Abstract
The desert locust (Schistocerca gregaria) has been an important agricultural pest at least since biblical times. Although the ecology, physiology and behaviour of this insect species have been well characterized, its biogeographical origins and evolutionary history are more obscure. Schistocerca gregaria occurs throughout Africa, the Middle East and Western Asia, but all other species in the genus Schistocerca are found in the New World. Because S. gregaria has the capacity for extreme long-distance movement associated with swarming behaviour, dispersal may have played an important role in determining current distribution patterns. Some authors have argued that S. gregaria is the product of an eastward trans-Atlantic dispersal from North America to Africa; others consider it more likely that the New World taxa are the product of westward dispersal from Africa. Here, we present a mitochondrial DNA phylogeny of Schistocerca species that supports the monophyly of New World species (including the Galapagos endemic Halmenus) relative to S. gregaria. In concert with observed patterns of molecular divergence, and in contrast to previous morphological studies, our analysis indicates a single trans-Atlantic flight from Africa to South America, followed by extensive speciation and ecological divergence in the New World.
Keywords: Schistocerca, desert locust, biogeography, molecular phylogeny, gregarious behaviour, swarming
1. Introduction
The desert locust, Schistocerca gregaria, exhibits behavioural and morphological phase polyphenism, such that at low population densities, individuals are solitarious and relatively benign, while at high population densities, individuals become gregarious, forming massive swarms that migrate long distances and decimate crops (Uvarov 1977; Pener 1991; Simpson et al. 1999; figure 1). When environmental conditions favour locust population growth, swarm formation can result from phenotypic changes that are associated with crowding (Sword et al. 2000; Bouaïchi & Simpson 2003) and mediated by physical contact (Roessingh et al. 1998; Simpson et al. 2001). The capacity of swarming locusts to travel long distances was amply demonstrated by the observed trans-Atlantic crossing of S. gregaria swarms in October 1988. Ships off the coast of Africa reported large numbers of insects flying west over the Atlantic, and a few days later desert locusts were collected as they landed in the Caribbean islands (Kevan 1989; Ritchie & Pedgley 1989; Rosenberg & Burt 1999). Although desert locusts had previously been observed hundreds of kilometres offshore, the observation of definitive trans-Atlantic travel has important ramifications for biogeographical scenarios.
Figure 1.
Nymphs of Schistocerca gregaria, solitarious phase on the left and gregarious phase on the right. Photograph by G. Sword.
The desert locust is the sole representative of the genus Schistocerca found in the Eastern Hemisphere, whereas some 50 species occur in the Western Hemisphere (Dirsh 1974; Harvey 1981; Amedegnato 1993; Song 2004). This distribution pattern is enigmatic, given that, with one exception (the endemic Galapagos genus Halmenus), Schistocerca belongs to a subfamily of grasshoppers (Cyrtacanthacridinae) that are geographically restricted to the Palearctic and Austral–Oriental regions (Dirsh 1974; Otte 1981). This biogeographical pattern suggested to some authors that Schistocerca most likely originated in the Old World, where almost all other genera in its subfamily are endemic, and later invaded the New World on one or more occasions, giving rise to the radiation of North and South American Schistocerca species (Kevan 1989; Ritchie & Pedgley 1989; Vickery 1989; Amedegnato 1993; Rosenberg & Burt 1999). In this view, the African Schistocerca lineage evolved first, and trans-Atlantic invasions of the New World were facilitated by the ability of these locusts to undertake extreme long-distance flights in very large swarms. This scenario has been called the ‘Old World Origin’ hypothesis for Schistocerca (Song 2004).
Alternatively, Scudder (1899) and Dirsh (1974) proposed that Schistocerca originated and diversified in the New World, and that a single lineage from the New World dispersed eastwards, across the Atlantic, to give rise to S. gregaria. Known as the ‘New World Origin’ hypothesis (Song 2004), this scenario was based on the apparent close relationship between S. gregaria and a subset of American locusts. For example, Dirsh (1974) argued that the desert locust was best considered a subspecies of the American bird locust, Schistocerca americana, which therefore indicated a recent dispersal from the New to the Old World. Similarly, Grunshaw et al. (1990) suggested that cuticular hydrocarbon profiles, showing a close relationship between S. gregaria and the South American Schistocerca cancellata, provided evidence for a recent dispersal from South America to Africa. Song (2004) presented the first comprehensive cladistic analysis of Schistocerca based on morphology, and found S. gregaria to be nested within a clade of ‘mobile’ New World Schistocerca species that included both S. americana and S. cancellata. Based on this result, Song supported the New World Origin hypothesis:
The most parsimonious explanation from the current phylogeny would be that the ancestral desert locust colonized the Old World from the New World.
(Song 2004, p. 1644)
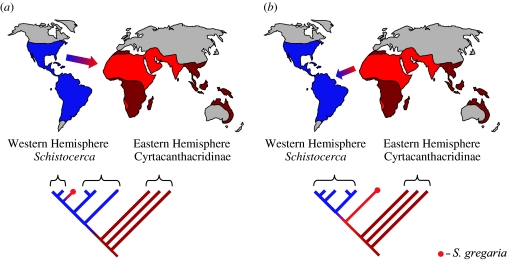
The most important test of the alternative Schistocerca origin biogeographical scenarios is the phylogenetic position of S. gregaria relative to other Schistocerca species (figure 2; see also Song 2004). Single or multiple west to east crossings (New World Origin hypothesis) predict that S. gregaria will be embedded within the New World clade and sister to the New World species or lineages that traversed the Atlantic (figure 2a). Similarly, multiple east to west crossings would result in S. gregaria being embedded within the New World clade and most closely related to the New World lineage that resulted from the most recent colonization. In contrast, only the hypothesis of a single east to west crossing predicts a phylogeny in which S. gregaria is basal within the genus Schistocerca and sister to a monophyletic New World clade (figure 2b). A related and important issue is the phylogenetic position of the Galapagos endemic Halmenus. Aside from Schistocerca, this genus is the only other member of the Cyrtacanthacridinae known from the New World. Thus, its phylogenetic position has considerable relevance for biogeographical scenarios. Chapman et al. (2000) suggested that Halmenus and Schistocerca were not closely related, while Song (2004) found the two genera to be sister taxa.
Figure 2.
Two biogeographical hypotheses to explain the distribution of species within the genus Schistocerca. Many species of Schistocerca are found in the Western Hemisphere, but only the desert locust (S. gregaria) is found in the Eastern Hemisphere. On the world map, the distribution of the desert locust is shown in light red and the distribution of Western Hemisphere species of Schistocerca is shown in blue. The dark red areas are occupied by species in other genera within the Cyrtacanthacridinae (which include the outgroups for our analysis). The trees show expected phylogenetic relationships based on the alternative hypotheses of (a) west to east or (b) east to west crossings of the Atlantic Ocean.
The phylogeny of Schistocerca has not been considered using molecular data. Here, we present the results of an investigation based on 1.7 kb of mitochondrial DNA (mtDNA). We use the data to infer the phylogenetic position of S. gregaria relative to the New World Schistocerca species, to determine the phylogenetic position of Halmenus, and to test the alternative biogeographical scenarios for the origin of the desert locust. We also comment on the implications of our phylogeny for the evolution of swarming in Schistocerca.
2. Material and methods
We obtained mtDNA sequence data from 45 individuals representing 18 species of Schistocerca, one species of Halmenus and four outgroup taxa from the subfamily Cyrtacanthacridinae (table 1); individuals from across the range of S. gregaria (Egypt, Eritrea, Mauritania, Namibia, Saudi Arabia and Sudan), and representing both subspecies (Schistocerca gregaria gregaria and Schistocerca gregaria flaviventris), were included to ensure that genetic variation within that species would be represented. Voucher specimens have been deposited in the Cornell University Insect Collections.
Table 1.
Collection information for Schistocerca specimens and outgroup taxa used in phylogenetic analysis. (Lot numbers in bold correspond to DNA sequences deposited in GenBank.)
| species name | Cornell Entomology Museum voucher no. | collecting locality | collection date | collected/contributed by | accession number |
|---|---|---|---|---|---|
| Genus Schistocerca | Lot 1251 | ||||
| S. americana | 1251-A1, A2 | Florida, USA | 09/92 | G. Sword | AY605943 |
| S. albolineata | 1251-B1, B2 | Arizona, USA | 09/92 | G. Sword | AY605937 |
| S. ceratiola | 1251-C1, C2 | Florida, USA | 09/92 | G. Sword | AY605947 |
| S. chinatiensis | 1251-D1, D2 | New Mexico, USA | 09/92 | G. Sword | AY605938 |
| S. damnifica | 1251-E1, E2 | Florida, USA | 09/92 | G. Sword | AY605936 |
| S. flavofasciata | 1251-F1, F2 | Brazil | 10/92 | F. Mello | AY605950 |
| S. lineata | 1251-G1, G2 | Texas, USA | 09/92 | G. Sword | AY605939 |
| S. literosa | 1251-H1, H2 | Galapagos | 05/92 | S. Peck | AY605949 |
| S. melanocera | 1251-I1, I2 | Galapagos | 05/92 | S. Peck | AY605948 |
| S. nitens | 1251-J1, J2 | Arizona, USA | 09/92 | G. Sword | AY605934 |
| S. obscura | 1251-K1, K2 | Florida, USA | 09/92 | G. Sword | AY605935 |
| S. pallens | 1251-L1, L2 | Barbados | 10/92 | G. Sword | AY605946 |
| S. piceifrons | 1251-M1, M2 | Mexico | 10/92 | G. Sword | AY605944 |
| S. rubiginosa | 1251-N1, N2 | Florida, USA | 09/92 | G. Sword | AY605940 |
| S. shoshone | 1251-O1, O2 | New Mexico, USA | 07/92 | G. Sword | AY605941 |
| S. cancellata | 1251-P1, P2 | Argentina | 05/96 | C. Lange, M. Cigliano | AY605945 |
| S. quisqueya | 1251-Q1, Q2 | Puerto Rico | H. Rowell | AY605942 | |
| S. gregaria gregaria | 1251-R1, R2 | Egypt (2) | 08/91 | W. Bowers | |
| R3 (DNA only) | Eritrea | D.-X. Zhang, G. Hewitt | |||
| R4 (DNA only) | Saudi Arabia | D.-X. Zhang, G. Hewitt | |||
| R5 (DNA only) | Sudan | D.-X. Zhang, G. Hewitt | AY605952 | ||
| R6 (DNA only) | Mauritania | D.-X. Zhang, G. Hewitt | AY605951 | ||
| S. g. flaviventris | 1251-S1, S2 | Namibia (2) | 02/02 | H. Ferenz, K. Seidmann | DQ309428 |
| Halmenus robustus | submitted | Galapagos | D. Otte | DQ309427 | |
| outgroups | |||||
| Acanthacris ruficornis | 1251-T1 | Nairobi, Kenya | 08/94 | M. Mungai | AY605954 |
| Cyrtacanthacris tatarica | 1251-U1 | Nairobi, Kenya | 08/94 | M. Mungai | AY605953 |
| Nomadacris succincta | 1251-V1 | Hong Kong, China | 09/94 | E. Easton | AY605955 |
| Valanga sp. | 1251-W1 | Queensland, Australia | 08/94 | M. Taylor | AY605956 |
DNA was isolated from femoral muscle using Qiagen (Valencia, CA) extraction columns. We sequenced in both directions two overlapping segments of mtDNA, resulting in ca 1.7 kb of sequence that included a portion of ND1, tRNA leucine, the large rRNA subunit (16S), tRNA valine and a portion of the small rRNA subunit (12S). We initially amplified an 840 bp portion of rRNA mtDNA using primers LR-J-13477 (5′-ATGTTTTTGATAAACAGGCG-3′) and SR-N-14275 (5′-AAGGTGGATTTGATAGTAAT-3′) (Dopman et al. 2002). A second ca 850 bp overlapping region was amplified with primers designed using Locusta migratoria mtDNA sequence downloaded from GenBank and then modified for Schistocerca. These primers were ND1-12342 (5′-ARRTAATTAGATATAAWAGGRATWGGYTG-3′) and 16S-R2 (5′-AACCAGCYATCTTWGAGATTACG-3′). PCR reactions (10 μl volume) contained 3 mM MgCl2, 0.2 mM dNTPs, 50 mM KCl, 20 mM Tris (pH 8.4), 2.5 ng of each primer and 1 U of Taq DNA polymerase (Gibco-BRL) and 1 μl of genomic DNA. PCR amplifications were performed using an OmniGene (Hybaid) thermal cycler; cycling conditions were 95 °C for 1 min, 50–52 °C for 1 min and 72 °C for 1 min. PCR products were incubated with 1 μl of exonuclease I and shrimp alkaline phosphatase (Exo/Sap) at 37 °C for at least 45 min. Exo/Sap reactions were terminated by a brief (10 min) incubation at 90 °C. Cleaned PCR products were sequenced on an ABI PRISM 377 automated sequencer using BigDye terminator labelling (Applied Biosystems). All individuals were sequenced with primers LR-J-13477 and SR-N-14275. A preliminary comparison of sequences from duplicate individuals of the same species revealed few if any sequence differences within species. Therefore, the full 1.7 kb of mtDNA sequence was generated for only one individual of each species, except for S. gregaria for which complete sequences were obtained from eight individuals.
Previous studies of S. gregaria using mtDNA sequences revealed the presence of multiple nuclear copies of some mitochondrial genes (Zhang & Hewitt 1996; Bensasson et al. 2000). To avoid problems posed by the presence of these paralogues, we selected a region of the mitochondrial genome previously found to be free of nuclear copies (Dopman et al. 2002). The mitochondrial gene regions we analysed produced unambiguous sequencing reads with no base-calling conflicts. Furthermore, our sequence results were found to be highly repeatable across multiple PCR amplifications for single individuals as well as for multiple conspecific samples. In addition, we cloned PCR products from six Schistocerca species, including S. gregaria, into the vector pCR 2.1 TOPO (Invitrogen, Carlsbad, CA). For each of the six PCR products we sequenced inserts from five positive colonies using M13 forward and reverse primers. In all cases, the sequences were identical to those from direct sequencing of the PCR products. These results are consistent with the absence of nuclear copies or the preferential amplification of the mitochondrial copy in our reactions.
Sequences were aligned using a freestanding Clustal-X alignment program (Thompson et al. 1997) that implements a delayed-alignment algorithm. Sensitivity of the phylogenetic topology to alignment parameters was systematically explored by varying gap penalties, gap extensions and weighting. The topology was found to be robust to such alterations and we used the following parameters: gapopen=10, gapext=2, maxdiv=30, transweight=0. Phylogenetic trees were generated using maximum parsimony and maximum likelihood as implemented by PAUP*4.0 (Swofford 2002) and Bayesian approaches as implemented by MrBayes 3.0 (Ronquist & Huelsenbeck 2003). Trees were rooted using sequences from four outgroup genera of cyrtacanthacridines. Parsimony settings were as follows: heuristic search, 1000 replicates of random taxon additions, TBR branch swapping, gaps treated as fifth base. Nodal support was assessed using 10 000 non-parametric bootstrap replicates of 10 random addition sequences. For maximum-likelihood (ML) analysis, a GTR+I+Γ model of sequence evolution was chosen using Modeltest 3.0 (Posada & Crandall 1998) under the Akaike Information Criterion. Additional model testing was done by an iterative approximation of ML values in PAUP* 4.0 following Rogers & Swofford (1998). This approach produced likelihood parameters for a GTR+I+Γ model similar to those obtained from Modeltest. Subsequent ML tree searches were carried out using these parameter estimates (with settings: Base=[0.3188 0.0936 0.1381], Nst=6, Rmat=[0.3967 4.3725 1.1017 0.0527 2.6570], Rates=gamma, Shape=0.5739, Pinvar=0.5584). The GTR+I+Γ model was also used in Bayesian analysis (settings: Nst=6, Rates=invgamma). Bayesian posteriors were obtained using a Markov chain Monte Carlo simulation. We ran four simultaneous chains for one million generations (sampling every 100 generations and using a ‘burn-in time’ of 500 000 generations). To assess stationarity, we repeated this search 10 times, with up to 107 generations, and obtained very stable posterior values.
A priori hypotheses of relationships were tested using Shimodaira–Hasegawa (SH) tests (Shimodaira & Hasegawa 1999), implemented in PAUP* 4.0 (Swofford 2002) for alternate likelihood trees. The ML tree was generated using the likelihood parameter estimates obtained from our iterative approximations for a GTR+I+Γ model. Likelihood scores for trees constrained to conform to a priori hypotheses were then generated using the same parameter values and compared to the ML tree. The fit of the dataset to a global molecular clock was tested and rejected using a likelihood ratio test (2LR=42.90, critical value=31.41, d.f.=20, p<0.05). We investigated the evolution of swarming with a parsimony mapping approach using Maclade (Maddison & Maddison 2005). Propensity to swarm was considered a two-state presence/absence character and optimized on the resolved phylogenies using both ACCTRAN and DELTRAN.
3. Results
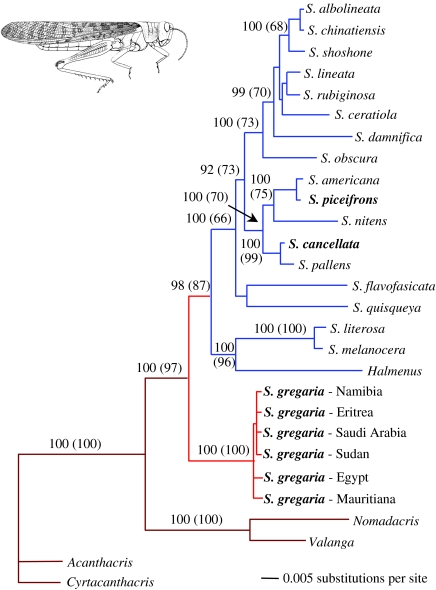
All phylogenetic methods recovered similar topologies with strong non-parametric bootstrap support and high posterior probabilities (figure 3). In all trees, and with high support, S. gregaria is the sister taxon to a monophyletic group that includes all the Western Hemisphere species of Schistocerca and Halmenus. Within this New World clade, lineages from the Galapagos (Halmenus, Schistocerca literosa and Schistocerca melanocera), Central America (Schistocerca quisqueya) and South America (Schistocerca flavofasciata) are basal to lineages primarily from North America. The Galapagos representatives form a monophyletic group, with Halmenus as the sister group of S. literosa and S. melanocera.
Figure 3.
Maximum-likelihood tree showing relationships among Western Hemisphere Schistocerca species and the basal position of S. gregaria, a pattern observed using all phylogenetic methods. The tree is rooted using outgroup taxa from the subfamily Cyrtacanthacridinae. Bayesian posterior probabilities and parsimony bootstrap values (in parentheses) are included for nodes of major biogeographical interest. Colours reflect the distributions of species as depicted in figure 2 (dark red, outgroups; light red, S. gregaria; blue, Western Hemisphere species of Schistocerca). Swarming taxa are indicated in bold. Image of S. gregaria after Dirsh (1965).
The finding that all New World cyrtacanthacridine species comprise a monophyletic group is consistent with the hypothesis of a single westwards trans-Atlantic dispersal event. Constraining the tree topology to make S. gregaria the sister taxon to S. americana, S. cancellata, Schistocerca piceifrons and Schistocerca pallens (consistent with an a priori hypothesis based on Song's morphological study) resulted in trees that were significantly less likely (SH test: Tree 1: 5549.52588, Tree 2: 5578.97631; p=0.017). Similarly, constraining Halmenus to be the sister taxon of Schistocerca, as per Song (2004), resulted in trees that were less likely but not significantly so (Tree 1: 5549.52588, Tree 2: 5558.78262; p=0.08).
Parsimony optimization of swarming behaviour suggests that the propensity to swarm has evolved on three separate occasions (in S. gregaria, S. cancellata and S. piceifrons) (figure 3). Alternatives, such as the evolution of swarming in S. gregaria and in the most recent common ancestor of S. cancellata and S. piceifrons, followed by its loss in S. pallens, Schistocerca nitens and S. americana, are less parsimonious (five steps, instead of three).
4. Discussion
Our molecular data unambiguously show that S. gregaria is not closely related to any particular subset of Western Hemisphere Schistocerca or Halmenus species. Rather, the desert locust is the sister taxon to all the New World cyrtacanthacridine species. This result allows us to reject the hypothesis that Schistocerca colonized Africa via a dispersal event from the New World (figure 2a). Given our phylogeny, for the New World Origin hypothesis to be correct, a Schistocerca ancestor would have had to disperse from the Old World to the New World (without leaving any relict lineages), and then back again to Africa to give rise to S. gregaria. This is an unparsimonious explanation of the available data. Furthermore, our results are inconsistent with a scenario of multiple trans-Atlantic crossings from Africa to the Western Hemisphere, which would require unjustified hypotheses of extinction of New World lineages. Biogeographical analysis using dispersal–vicariance analysis (Ronquist 1996) confirms that these scenarios are unparsimonious. Instead, our results are most consistent with the Old World Origin hypothesis that Schistocerca originated in Africa, and a single trans-Atlantic crossing of the ancestral S. gregaria lineage produced the monophyletic Schistocerca/Halmenus species of the Americas (figures 2b and 3).
An alternative explanation for the observed biogeographical pattern is that the genus Schistocerca originated in Gondwanaland, and Cretaceous vicariance associated with the separation of Africa and South America produced the current disjunction of S. gregaria on one side of the Atlantic, and the rest of the Schistocerca/Halmenus species on the other. The Gondwanan origin hypothesis is unlikely for three reasons. First, all other genera of the subfamily Cyrtacanthacridinae are distributed in the Eastern Hemisphere. If Schistocerca had a Gondwanan origin, we would expect other cyrtacanthacridine genera to also be distributed in both Africa and the Americas. Instead, most subfamilies of the acridid grasshoppers have either exclusively Eastern or Western Hemisphere distributions (Otte 1981), suggesting that much of the diversification of Acrididae (including the origin of Schistocerca) occurred after Africa and South America were separated. Second, fossil evidence (summarized in Song 2004) also suggests that Schistocerca evolution post-dates the Cretaceous. The oldest acridid fossils are known from the Oligocene and Miocene (Lewis 1974, 1976). Finally, the molecular data, which indicate 3–4% mtDNA sequence divergence between S. gregaria and Western Hemisphere members of the genus, are inconsistent with a Gondwanan origin. We were unable to obtain a precise date for this divergence because of rate heterogeneity. Although methods for relaxing molecular clock models and using ‘local’ clocks are available (Thorne et al. 1998; Kishino et al. 2000; Sanderson 2003), the lack of reasonable calibration points for these locusts prevents our use of these approaches. However, insect mtDNA substitution rates are typically considered to be ca 1.1% per million years per lineage (Brower 1994), and even rates an order of magnitude lower than this estimate would suggest that the origin of Schistocerca must post-date the break up of Gondwanaland.
Within the New World Schistocerca/Halmenus clade, the first lineages to diverge are (i) Halmenus, S. literosa and S. melanocera from the Galapagos and (ii) S. quisqueya from Central America and S. flavofasciata from South America. We suggest that an invading population from Africa probably arrived in the broad region of the Caribbean/Central America/northern South America, and very soon thereafter colonized the Galapagos islands. The relationships of Halmenus, which includes four species of brachypterous Galapagos endemics, have been controversial. Dirsh (1974, p. 22) suggested that Halmenus represented a ‘relic of the ancestral stock of Schistocerca’, and noted that the phallic complex was nearly identical to that of Schistocerca. Amedegnato (1993, p. 70) noted ‘Halmenus… from the Galapagos is an immediate derivative of Schistocerca (insular brachypterism)’. However, based on cuticular hydrocarbons, Chapman et al. (2000, p. 579) found ‘no support for a close taxonomic link between Halmenus and Schistocerca’, while Song (2004) placed Halmenus as the sister group to Schistocerca. In our molecular phylogeny, Halmenus is most closely related to the two Galapagean Schistocerca representatives, providing a reasonable explanation for its origin. We propose that an early New World Schistocerca lineage colonized the Galapagos, and, subsequently, gave rise to the brachypterous Halmenus. This is a simpler biogeographical explanation than earlier proposals that invoke two independent colonizations. Our phylogenetic result also explains an interesting, but previously unexplained, aspect of Halmenus cuticular hydrocarbons. Chapman et al. (2000) noted that Halmenus shares with Galapagos Schistocerca relative proportions of n-alkanes not seen in other species. Based on our phylogeny, we interpret this character as a synapomorphy for Galapagos Schistocerca and Halmenus.
Song (2004), based on his morphological analysis, found the closest cyrtacanthacridine outgroups of Schistocerca and Halmenus to be distributed in Australia and Asia. Thus, he proposed a trans-Pacific dispersal (Asia to the New World), followed by a trans-Atlantic dispersal (New World to Africa). We have shown that our phylogeny contradicts the latter part of this scenario. Additionally, the sister group relationship between the monophyletic New World cyrtacanthacridines and African S. gregaria obviates the need for a trans-Pacific dispersal hypothesis. In our analysis, the closest relatives of Schistocerca were found to be Valanga (Indo-Pacific) and Nomadacris (Africa, Asia, Australia), which is consistent with an African origin for Schistocerca. We note that both Song's (2004) and our study include only a fraction (10 genera and 4 genera, respectively) of the 30+ cyrtacanthacridine outgroup genera. We have begun exploring the possible effects of different outgroup selection on our topology by including more distantly related taxa, such as L. migratoria, and found it to be robust to such changes. However, further studies that incorporate more detailed taxon sampling of cyrtacanthacridines and additional sources of data (e.g. nuclear DNA) should help to clarify the biogeography of the many African, Asian and Australian taxa, and confirm the interpretations presented here.
Based on the molecular phylogeny, distribution and estimated age of Schistocerca divergence, we infer the following scenario: Schistocerca originated in north Africa, where arid habitats and extreme environmental heterogeneity favoured the evolution of swarming (Uvarov 1977). Swarming behaviour facilitated a single trans-Atlantic crossing, leading to the colonization of Central and South America, the Galapagos, and, subsequently, North America. For a number of reasons, swarming locusts are one of the few insects that could manage a flight across the Atlantic Ocean: they occur in enormous numbers (greater than 108), and fly during the day when thermal upcurrents can carry them to very high altitudes (2000 m), enabling transport by high velocity winds (Uvarov 1977; Steedman 1990). Most Schistocerca species in the Americas do not exhibit a gregarious phase, and do not form swarms. This helps to explain an obvious aspect of Schistocerca phylogeny—the disparity in species diversity between the Eastern and Western Hemisphere lineages. The long-distance migration of S. gregaria during swarming promotes gene flow across the vast range of the species, thereby reducing the potential for isolation and speciation (Ibrahim et al. 2000). In contrast, most species in the Americas do not swarm, are more sedentary, and may be more prone to geographical isolation and speciation (Dopman et al. 2002; Sword 2003). Availability of a diversity of novel food resources may also have contributed to the radiation of New World species—several taxa are associated with specific host plants (Hubbell 1928; Dirsh 1974; Sword & Chapman 1994; Sword & Dopman 1999).
The diversification of Schistocerca in the Americas constitutes a large-scale natural experiment on the evolution of locust phase traits. The expression of density-dependent behavioural, physiological and morphological phase polyphenism correlates with swarming in locusts (Uvarov 1977; Pener 1991; Simpson et al. 1999), but the cause and effect relationship between these phenomena can be difficult to establish (Sword 2003). Much like the desert locust, a few species of American Schistocerca, including S. piceifrons from Central America and S. cancellata from South America, also express extreme density-dependent phenotypic changes and regularly swarm (Dirsh 1974; Harvey 1981). Mapping swarming on the Schistocerca phylogeny suggests three independent origins of this character. However, it is important to note that locust swarm formation is likely to result from a complex interaction between local environmental factors, such as habitat stability and underlying genetic traits, such as the expression of phase polyphenism (Sword 2002, 2003; Babah & Sword 2004). Some non-swarming Schistocerca species may retain genetic variation for the expression of important density-dependent traits, but inhabit environments that are not conducive to swarm formation. Other non-swarming species may exist in the appropriate environments, but no longer retain the genetic capacity to become swarming locusts. Future work using Schistocerca as a model system will help to disentangle the role of genotype and environment in locust swarm formation.
Acknowledgments
We thank the following people for collecting and sending specimens: W. Bowers, M. Cigliano, E. Easton, H. Ferenz, G. Gade, A. Joern, C. Lange, M. Lecoq, F. Mello, K. Milton, M. Mungai, D. Otte, S. Peck, J. Ribeiro, K. Seidelman, H. Song, M. Taylor and W. Wcislo. Thanks also to D.-X. Zhang and G. Hewitt for providing DNA samples from geographical populations of S. gregaria and to S. Bogdanowicz and E. Dopman for advice and help with the molecular work. S. Khattak generously assisted with figure preparation. Funding was provided by a National Geographic grant to R.F.C. and NSF grants to R.G.H.
We dedicate this paper to the memory of Reg Chapman, who provided the initial impetus for the work but sadly passed away before it was completed.
These authors contributed equally to this work.
References
- Amedegnato C. African–American relationships in the acridians (Insecta, Orthoptera) In: George W, Lavocat R, editors. The Africa–South America connection. Clarendon Press; Oxford, UK: 1993. pp. 59–75. [Google Scholar]
- Babah M.A.O, Sword G.A. Linking locust gregarization to local resource distribution patterns across a large spatial scale. Environ. Entomol. 2004;33:1577–1583. [Google Scholar]
- Bensasson D, Zhang D, Hewitt G.M. Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes. Mol. Biol. Evol. 2000;17:406–415. doi: 10.1093/oxfordjournals.molbev.a026320. [DOI] [PubMed] [Google Scholar]
- Bouaïchi A, Simpson S.J. Density-dependent accumulation of phase characteristics in a natural population of the desert locust Schistocerca gregaria. Physiol. Entomol. 2003;28:25–31. 10.1046/j.1365-3032.2003.00317.x [Google Scholar]
- Brower A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.F, Espelie K.E, Peck S.B. Cuticular hydrocarbons of grasshoppers from the Galapagos Islands, Ecuador. Biochem. Syst. Ecol. 2000;28:579–588. doi: 10.1016/s0305-1978(99)00094-0. 10.1016/S0305-1978(99)00094-0 [DOI] [PubMed] [Google Scholar]
- Dirsh V.M. Cambridge University Press; Cambridge, UK: 1965. The African genera of Acridoidea. [Google Scholar]
- Dirsh V.M. W. Junk; The Hague, The Netherlands: 1974. Genus Schistocerca (Acridomorpha, Insecta) [Google Scholar]
- Dopman E.B, Sword G.A, Hillis D.M. The importance of the ontogenetic niche in host-associated divergence: evidence from a generalist grasshopper. Evolution. 2002;56:731–740. doi: 10.1111/j.0014-3820.2002.tb01384.x. [DOI] [PubMed] [Google Scholar]
- Grunshaw J.P, Guermouche H, Guermouche S, Jago N.D, Jullien R, Knowles E, Perez F. Chemical taxonomic studies of cuticular hydrocarbons in locusts of the Schistocerca americana complex (Acrididae: Cyrtacanthacridinae): chemical relationships between New World and Old World species. J. Chem. Ecol. 1990;16:2835–2858. doi: 10.1007/BF00979477. 10.1007/BF00979477 [DOI] [PubMed] [Google Scholar]
- Harvey A.W. A reclassification of the Schistocerca americana complex (Orthoptera: Acrididae) Acrida. 1981;10:61–77. [Google Scholar]
- Hubbell T.H. A new shrub-inhabiting species of Schistocerca from central Florida. Occas. Pap. Mus. Zool. Univ. Mich. 1928;197:1–10. [Google Scholar]
- Ibrahim K.M, Sourrouille P, Hewitt G.M. Are recession populations of the desert locust (Schistocerca gregaria) remnants of past swarms? Mol. Ecol. 2000;9:783–791. doi: 10.1046/j.1365-294x.2000.00932.x. 10.1046/j.1365-294x.2000.00932.x [DOI] [PubMed] [Google Scholar]
- Kevan D.K.M. Transatlantic travelers. Antenna. 1989;13:12–15. [Google Scholar]
- Kishino H, Thorne J.L, Bruno W.J. Performances of a divergence time estimation method under a probabilistic model of rate evolution. Mol. Biol. Evol. 2000;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- Lewis S.E. Four specimens of fossil grasshoppers (Orthoptera: Caelifera) from the Ruby River basin (Oligocene) of Southwestern Montana. Ann. Entomol. Soc. Am. 1974;67:523–524. [Google Scholar]
- Lewis S.E. A new specimen of fossil grasshopper from the Ruby River basin (Oligocene) of Southwestern Montana. Ann. Entomol. Soc. Am. 1976;69:120. [Google Scholar]
- Maddison D.R, Maddison W.P. Sinauer Associates; Sunderland, MA: 2005. MacClade ver 4.0. [Google Scholar]
- Otte D. Harvard University Press; Cambridge, MA: 1981. The North American grasshoppers, vol. 1—Acrididae: Gomphocerinae and Acridinae. [Google Scholar]
- Pener M.P. Locust phase polymorphism and its endocrine relations. Adv. Insect Physiol. 1991;23:1–79. [Google Scholar]
- Posada D, Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Ritchie M, Pedgley D.E. Desert locusts cross the Atlantic. Antenna. 1989;13:10–12. [Google Scholar]
- Roessingh P, Bouaïchi A, Simpson S.J. Effects of sensory stimuli on the behavioral phase state of the desert locust, Schistocerca gregaria. J. Insect Physiol. 1998;44:883–893. doi: 10.1016/s0022-1910(98)00070-5. 10.1016/S0022-1910(98)00070-5 [DOI] [PubMed] [Google Scholar]
- Rogers J.S, Swofford D.L. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst. Biol. 1998;47:77–89. doi: 10.1080/106351598261049. 10.1080/106351598261049 [DOI] [PubMed] [Google Scholar]
- Ronquist, F. 1996 DIVA ver. 1.1. Computer program and manual available by anonymous FTP from Uppsala University (ftp.uu.se or ftp.systbot.uu.se).
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rosenberg J, Burt P.J.A. Windborne displacements of Desert Locusts from Africa to the Caribbean and South America. Aerobiologia. 1999;15:161–175. 10.1023/A:1007529617032 [Google Scholar]
- Sanderson M.J. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. 10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Scudder S.H. The orthopteran genus Schistocerca. Proc. Am. Acad. Arts Sci. 1899;34:439–476. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Simpson S.J, McCaffery A.R, Hägele B.F. A behavioural analysis of phase change in the desert locust. Biol. Rev. Camb. Phil. Soc. 1999;74:461–480. 10.1017/S000632319900540X [Google Scholar]
- Simpson S.J, Despland E, Hägele B.F, Dodgson T. Gregarious behavior in desert locusts is evoked by touching their back legs. Proc. Natl Acad. Sci. USA. 2001;98:3895–3897. doi: 10.1073/pnas.071527998. 10.1073/pnas.071527998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. On the origin of the desert locust Schistocerca gregaria (Forskål) (Orthoptera: Acrididae: Cyrtacanthacridinae) Proc. R. Soc. B. 2004;271:1641–1648. doi: 10.1098/rspb.2004.2758. 10.1098/rspb.2004.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steedman A. 3rd edn. Natural Resources Institute; Chatham: 1990. Locust handbook. [Google Scholar]
- Swofford D.L. Sinauer Press; Sunderland, MA: 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. [Google Scholar]
- Sword G.A. A role for phenotypic plasticity in the evolution of aposematism. Proc. R. Soc. B. 2002;269:1639–1644. doi: 10.1098/rspb.2002.2060. 10.1098/rspb.2002.2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sword G.A. To be or not be a locust? A comparative analysis of behavioral phase change in nymphs of Schistocerca americana and S. gregaria. J. Insect Physiol. 2003;49:709–717. doi: 10.1016/s0022-1910(03)00092-1. 10.1016/S0022-1910(03)00092-1 [DOI] [PubMed] [Google Scholar]
- Sword G.A, Chapman R.F. Monophagy in a polyphagous grasshopper. Entomol. Exp. Appl. 1994;73:255–264. [Google Scholar]
- Sword G.A, Dopman E. Developmental specialization and geographic structure of host plant use in a polyphagous grasshopper, Schistocerca emarginata (=lineata) (Orthoptera: Acrididae) Oecologia. 1999;120:437–445. doi: 10.1007/s004420050876. 10.1007/s004420050876 [DOI] [PubMed] [Google Scholar]
- Sword G.A, Simpson S.J, El Hadi O.T.M, Wilps H. Density-dependent aposematism in the desert locust. Proc. R. Soc. B. 2000;267:63–68. doi: 10.1098/rspb.2000.0967. 10.1098/rspb.2000.0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne J.L, Kishino H, Painter I.S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Uvarov B.P. Grasshoppers and locusts. vol. 2. Centre for Overseas Pest Research; London: 1977. [Google Scholar]
- Vickery V.R. The biogeography of Canadian Grylloptera and Orthoptera. Can. Entomol. 1989;121:389–424. [Google Scholar]
- Zhang D, Hewitt G.M. Highly conserved nuclear copies of the mitochondrial control region in the desert locust Schistocerca gregaria and some implications for population studies. Mol. Ecol. 1996;5:295–300. doi: 10.1111/j.1365-294x.1996.tb00317.x. 10.1046/j.1365-294X.1996.00078.x [DOI] [PubMed] [Google Scholar]