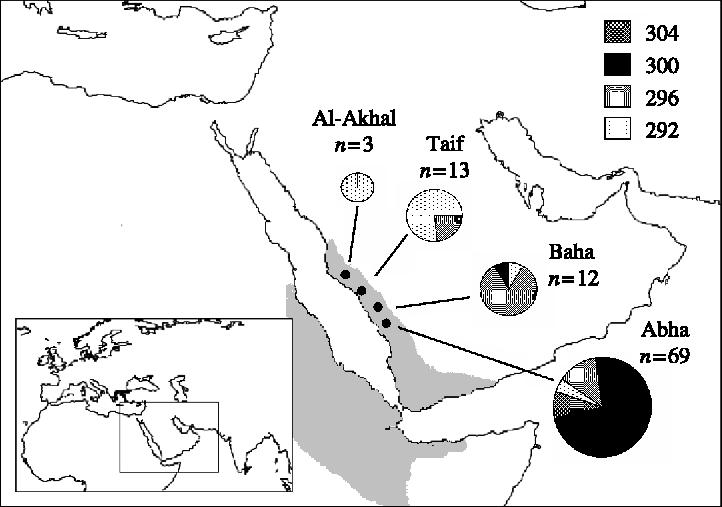
Location of populations sampled, number of males genotyped and allele frequency pie charts for the Y-linked microsatellite, DYS576. Key shows allele sizes in bp. Shaded areas show the approximate distribution of hamadryas baboons.
(a) Sex-biased dispersal
We analysed previously published data from 16 autosomal microsatellite and three autosomal protein loci to investigate sex-biased dispersal. Individuals were typed at an average of 17 autosomal loci (population averages: Abha, 16.8; Baha, 18.5; Taif, 18.6; Al-Akhal, 18.0) and details of loci and genotyping methods can be found in Hammond et al. (submitted). The Al-Akhal population was omitted from these analyses (but included in the ‘sex-biased gene flow’ analyses, see below) since there were no adult females sampled from this population.
We investigated whether dispersal was sex-biased following methods described by Goudet et al. (2002) and implemented in Fstat v.2.9 (Goudet 2001). We estimated: Fis, the level of inbreeding within a population relative to the whole sample; Fst, the proportion of genetic variation among populations (Weir & Cockerham 1984); r, the average relatedness of individuals within a population relative to the whole sample (where r=2Fst/(1+Fit)), and mAIc and vAIc which are, respectively, the mean and the variance of the assignment index (Favre et al. 1997; Mossman & Waser 1999). The p values were estimated using 10 000 randomizations. Fst, r and mAIc are expected to be higher in the philopatric sex, whereas Fis and vAIc should be lower (see Goudet et al. 2002 for further explanation).
Both a ‘pre-dispersal’ and ‘post-dispersal’ set of males and females were tested. The pre-dispersal set (total n=53 females; n=70 males; table 1) comprised male and female infants and young juveniles, the latter aged approximately 3 years or less. Our assumption is that individuals of both sexes in our pre-dispersal set are too young to have dispersed from their natal group, and so this analysis represents a null control. Our assumption that this age cohort is pre-dispersal is supported by parentage analysis, which showed that the majority of offspring of this age had parents in the focal offspring's OMU and so had not dispersed from their natal group (Hammond et al. submitted). As our prediction in the pre-dispersal set was no difference between males and females we used two-sided tests. The post-dispersal set comprised adult males and females (total n=64 females; n=26 males; table 2). In this case, we set males as the philopatric sex and used one-sided tests since our hypothesis was that dispersal in hamadryas baboons is female biased. The post-dispersal sample from Abha was heavily female biased (n=52 females versus n=16 males, table 2) in contrast to the pre-dispersal set which was mildly male biased (n=46 females versus n=60 males, table 1). Although simulations suggest that sex-biased sampling is of little consequence (Jerome Goudet, personal communication), others have suggested that it may be important (Bekkevold et al. 2004). We checked the effects of sex bias in the Abha sample by repeating the post-dispersal analysis on a series of 10 re-sampled datasets in which 16 adult females were randomly sampled without replacement from the total Abha sample of 52 adult females.
Table 1.
‘Pre-dispersal’ sex-biased dispersal analysis. (‘Pre-dispersal’ individuals were male or female infants or juveniles aged approximately 3 years or less. See methods section for definitions of Fis, Fst, r, mAIc, vAIc; p values are for two-sided tests based on 10 000 randomizations using Fstat; n/a=non-applicable.)
| number of individuals per population | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fis | Fst | r | mAIc | vAIc | Abha | Baha | Taif | |
| prediction | F=M | F=M | F=M | F=M | F=M | |||
| females | −0.006 | 0.167 | 0.287 | −0.071 | 15.123 | 46 | 3 | 4 |
| males | 0.034 | 0.148 | 0.252 | 0.054 | 14.114 | 60 | 4 | 6 |
| overall | 0.016 | 0.153 | 0.262 | n/a | n/a | |||
| p value | 0.284 | 0.457 | 0.351 | 0.851 | 0.826 | |||
Table 2.
‘Post-dispersal’ sex-biased dispersal analysis. (‘Post-dispersal’ individuals were adult males and females. See methods section for definitions of Fis, Fst, r, mAIc, vAIc; p values are for one-sided tests based on 10 000 randomizations using Fstat; n/a=non-applicable.)
| number of individuals per population | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fis | Fst | r | mAIc | vAIc | Abha | Baha | Taif | |
| prediction | F>M | F<M | F<M | F<M | F>M | |||
| females | 0.063 | 0.112 | 0.191 | −0.414 | 12.083 | 52 | 7 | 5 |
| males | −0.029 | 0.200 | 0.339 | 1.018 | 10.742 | 16 | 4 | 6 |
| overall | 0.037 | 0.146 | 0.249 | n/a | n/a | |||
| p value | 0.029 | 0.009 | 0.005 | 0.041 | 0.585 | |||
(b) Sex-biased gene flow
If gene flow is female biased we predict that Fst estimated from paternally inherited markers should be higher than that estimated from biparentally and maternally inherited markers, even when low male effective size caused by polygyny is taken into account (Laporte & Charlesworth 2002). We tested this by comparing the population genetic structure (Fst) of a human-derived Y-linked microsatellite (DYS576), 279 bp of mtDNA d-loop sequence and the 19 autosomal loci described above. DYS576 was genotyped in a sample of 97 males (Abha, n=69; Baha, n=12; Taif, n=13; Al-Akhal, n=3). This was the only polymorphic locus from a panel of seven microsatellites that were tested as part of a survey of Y chromosome polymorphism in Saudi Arabian hamadryas baboons (Lawson Handley et al. submitted). We tested whether allele frequencies for DYS576 differed among populations using exact tests, with the program R×C (Miller 1997a). Weir & Cockerham's (1984) estimate of Fst was calculated over all populations in Fstat for each of the three genomic regions (Y, mtDNA and autosomes). For mtDNA, Fst was calculated based on haplotype frequencies for a sample of 107 males and females (Abha, n=72; Baha, n=14; Taif, n=15; Al-Akhal, n=6) as described in Winney et al. (2004). For autosomal loci we calculated Fst for all available individuals (n=298) and for the sub-sample of 97 males that were genotyped at DYS576. Autosomal Fst was averaged over all loci, and the 99% confidence limits of the mean estimated by bootstrapping over loci (Goudet 2001). In addition, we calculated average Nei's genetic distance (Nei 1972; Nei 1978) for the Y, mtDNA and autosomal data (for the male only dataset) using the programme TFPGA (Miller 1997b).
3. Results
(a) Sex-biased dispersal
In our pre-dispersal analysis we found, as predicted, no significant difference between males and females in estimates of population structure (Fst and r), assignment index (mAIc and vAIc) or inbreeding (Fis; table 1). The genotypes of the pre-dispersal set, therefore, represent a random assortment of alleles from dispersing and non-dispersing parents.
In our post-dispersal analysis, in agreement with predictions based on FBD, we found that Fst and r were significantly lower in adult females than in adult males (both p<0.01, table 2), therefore, males show greater genetic structure among populations and are more related within populations. mAIc was negative for females and positive for males (p<0.05, table 2), hence males have a greater probability of being residents than do females. Fis was positive for females but negative for males, a pattern compatible with greater heterozygote deficit due to the Wahlund effect (since the sample is a mixture of residents and immigrants) in the dispersing sex (p<0.05). Although non-significant (p>0.1), variance in assignment index (vAIc) was larger for females than for males, and, therefore, consistent with FBD, even though the more than threefold greater number of females in the Abha sample would tend to bias a measure of variance in the opposite direction.
Equalizing the number of males and females in the Abha post-dispersal sample did not alter the overall results of our tests. In all 10 sub-sampled datasets Fst and relatedness were significantly higher in males compared to females (Fst mean p=0.016, range, 0.006–0.034; r mean p=0.010, range, 0.001–0.025). Fis was consistently positive in females and mAIc was consistently negative in females, although the mean p-value was, in both cases, just above formal significance (Fis mean p=0.051, range, 0.004–0.164; mAIc mean p=0.087, range, 0.009–0.230). The lack of overall significance for Fis and mAIc most likely arose because statistical power was lower in tests using the sub-sampled dataset. vAIc was not significant in any of the tests (mean p=0.598, range, 0.151–0.844).
(b) Sex-biased gene flow
Allele frequencies for the Y-linked microsatellite DYS576 (figure 1) were highly significantly different among the four populations (exact test p<0.0001), indicating strong genetic structure. Fst for DYS576 was much higher (Fst=0.541) than the average Fst for autosomal loci, when the latter was calculated both for the total sample (Fst=0.148; 99% confidence limits calculated by bootstrapping over loci: upper=0.224, lower=0.080) and for the set of males typed at DYS576 (Fst=0.145; 99% confidence limits: upper=0.232, lower=0.071), and it was also higher than Fst based on mtDNA haplotype frequencies (Fst=0.124). Nei's genetic distance (Nei 1972; Nei 1978) was highest for the Y chromosome (D=1.60), next highest for mtDNA (D=0.71) and lowest averaged over the 19 autosomal loci (D=0.23).
4. Discussion
We predicted greater genetic structuring at autosomal loci in males than in females post-dispersal if dispersal is female biased. Consistent with this prediction, we found significant differences between females and males in four out of five tests (all but vAIc) for our post-dispersal sample set, and that these were in the direction expected if dispersal is female biased (table 2). In contrast, we found no differences between the sexes in any tests for our pre-dispersal sample set (table 1). The power to detect sex-biased dispersal is positively correlated with bias intensity, sampling intensity and genetic variation per locus (Goudet 2001). The significance of our analyses, despite incomplete sampling and the relatively low genetic variation in our markers (Hammond et al. submitted), suggests that dispersal is heavily female biased in hamadryas baboons. Furthermore, the power to detect sex-bias declines faster as dispersal rate increases for vAIc than for Fst, r, Fis and mAIC (Goudet et al. 2002). The non-significance of vAIc compared to the significance of the other four statistics, therefore, suggests that the overall rate of dispersal is relatively high. Finally, our conclusion of FBD was not a result of female-biased sampling in Abha, as equalizing the sex ratio had little influence on the results. The analyses are, therefore, robust to sex-biased sampling and are highly consistent with dispersal being female biased.
Our results also support the prediction of greater population structure for paternal rather than maternal or biparental markers if gene flow is female biased. We found a high level of differentiation among populations for the Y-linked microsatellite (figure 1), and Fst for this locus was about four times higher than the average over 19 autosomal loci and for mtDNA. The ratio of Fst for these three markers is similar to that predicted theoretically when gene flow is female biased and males have a higher variance in reproductive success than females, as in species with polygynous mating systems (see fig. 3, panel C in Laporte & Charlesworth 2002). Our data fit less well with Fst ratios predicted for polygynous mating systems when dispersal is not sex biased (see fig. 3, panel A in Laporte & Charlesworth 2002). We must note that our estimate of Fst for the Y is based on only one microsatellite locus and additional variable loci on the Y would increase haplotype diversity. As Fst is sensitive to allelic diversity (Charlesworth 1998) our Y Fst estimate may decline by some unknown amount with additional Y haplotypes, however, Nei's genetic distance for the Y microsatellite, a measure of differentiation which is less sensitive than Fst to allelic diversity, was twice that for mtDNA and almost seven times greater than for the autosomal loci.
We conclude that both instantaneous dispersal and historical gene flow are female biased in hamadryas baboons. This conclusion contradicts the reports that categorize hamadryas dispersal as non sex biased (Greenwood 1980; Dobson 1982; Clutton-Brock 1989), or female biased at only a local level (Sigg et al. 1982; Abbeglen 1984; Henzi & Barrett 2003; Yamane et al. 2003). But they are consistent with other detailed field observations, which indicate that although young males may temporarily transfer into other clans or bands, they return to their natal clan to breed (Sigg et al. 1982). Females, on the other hand, disperse initially to adjacent OMUs within their natal clan, but (as illustrated also by our data) many eventually breed far from their natal group because secondary transfer to a different clan or band is common and return to the natal group is rare (e.g. Stammbach 1987).
Combined genetic and field data show that the hamadryas is one of the few exceptions to the general mammalian pattern of philopatric females and dispersing males. The hamadryas is also the only baboon subspecies with FBD, so this trait has evolved recently from the ancestral state of male-biased dispersal. Why does the direction of sex-biased dispersal in hamadryas baboons differ from all other baboons and most other mammals?
In terms of ultimate explanations for FBD, a polygynous mating system, such as that found in harem defending hamadryas baboons, predicts male-biased dispersal if competition among related males for mates (LMC) is higher than competition among related females for resources (LRC). Behavioural data have shown that males within clans do compete with one another for mates, and older males often lose breeding females to males within their clan (Abbeglen 1984), so LMC is likely to occur. In spite of this, our results contradict this prediction. FBD is predicted by a simple cost of inbreeding model (Waser et al. 1986) if the costs of inbreeding are greater to females than to males, as is likely the case in polygynous species where females invest more than males in offspring production and consequently have more to lose by mating with close relatives. Previous authors have interpreted FBD in hamadryas baboons as a means of preventing inbreeding, because all females leave their natal unit before reaching adulthood and so close genetic relatives of the opposite sex seldom reside in the same group as adults (Sigg et al. 1982). The simple inbreeding avoidance model, however, assumes that there is no LMC (Perrin & Mazalov 1999), which, as argued above, seems unlikely in hamadryas baboons. Furthermore, inbreeding avoidance does not explain secondary transfers of females, where females move social groups after they have left their natal group (Sigg et al. 1982).
In more proximate terms, behavioural observations (Sigg et al. 1982), and experiments where males and females were transferred between bands (Kummer 1968, Abbeglen 1984), have demonstrated that females are readily accepted into a new OMU, whereas males are usually forcefully ejected, and even if accepted do not get access to females. Philopatric males may also benefit by increased cooperation with male kin and observations have shown that male–male interaction within clans is more frequent than between clans. Furthermore, there is some evidence that males within clans cooperate to prevent males from other clans or bands abducting females (Sigg et al. 1982). During aggressive encounters between clans and bands females may become separated from their OMU and be abducted by males from other clans or bands (Abbeglen 1984). Indeed, adoption or abduction of females by males from outside their natal group could be an important proximate explanation for female dispersal in hamadryas baboons (Pusey & Packer 1987).
There is a close tie between the social organization of hamadryas baboons and the costs and benefits of female dispersal and male philopatry (Sterck et al. 1997). The explanations of the lower costs to female dispersal and the higher benefits to male philopatry raise further questions about what selective forces have lead to the evolution of the complex, hierarchical and unique social organization found in hamadryas baboons. Generally, sex-biased dispersal and social organization are more likely to coevolve (Sterck et al. 1997), than to evolve in isolation from one another, and models that integrate both sex-biased dispersal and social organization are therefore needed. Finally, the rare combination of polygyny and FBD found in hamadryas baboons are traits also shared by African great apes (Pusey & Packer 1987; Stokes et al. 2003) and many human societies (Seielstad et al. 1998). These common social features highlight the importance of hamadryas baboons in comparative studies of the evolution of human and ape societies.
Acknowledgments
The authors would like to thank Sylvain Biquand, Veronique Biquand and Ahmed Boug for collecting the samples used in this study, Anabelle Reber and Guillaume Emaresi for their enthusiastic contributions during their diploma work, and Jérôme Goudet for advice on the analysis. We would also like to thank two anonymous reviewers and the editor for helpful comments on a previous version of the MS. Financial support for this project was obtained from the University of Lausanne 2ème cycle funds for teaching.
Footnotes
Both authors contributed equally to this work.
Present address: Department of Genetics, University of Cambridge, Downing Street, Cambridge CB2 3EH, UK.
References
- Abbeglen J.J. Bucknell University Press; Lewisburg, PA: 1984. On socialization in hamadryas baboons. [Google Scholar]
- Alberts S.C, Altmann J. Balancing costs and opportunities—dispersal in male baboons. Am. Nat. 1995;145:279–306. doi:10.1086/285740 [Google Scholar]
- Banks S.C, Skerratt L.F, Taylor A.C. Female dispersal and relatedness structure in common wombats (Vombatus ursinus) J. Zool. 2002;256:389–399. doi:10.1017/S0952836902000432 [Google Scholar]
- Bekkevold D, Hansen M.M, Mensberg K.-L.D. Genetic detection of sex-specific dispersal in historical and contemporary populations of anadromous brown trout Salmo trutta. Mol. Ecol. 2004;13:1707–1712. doi: 10.1111/j.1365-294X.2004.02156.x. doi:10.1111/j.1365-294X.2004.02156.x [DOI] [PubMed] [Google Scholar]
- Bengtsson B.O. Avoiding inbreeding: at what cost? J. Theor. Biol. 1978;73:439–444. doi: 10.1016/0022-5193(78)90151-0. doi:10.1016/0022-5193(78)90151-0 [DOI] [PubMed] [Google Scholar]
- Bradley B.J, Doran-Sheehy D.M, Lukas D, Boesch C, Vigilant L. Dispersed male networks in western gorillas. Curr. Biol. 2004;14:510–513. doi: 10.1016/j.cub.2004.02.062. doi:10.1016/j.cub.2004.02.062 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Clarke A, Saether B, Roskaft E. Sex biases in avian dispersal: a reappraisal. Oikos. 1997;79:429–438. [Google Scholar]
- Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- Clutton-Brock T.H. Female transfer and inbreeding avoidance in social mammals. Nature. 1989;337:70–72. doi: 10.1038/337070a0. doi:10.1038/337070a0 [DOI] [PubMed] [Google Scholar]
- Dobson F.S. Competition for mates and predominant juvenile male dispersal in mammals. Anim. Behav. 1982;30:1183–1192. [Google Scholar]
- Favre L, Balloux F, Goudet J, Perrin N. Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field and microsatellite patterns. Proc. R. Soc. B. 1997;269:127–132. doi: 10.1098/rspb.1997.0019. doi:10.1098/rspb.1997.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriere R, Belthoff J, Olivieri I, Krackow S. Evolving dispersal: where to go next? Trends Ecol. Evol. 2000;15:5–7. doi: 10.1016/s0169-5347(99)01757-7. doi:10.1016/S0169-5347(99)01757-7 [DOI] [PubMed] [Google Scholar]
- Fontanillas P, Petit E, Perrin N. Estimating sex-specific dispersal rates with autosomal markers in hierarchically structured populations. Evolution. 2004;58:886–894. doi: 10.1111/j.0014-3820.2004.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fraser D.J, Lippe C, Bernatchez L. Consequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis) Mol. Ecol. 2004;13:67–80. doi: 10.1046/j.1365-294x.2003.02038.x. doi:10.1046/j.1365-294X.2003.02038.x [DOI] [PubMed] [Google Scholar]
- Goudet J. University of Lausanne; Lausanne: 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) Available from http://www.unil.ch/dee/page6767_en.html. [Google Scholar]
- Goudet J, Perrin N, Waser P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 2002;11:1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. doi:10.1046/j.1365-294X.2002.01496.x [DOI] [PubMed] [Google Scholar]
- Greenwood P.J. Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 1980;28:1140–1162. [Google Scholar]
- Hamilton W.D. Extraordinary sex ratios. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- Hammond, R. L., Winney, B. J., Macasero, W., Flores, B., Boug, A., Scheffrahn, W., Biquand, V., Biquand, S. & Bruford, M. W. Submitted. Kin structure in Arabian hamadryas baboons, Papio hamadryas hamadryas Behav. Ecol. Sociobiol.
- Hapke A, Zinner D, Zischler H. Mitochondrial DNA variation in Eritrean hamadryas baboons (Papio hamadryas hamadryas): life history influences population genetic structure. Behav. Ecol. Sociobiol. 2001;50:483–492. doi:10.1007/s002650100393 [Google Scholar]
- Harvey P.H, Pagel M.D. Oxford series in ecology and evolution. Oxford University Press; Oxford, UK: 1991. The comparative method in evolutionary biology. [Google Scholar]
- Henzi P, Barrett L. Evolutionary ecology, sexual conflict, and behavioural differentiation among baboon populations. Evol. Anthropol. 2003;12:217–230. doi:10.1002/evan.10121 [Google Scholar]
- Koenig W.D, van Vuren D, Hooge P.N. Detectability, philopatry, and the distribution of dispersal distances in vertebrates. Trends Ecol. Evol. 1996;11:514–517. doi: 10.1016/s0169-5347(96)20074-6. doi:10.1016/S0169-5347(96)20074-6 [DOI] [PubMed] [Google Scholar]
- Kummer H. Karger; Basel: 1968. Social organisation of hamadryas baboons: a field study. [Google Scholar]
- Laporte V, Charlesworth B. Effective population size and population subdivision in demographically structured populations. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley, L. J., Hammond, R. L., Emaresi, G., Reber, A. & Perrin, N. Submitted. Low Y chromosome polymorphism in Saudi-Arabian hamadryas baboons (Papio hamadryas hamadryas). Heredity [DOI] [PubMed]
- Marks J.S, Redmond R.L. Parent–offspring conflict and natal dispersal in birds and mammals: comments on the Oedipus hypothesis. Am. Nat. 1987;129:158–164. doi:10.1086/284627 [Google Scholar]
- Miller M.P. Department of Biological Sciences, North Arizona University; Flagstaff, AZ: 1997a. R×C: a program for the analysis of contingency tables. [Google Scholar]
- Miller M.P. Department of Biological Sciences, Northern Arizona University; Flagstaff, AZ: 1997b. Tools for population genetic analyses (TFPGA) 1.3: a Windows program for the analysis of allozyme and molecular population genetic data. [Google Scholar]
- Moore J, Ali R. Are dispersal and inbreeding avoidance related? Anim. Behav. 1984;32:94–112. [Google Scholar]
- Mossman C.A, Waser P.M. Genetic detection of sex-biased dispersal. Mol. Ecol. 1999;8:1063–1067. doi: 10.1046/j.1365-294x.1999.00652.x. doi:10.1046/j.1365-294x.1999.00652.x [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Am. Nat. 1972;106:283–292. doi:10.1086/282771 [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer C. Inter-troop transfer and inbreeding avoidance in Papio anubis. Anim. Behav. 1979;27:1–36. doi: 10.1016/0003-3472(79)90127-1. doi:10.1016/0003-3472(79)90126-X [DOI] [PubMed] [Google Scholar]
- Perrin N, Mazalov V. Dispersal and inbreeding avoidance. Am. Nat. 1999;154:282–292. doi: 10.1086/303236. doi:10.1086/303236 [DOI] [PubMed] [Google Scholar]
- Perrin N, Mazalov V. Local competition, inbreeding, and the evolution of sex-biased dispersal. Am. Nat. 2000;155:116–127. doi: 10.1086/303296. doi:10.1086/303296 [DOI] [PubMed] [Google Scholar]
- Phillips-Conroy J.E, Jolly C.J, Nystrom P, Hemmalin H.A. Migration of male hamadryas baboons into anubis groups in the Awash national park, Ethiopia. Int. J. Primatol. 1992;13:455–476. [Google Scholar]
- Prugnolle F, de Meeus T. Inferring sex-biased dispersal from population genetic tools: a review. Heredity. 2002;88:161–165. doi: 10.1038/sj.hdy.6800060. doi:10.1038/sj.hdy.6800060 [DOI] [PubMed] [Google Scholar]
- Pusey A.E. Sex-biased dispersal and inbreeding avoidance in mammals. Trends Ecol. Evol. 1987;2:295–299. doi: 10.1016/0169-5347(87)90081-4. doi:10.1016/0169-5347(87)90081-4 [DOI] [PubMed] [Google Scholar]
- Pusey A, Packer C. Dispersal and philopatry. In: Smuts B.B, Cheney D.L, Seyfarth R.M, Wrangham R.W, Struhsaker T.T, editors. Primate societies. University of Chicago Press; Chicago, IL: 1987. pp. 250–266. [Google Scholar]
- Seielstad M.T, Minch E, Cavalli-Sforza L.L. Genetic evidence for a higher female migration rate in humans. Nat. Genet. 1998;20:278–280. doi: 10.1038/3088. doi:10.1038/3088 [DOI] [PubMed] [Google Scholar]
- Sigg H, Stolba A, Abegglen J.-J, Dasser V. Life history of hamadryas baboons: physical development, infant mortality, reproductive parameters and family relationships. Primates. 1982;23:473–487. [Google Scholar]
- Stammbach E. Desert, forest and montane baboons: multilevel-societies. In: Smuts B.B, Cheney D.L, Seyfarth R.M, Wrangham R.W, Struhsaker T.T, editors. Primate societies. The University of Chicago Press; Chicago, IL: 1987. pp. 112–120. [Google Scholar]
- Sterck E.H.M, Watts D.P, van Schaik C.P. The evolution of female social relationships in nonhuman primates. Behav. Ecol. Sociobiol. 1997;41:291–309. doi:10.1007/s002650050390 [Google Scholar]
- Stokes E.J, Parnell R.J, Olejniczak C. Female dispersal and reproductive success in wild western lowland gorillas (Gorilla gorilla gorilla) Behav. Ecol. Sociobiol. 2003;54:329–339. doi:10.1007/s00265-003-0630-3 [Google Scholar]
- Waser P.M, Austad S.N, Keane B. When should animals tolerate inbreeding. Am. Nat. 1986;128:529–537. doi:10.1086/284585 [Google Scholar]
- Weir B.S, Cockerham C.O. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Winney B.J, Hammond R.L, Macasero W, Flores B, Bourg A, Biquand V, Biquand S, Bruford M.W. Crossing the Red sea: phylogeography of the hamadryas baboon, Papio hamadryas hamadryas. Mol. Ecol. 2004;13:2819–2827. doi: 10.1111/j.1365-294X.2004.02288.x. doi:10.1111/j.1365-294X.2004.02288.x [DOI] [PubMed] [Google Scholar]
- Wolff J.O. More on juvenile dispersal in mammals. Oikos. 1994;71:349–352. [Google Scholar]
- Yamane A, Shotake T, Mori A, Boug A.I, Iwamoto T. Extra-unit paternity of hamadryas baboons (Papio hamadryas) in Saudi Arabia. Ethol. Ecol. Evol. 2003;15:379–387. [Google Scholar]


