Abstract
In addition to their nuclear genome, the vast majority of eukaryotes harbour cytoplasmic genomes, e.g. in mitochondria or chloroplasts. In the majority of cases, these cytoplasmic genomes are transmitted maternally only, leading to selective pressures divergent from those that act on nuclear genes. In particular, cytoplasmic genes, which reduce the fitness of males that carry them, but have no fitness effect in females, are believed to be selectively neutral. Here, we go a step further and argue that in outbreeding populations (i.e. populations with inbreeding avoidance), ‘spiteful’ cytoplasmic elements that reduce the number of offspring produced by males are in fact selected for. We study this process by means of a stochastic model, analysing both the probability of spread and the impact that such a spiteful cytotype can have on population dynamics. Our results demonstrate that the probability of spread of the spiteful cytotype can be several times higher in outbreeding than in panmictic populations. Spread and fixation of the spiteful cytotype can lead to different qualitative effects on the population dynamics, including extinction, decreased or increased stable population size. We discuss our results in respect to cytoplasmically induced male infertility and cytoplasmic incompatibility.
Keywords: cytoplasmic incompatibility, infertility, mitochondria, selfish genetic elements, spite, Wolbachia
1. Introduction
According to their mode of inheritance, eukaryotic genomes can be divided into two fractions. Whereas the majority of genes are located in the nucleus and usually show Mendelian inheritance, a smaller fraction of genes are located in the cytoplasm and are passed on predominantly through the maternal line. These cytoplasmic genetic elements include mitochondria, chloroplasts and inherited micro-organisms.
The different modes of inheritance give rise to diverging selective pressures that act on nuclear and cytoplasmic genes: whereas nuclear genes are selected to produce healthy and sexually successful males, the production of males is wasteful from the cytoplasmic genes' ‘point of view’. This gives rise to various manifestations of intragenomic conflict (Cosmides & Tooby 1981), the empirical reality of which is corroborated by a wealth of phenotypes induced by cytoplasmic elements, including cytoplasmic male sterility induced by mitochondria in plants (Saumitou-Laprade et al. 1994), and cytoplasmic incompatibility (CI), male-killing and feminization induced by intracellular bacteria in arthropods (reviewed in Stouthamer et al. 1999).
While it has been recognized that cytoplasmic genetic elements with deleterious effects in males are selectively neutral (Frank & Hurst 1996), we here present the thesis that mutations with deleterious effects can even spread through natural selection. Consider an outbreeding population, that is, one in which individuals mate with their kin less often than expected by chance. Consider further a mutant cytoplasmic allele that causes its male carrier to produce fewer offspring than wild-type males. Mating of such a male with a female can then be expected to also reduce the number of offspring this female produces. Thus, the average offspring production of all females unrelated to the focal male is reduced, and therefore the relative fitness of the females closely related to that male is increased.
This argument can also be put in terms of ‘spite’, defined as an action that decreases other individuals' direct fitness while not increasing the actor's fitness. In theory, spiteful behaviour can increase the indirect fitness of the actor, and thus, be selectively favoured (Hamilton 1970). The crucial necessary condition for selection for spite is that on average, the recipient is less related to the actor than a randomly chosen member of the population, which is equivalent to a negative coefficient of relatedness R (e.g. Grafen 1985). Outbreeding provides a pre-existing system of interactions between negatively related partners that may enable spiteful cytoplasmic elements in males to evolve: by inducing spite towards females that have a higher probability of bearing the wild-type allele than close relatives, a mutant cytoplasmic allele can be expected to be selected for.
Outbreeding is defined as mating between partners that are less strongly related than randomly chosen members of the population. Outbreeding is often thought of as an adaptation to inbreeding depression and/or heterosis and can be the result of several mechanisms. For example, sex-biased dispersal before reproduction is commonly found in mammals and birds (e.g. Cockburn et al. 1985, 2003; Wolff et al. 1988; Clutton-Brock 1989; Wolff 1992). Behavioural avoidance of mating with kin is another trait that leads to outbreeding if mating is otherwise random. Behavioural inbreeding avoidance (including females not entering oestrous) is particularly common in mammals (Hoogland 1981, 1991; Dobson et al. 1997; Ishida et al. 2001; Tai et al. 2002), but has also been demonstrated in skinks (Stow & Sunnucks 2004), crickets (Simmons 1989, 1991) and mites (Enigl & Schausberger 2004). Finally, inbreeding avoidance may also occur after copulation. Examples include failure of females mated with close kin to fertilize their eggs, found in two cactophilic Drosophila species (reviewed in Markow 1997), as well as reduced sperm competitive ability of related compared to unrelated males reported in a lizard (Olssen et al. 1996), Drosophila melanogaster (Mack et al. 2002) and crickets (Stockley 1999; Bretman et al. 2004). Given that it is difficult to detect inbreeding avoidance, it may be inferred from these examples that outbreeding is not rare in natural populations.
Here, we develop an individual based stochastic model by means of which we study the spread of a spiteful cytoplasmic element (‘S-cytotype’). We assume that this S-cytotype, when present in males, results in increased mortality among the offspring of these males. First, we analyse how likely the invasion of a mutant S-cytotype is. We show that the S-cytotype can indeed be selected for and derive approximations for its selective advantage and its probability of fixation in a population with sibmating avoidance. Second, we analyse the impact that the spread of the S-cytotype has on the population dynamics. This is important to determine the conditions when spread of a S-cytotype leads to extinction of the population, but also reveals other interesting effects the S-cytotype may have on the population dynamics.
2. The model
Our approach is an individual based stochastic model with discrete, non-overlapping generations. We consider two different cytotypes, the ‘normal’, wild-type cytotype (N-cytotype) and the spiteful, mutant cytotype (S-cytotype). Within one generation, events occur as follows. Each female in the population mates either with a randomly chosen male (panmixis scenario), or with a randomly chosen male other than one of her brothers (sibmating avoidance scenario, outbreeding). Each female then produces m male and m female eggs, all of which inherit their mother's cytotype. While eggs with an N-cytotype father then develop into juveniles, eggs with an S-type father die with a probability λ due to a spiteful modification of their father's sperm. In the next step, juveniles die with a probability μ, regardless of their cytotype. Finally, frequency dependent mortality occurs: immature adults die with a probability Nim/K, where Nim is the total population size of immature adults, and K is the carrying capacity of the population.
3. Invasion of the S-cytotype
How likely is the invasion and fixation of a newly arisen S-cytotype in a population? We will first present some analytic considerations that can serve as approximations for the stochastic model. In a panmictic population, the S-cytotype is neither favoured nor disfavoured by natural selection, so that its frequency is solely determined by genetic drift. We denote by Nf the number of adult females in the population and assume that this number remains constant during the spread of the S-cytotype to keep the calculations simple (but see §4). The probability of spread and fixation of a newly mutated S-cytotype in a panmictic population is then given by u=1/Nf (Hartl & Clark 1997).
In a population with sibmating avoidance, the S-cytotype is selectively favoured. Consider a population with Nm males, a fraction p of which bear the S-cytotype. A female with the S-cytotype that has k brothers (among the pNm males bearing the S-cytotype) will on average have a relative offspring production of
| 3.1 |
Likewise, an N-cytotype female with k brothers will produce a relative number of
| 3.2 |
offspring. Note that for both types of females the offspring production is reduced and decreases with increasing frequency p of the S-cytotype, because both types of females mate with S-cytotype bearing males. Also, while wS(k, p) increases with increasing number k of brothers of the female, wN(k, p) decreases with increasing k.
We will now assume that each reproducing female in the population has exactly one reproducing brother in the population, as can be expected on average with a 1 : 1 sex ratio. The selective advantage of the S-cytotype relative to the N-cytotype can then be calculated as
| 3.3 |
It can be seen from this formula that the selective advantage s of the S-cytotype decreases with increasing population size, which is a general result deriving from the theory of spite (Hamilton 1970). In addition, equation 3.3 shows that s increases with increasing frequency of the S-cytotype, yielding positive frequency dependence. Nevertheless, the S-cytotype is favoured over the whole range of frequencies, which becomes clear when considering .
We now apply theory developed by Kimura (1962) to obtain an approximation for the probability of fixation of an S-cytotype. We will assume that the frequency of the S-cytotype in males always equals the frequency in females and denote both by p. Also, we will assume Nf=Nm, but retain these two variables for more clarity. Note that both of these assumptions are in accord with what we can expect on average in the stochastic model.
In general, the probability of fixation of an allele with initial frequency p0 is estimated by
| 3.4 |
where
| 3.5 |
(Kimura 1962). In this formula, Mδp and Vδp denote the mean and the variance in the change of the frequency p of the S-cytotype. These quantities can be approximated by
| 3.6a |
| 3.6b |
(Note that the effective population size with respect to a cytoplasmic gene equals the number of reproducing females in the population.) After solving the integrals, we get
| 3.7 |
and
| 3.8 |
Thus, for a newly arisen S-mutant in a population with sibmating avoidance we obtain a fixation probability of
| 3.9 |
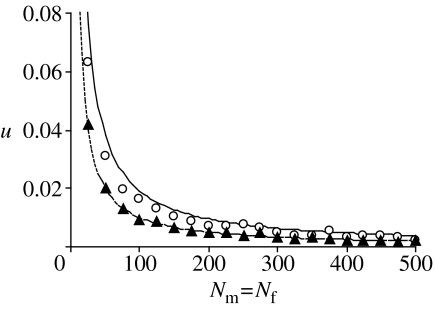
It should be emphasized that this result can only be a coarse approximation because it relies on several simplifying assumptions. Figure 1 shows how the estimates u for the panmictic and the sibmating avoidance scenario depend upon the population size. As can be seen, the fixation probability increases in both cases with decreasing population size.
Figure 1.
Fraction of simulations out of 100 000, where the S-cytotype could invade the population and became fixed in the panmixis (filled triangles) and the outbreeding (open circles) scenario. The dashed and solid lines show the analytic approximations for panmixis and outbreeding, respectively. Parameters take the values m=5, μ=0.5, λ=0.33. The parameter K is varied such as to lead to the stable population size as given on the x-axis when the N-cytotype is fixed.
Figure 1 also shows simulation results of the stochastic model. For each data point, 100 000 simulations have been run. Parameters and starting conditions were chosen such as to match the assumption of the above derivation of u as closely as possible. In particular, in each simulation, initially one sibling pair (sister and brother) had the S-cytotype so that selection commences in the first generation. Moreover, the parameters m, μ and λ chosen result in a relatively stable population size, even when the S-cytotype spreads (see §4). Comparing the proportions of simulations were the S-cytotype became fixed in the population, it can be seen that while the estimate for the fixation probability in a panmictic population matches closely the simulation results, the approximation for u in a population with sibmating avoidance slightly overestimates the simulation results. Nevertheless, it is notable that even for the moderate mortality among offspring from S-cytotype fathers chosen (λ=0.33), the probability of fixation is consistently about twice as high in the sibmating avoidance scenario than in the panmixis scenario.
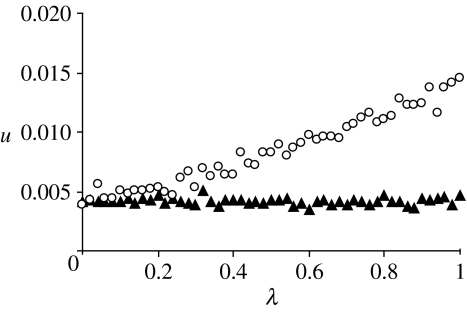
For figure 2, the embryo mortality λ that ensues from a paternal S-cytotype was varied. In each simulation, we started with a population were all individuals had the N-cytotype; we assumed that the population was at its equilibrium size (see §4), with equal numbers of females and males. We then ‘mutated’ one of the females to an S-cytotype female and simulated until one of the cytotypes became fixed or the population went extinct. For each combination of parameters, we ran 106 simulations. The plot shows that in an outbreeding population, the probability of invasion of the S-cytotype increases with increasing λ, i.e. with increasing spitefulness of the S-cytotype. Again, this is in accord with the above considerations on selection for the S-cytotype.
Figure 2.
Fraction of simulations out of 1 000 000 where the S-cytotype could invade the population and became fixed in the panmixis (filled triangles) and the outbreeding scenario (open circles). Each simulation was run until one of the cytotypes became fixed in the population or until the population went extinct. Note that extinction of the population before the S-cytotype was fixed was also counted as successful invasion, because extinction is extremely unlikely with low frequencies of the S-cytotype. Parameters take the values m=5, μ=0.5, K=2000, leading to a stable total population size of when the N-cytotype is fixed.
4. Population dynamics
Having discussed the likelihood of an invasion of the S-cytotype, we will now try to understand what impact such an invasion will have on the population dynamics. We would like to stress that due to the particularities of our model assumptions on life history and density dependence, the following treatment does not claim to make any quantitative predictions for real populations. Rather, it is meant to demonstrate qualitatively the variety of effects that are possible even under the simple assumptions made. We will first attempt to gain insight into the population dynamics of the model by interpreting the mortality probabilities as deterministic mortalities, and then proceed to results of computer simulations of the stochastic model.
Consider first the case where all individuals in the population have the N-cytotype. We denote by N the number of mature females in the population at a given generation. The expected number of mature females in the following generation is then given by
| 4.1 |
Note that this recursion equation is essentially the well-known logistic equation. Solving the equation for N yields the non-trivial equilibrium population size
| 4.2 |
Obviously, this equilibrium is positive for only, otherwise, the population will go extinct. Taking advantage of previous results on the logistic equation (May 1974), it can further be shown that for , is a globally stable equilibrium (i.e. when starting from any population size , the system will converge to ). For , the system will converge to cycles with periods 2i, with i taking increasing integer values, while for , chaotic behaviour of the system will prevail.
Consider now the case where all individuals in the population bear the S-cytotype. All offspring then suffer increased mortality, and equation (4.1) becomes:
| 4.3 |
The equilibrium population size can now be calculated as:
| 4.4 |
and it can be seen that the new condition for this equilibrium to exist (i.e. be positive), can be considerably more restrictive than for the case that all individuals bear the N-cytotype. In other words, a population that can stably exist with the N-cytotype may go extinct when the S-cytotype is fixed.
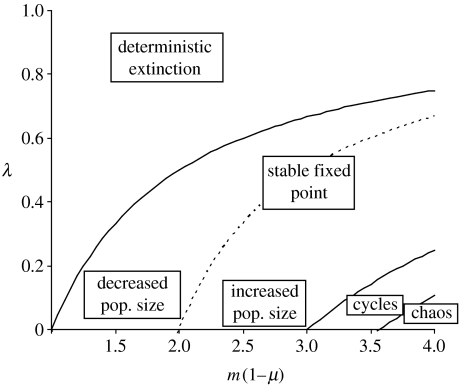
When is fulfilled, the population can again converge to a stable equilibrium or a stable cycle, or exhibit chaotic behaviour. Interestingly, a population fixed for the N-cytotype that behaves chaotically may converge to stable cycles or a stable equilibrium when the S-cytotype becomes fixed. Also, the equilibrium population size may be higher when all individuals bear the S-cytotype than when all individuals bear the N-cytotype. These different outcomes of population dynamics depending on the parameters m, μ and λ are summarized in figure 3, which has been derived analytically using well-known results of the logistic equation (May 1974).
Figure 3.
Different qualitative population dynamics depending on the parameters m, μ and λ when the population is fixed for the S-cytotype. The term m(1−μ) corresponds to the intrinsic growth factor in the standard logistic equation. The three regions on the x-axis (λ=0) correspond to the different types of dynamics with the N-cytotype fixed. Fixation of the S-cytotype in the population (with λ>0) can lead to deterministic population extinction, decreased or increased stable equilibrium population size when the population was at equilibrium population size with the N-type, transition from cyclic behaviour to a stable equilibrium and from chaos to cycles or stable equilibrium.
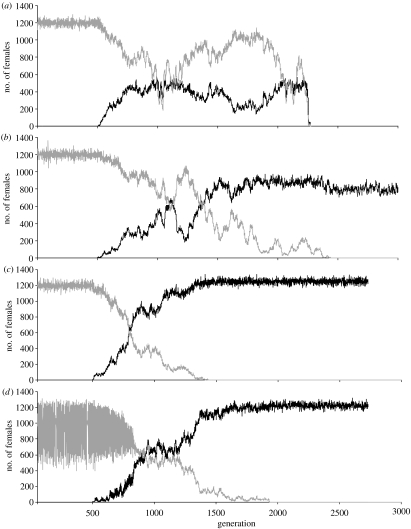
For selected parameter values, we have confirmed the above considerations in the stochastic model (figure 4). For each plot, we simulated a population fixed for the N-cytotype for 500 generations, after which we introduced a single female mutant with the S-cytotype. It can be seen that spread and fixation of the S-cytotype can lead to population extinction (figure 4a), decreased, but stable population size (figure 4b), increased stable population size (figure 4c) or stabilization of population size (figure 4d).
Figure 4.
Examples for transitions in population dynamics caused by invasion of the S-cytotype. The grey curves show the numbers of females with the N-cytotype, while S-cytotype females are shown in black. In each plot, the dynamics of a population with only N-cytotype individuals was simulated for 500 generations, after which a single female mutant with the S-cytotype was introduced. In all simulations, sibmating avoidance is assumed. Parameters take the values K=10 000, m=5; (a–c) μ=0.5 or (d) μ=0.3; (a) λ=0.9, (b) λ=0.5, (c) λ=0.2, (d) λ=0.5.
5. Discussion
We have developed a model that involved a spiteful cytoplasmic genetic element that is inherited maternally only, but induces increased mortality in the offspring of males that carry it. By combining analytical considerations and simulation results, we have demonstrated that such an element can be selected for in populations where individuals avoid mating with their siblings. We have also shown that while extinction of the population is a possible result of the invasion of the spiteful cytoplasmic element, it may also lead to an increased population size and can have a stabilizing effect on the population dynamics. Taking these results together, our model suggests two possibilities that may seem paradoxical on first sight: (i) a strongly deleterious genetic element can be selected for and (ii) while detrimental for each individual that carries it, it may increase the fitness of the population.
The action of the spiteful cytotype was modelled by assuming an increased embryonic mortality in offspring of males that carry the spiteful element. However, several other phenotypes in males induced by cytoplasmic genes can be expected to lead to the same results. For example, increased mortality among the offspring of fathers bearing the spiteful cytotype may also result from decreased or absent paternal care, or from infanticide. As another example, consider a cytotype that induces a decrease in the fertility of its male host. Females that mate with such a male will on average produce fewer offspring than if they mate with normal males. This is in particular true for monogamous (or polygynous) species. More precisely, the fewer males an average females mates with during her lifetime, the stronger we expect the selection for a cytoplasmic gene that reduces male fertility to be in an outbreeding population.
To date, a ‘spiteful cytotype’ as assumed in our model has not been found in any species. There are, however, at least two closely related phenomena that guide us to think that the notion of such cytotypes is realistic. First, increasing evidence has accumulated in recent years that in humans, mutations in mitochondrial DNA exist that have detrimental effects on male fertility (Ruiz-Pesini et al. 2000; O'Connell et al. 2002; Spiropoulos et al. 2002; St. John et al. 2005). While previously, such mutations were seen as selectively neutral (Frank & Hurst 1996), the model developed here, along with the above remarks on infertility in monogamous species, suggests that such mitochondria may in fact be weakly selectively advantageous.
Second, a phenotype very similar to that of the hypothesized S-cytotype termed cytoplasmic incompatibility (CI) is widespread in arthropods. CI is induced by intracellular bacteria of the genera Wolbachia and Cardinium (Yen & Barr 1971; Hunter et al. 2003). In CI, the sperm of infected males is modified by the bacteria in a detrimental way. As a result, the offspring of such males show an increased mortality (up to 100%) unless the same or a similar strain of bacteria is present in the egg, in which case the modification is ‘rescued’ and the offspring survive (Laven 1951; Hoffmann et al. 1986). CI has previously been discussed in the context of spite (Hurst 1991; Foster et al. 2001; Gardner & West 2004). In terms of the modification-rescue system, the S-cytotype can be viewed as a ‘mod-only’ strain, the existence of which has been hypothesized but not proven in Wolbachia (Charlat & Merçot 2001). Clearly, because of the additional rescue function, CI is a much more efficient way of enhancing the fitness of a cytoplasmic genetic element than mod-only. However, the S-cytotype considered in this paper can be expected to evolve more easily, and the extant CI-inducing bacteria may well have gone through a stage in which they were not able to rescue their own modification. Thus, our model also provides a hypothesis on the evolution of CI.
To keep the model as simple as possible, we have assumed complete avoidance of matings between siblings. It could be argued that sibmating avoidance is unlikely to be absolute in most species in which it occurs, and hence, that the selection for the spiteful cytotype is weaker than predicted by our model. However, it should be noted that partial inbreeding avoidance can also extend to more distantly related relatives as well as parents or offspring in age-structured populations, which may enhance outbreeding beyond the rate assumed in our model.
Another type of inbreeding avoidance is avoidance of selfing in hermaphrodite species. This is of great interest in particular because low selfing rates or even complete selfing avoidance are very commonly found in plants, and also in most hermaphrodite animal taxa (reviewed in Jarne & Charlesworth 1993). Avoidance of selfing can be expected to have the same effect in our model as sibmating avoidance in dioecious species. This is because from the cytoplasm's viewpoint, a hermaphrodite is essentially the same as a pair of siblings in the context of reproduction. Moreover, at least one of the mechanisms of selfing avoidance—self-incompatibility—can be expected to also lead to incompatibility between closely related individuals, thus resulting in even higher outbreeding rates than with mere selfing avoidance.
In his seminal paper, Hamilton (1970) argued that spite would not be common in natural populations for three reasons. First, ‘all actions do cost something’. This is certainly not true for the mechanism we have studied, because the direct fitness of a maternally inherited gene in males is zero anyway and cannot be further reduced (see also Hurst 1991; Foster et al. 2001). Second, organisms would not be able to sufficiently distinguish between positively and negatively related members of the populations, so that even when close kin can be recognized, the rest of the population would on average be only slightly negatively related to the actor. In our model, inbreeding avoidance provides a natural, pre-existing mechanism by which kin is distinguished from non-kin. For some of the mechanisms that can lead to outbreeding—e.g. dispersal of one sex and self-incompatibility in plants—no cognitive recognition mechanisms are required. However, it is true that selection for a spiteful cytotype will be weak, especially in large populations. Finally, Hamilton (1970) suggested that the small populations required for spite to be substantially selected for would readily go extinct once the gene causing spite spreads. This notion is not confirmed by the model studied in this paper. Rather, we demonstrated that the spread of the spiteful cytotype can stabilize the population and also lead to an increased population size. Also, instead of one small population, a large population subdivided in many small subpopulations may also favour the spread of the spiteful cytotype. As recent theoretical work has demonstrated, spite can evolve in such a scenario especially with soft selection, i.e. local regulation of subpopulation size (reviewed in Gardner & West 2004).
An important aspect that is likely to strongly reduce the incidence of spiteful cytoplasmically induced phenotypes is selection acting on nuclear genes. Since the spiteful cytoplasm reduces the fitness of males that carry it and of females that mate with such males, two types of resistance gene can be expected to evolve. First, nuclear genes will be selected, which suppress the phenotype of the spiteful cytoplasm in males. Second, genes will be selected for which ameliorate the negative effects caused by males bearing the spiteful cytotype. (Interestingly, cytoplasmic genes are also selected for amelioration, which may lead to a modification-rescue system like CI). Because of these selection pressures, many spiteful cytoplasmic elements may be fixed in natural populations, but the phenotype is not expressed because resistance genes are also fixed. Thus, similar to cytoplasmic male sterility, CI or meiotic drive, crosses between different populations or species may be a promising way of detecting spiteful cytotypes as hypothesized in this paper.
Acknowledgements
We wish to thank Dr Greg Hurst, Lorenzo Zanette and Dr Andy Gardner for helpful comments on the manuscript, and acknowledge funding from the UCL Graduate School to J.E.
References
- Bretman A, Wedell N, Tregenza T. Molecular evidence of post-copulatory inbreeding avoidance in the field cricket Gryllus bimaculatus. Proc. R. Soc. B. 2004;271:159–164. doi: 10.1098/rspb.2003.2563. 10.1098/rspb.2003.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlat S, Merçot H. Wolbachia, mitochondria and sterility. Trends Ecol. Evol. 2001;16:431–432. 10.1016/S0169-5347(01)02224-8 [Google Scholar]
- Clutton-Brock T.H. Female transfer and inbreeding avoidance in social mammals. Nature. 1989;337:70–73. doi: 10.1038/337070a0. 10.1038/337070a0 [DOI] [PubMed] [Google Scholar]
- Cockburn A, Scott M.P, Scotts D.J. Inbreeding avoidance and male-biased natal dispersal in Antechinus spp. (Marsupialia, Dasyuridae) Anim. Behav. 1985;33:908–915. [Google Scholar]
- Cockburn A, Osmond H.L, Muller R.A, Green D.J, Double M.C. Divorce, dispersal and incest avoidance in the cooperatively breeding superb fairy-wren Malurus cyaneus. J. Anim. Ecol. 2003;72:189–201. 10.1046/j.1365-2656.2003.00694.x [Google Scholar]
- Cosmides L.M, Tooby J. Cytoplasmic inheritance and intragenomic conflict. J. Theor. Biol. 1981;89:83–129. doi: 10.1016/0022-5193(81)90181-8. 10.1016/0022-5193(81)90181-8 [DOI] [PubMed] [Google Scholar]
- Dobson F.S, Chesser R.K, Hoogland J.L, Sugg D.W, Foltz D.W. Do black-tailed prairie dogs minimize inbreeding? Evolution. 1997;51:970–978. doi: 10.1111/j.1558-5646.1997.tb03677.x. [DOI] [PubMed] [Google Scholar]
- Enigl M, Schausberger P. Mate choice in the predaceous mite Phytoseiulus persimilis: evidence of self-referent phenotype matching? Entomol. Exp. Appl. 2004;112:21–28. 10.1111/j.0013-8703.2004.00175.x [Google Scholar]
- Foster K.R, Wenseleers T, Ratnieks F.L.W. Spite: Hamilton's unproven theory. Ann. Zool. Fenn. 2001;38:229–238. [Google Scholar]
- Frank S.A, Hurst L.D. Mitochondria and male disease. Nature. 1996;383:224. doi: 10.1038/383224a0. 10.1038/383224a0 [DOI] [PubMed] [Google Scholar]
- Gardner A, West S.A. Spite and the scale of competition. J. Evol. Biol. 2004;17:1195–1203. doi: 10.1111/j.1420-9101.2004.00775.x. 10.1111/j.1420-9101.2004.00775.x [DOI] [PubMed] [Google Scholar]
- Grafen A. A geometric view of relatedness. In: Dawkins R, Ridley M, editors. Oxford surveys in evolutionary biology. Oxford University Press; Oxford: 1985. pp. 28–89. [Google Scholar]
- Hamilton W.D. Selfish and spiteful behaviour in an evolutionary model. Nature. 1970;228:1218–1220. doi: 10.1038/2281218a0. 10.1038/2281218a0 [DOI] [PubMed] [Google Scholar]
- Hartl D.L, Clark A.G. Sinauer Associates, Inc; Sunderland, MA: 1997. Principles of population genetics. [Google Scholar]
- Hoffmann A.A, Turelli M, Simmons G.M. Unidirectional incompatibility between populations of Drosophila simulans. Evolution. 1986;40:692–701. doi: 10.1111/j.1558-5646.1986.tb00531.x. [DOI] [PubMed] [Google Scholar]
- Hoogland J.L. Prairie dogs avoid extreme inbreeding. Science. 1981;215:1639–1641. doi: 10.1126/science.215.4540.1639. [DOI] [PubMed] [Google Scholar]
- Hoogland J.L. Levels of inbreeding among prairie dogs. Am. Nat. 1991;139:591–602. 10.1086/285345 [Google Scholar]
- Hunter M.S, Perlman S.J, Kelly S.E. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc. R. Soc. B. 2003;270:2185–2190. doi: 10.1098/rspb.2003.2475. 10.1098/rspb.2003.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L.D. The evolution of cytoplasmic incompatibility or when spite can be successful. J. Theor. Biol. 1991;148:269–277. doi: 10.1016/s0022-5193(05)80344-3. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Yahara T, Kasuya E, Yamane A. Female control of paternity during copulation: inbreeding avoidance in feral cats. Behaviour. 2001;138:235–250. 10.1163/15685390151074401 [Google Scholar]
- Jarne P, Charlesworth D. The Evolution of the selfing rate in functionally hermaphroditic plants and animals. Annu. Rev. Ecol. Syst. 1993;24:441–466. 10.1146/annurev.es.24.110193.002301 [Google Scholar]
- Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven H. Crossing experiments with Culex strains. Evolution. 1951;5:370–375. [Google Scholar]
- Mack P.D, Hammock B.A, Promislow D.E.L. Sperm competitive ability and genetic relatedness in Drosophila melanogaster: similarity breeds contempt. Evolution. 2002;56:1789–1795. doi: 10.1111/j.0014-3820.2002.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Markow T.A. Assortative mating in Drosophila. Proc. Natl Acad. Sci. USA. 1997;97:7756–7760. doi: 10.1073/pnas.94.15.7756. 10.1073/pnas.94.15.7756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R.M. Biological populations with nonoverlapping generations: stable points, stable cycles, and chaos. Science. 1974;186:645–647. doi: 10.1126/science.186.4164.645. [DOI] [PubMed] [Google Scholar]
- O'Connell M, McClure N, Lewis S.E.M. A comparison of mitochondrial and nuclear DNA status in testicular sperm from fertile men and those with obstructive azoospermia. Hum. Reprod. 2002;17:1571–1577. doi: 10.1093/humrep/17.6.1571. 10.1093/humrep/17.6.1571 [DOI] [PubMed] [Google Scholar]
- Olssen M, Shine R, Madsen Th. Sperm selection by females. Nature. 1996;383:585. 10.1038/383585a0 [Google Scholar]
- Ruiz-Pesini E, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Hum. Genet. 2000;67:682–696. doi: 10.1086/303040. 10.1086/303040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumitou-Laprade P, Cuguen J, Vernet P. Cytoplasmic male sterility in plants: molecular evidence and the nucleocytoplasmic conflict. Trends Ecol. Evol. 1994;9:431–435. doi: 10.1016/0169-5347(94)90126-0. 10.1016/0169-5347(94)90126-0 [DOI] [PubMed] [Google Scholar]
- Simmons L.W. Kin recognition and its influence on mating preference in the field cricket Gryllus bimaculatus (Degeer) Anim. Behav. 1989;38:68–77. [Google Scholar]
- Simmons L.W. Female choice and the relatedness of mates in the field cricket, Gryllus bimaculatus. Anim. Behav. 1991;41:493–501. [Google Scholar]
- Spiropoulos J, Turnbull D.M, Chinnery P.F. Can mitochondrial DNA mutations cause sperm dysfunction? Mol. Hum. Reprod. 2002;8:719–721. doi: 10.1093/molehr/8.8.719. 10.1093/molehr/8.8.719 [DOI] [PubMed] [Google Scholar]
- St John J.C.S, Jokhi R.P, Barratt C.L.R. The impact of mitochondrial genetics on male infertility. Int. J. Androl. 2005;28:65–73. doi: 10.1111/j.1365-2605.2005.00515.x. 10.1111/j.1365-2605.2005.00515.x [DOI] [PubMed] [Google Scholar]
- Stockley P. Sperm selection and genetic incompatibility: does relatedness of mates affect male success in sperm competition? Proc. R. Soc. B. 1999;266:1663–1669. 10.1098/rspb.1999.0829 [Google Scholar]
- Stouthamer R, Breeuwer J.A.J, Hurst G.D.D. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. 10.1146/annurev.micro.53.1.71 [DOI] [PubMed] [Google Scholar]
- Stow A.J, Sunnucks P. Inbreeding avoidance in Cunningham's skinks (Egernia cunninghami) in natural and fragmented habitat. Mol. Ecol. 2004;13:443–447. doi: 10.1046/j.1365-294x.2003.02060.x. 10.1046/j.1365-294X.2003.02060.x [DOI] [PubMed] [Google Scholar]
- Tai F.D, Sun R.Y, Wang T.Z. Does low fecundity reflect kin recognition and inbreeding avoidance in the mandarin vole (Microtus mandarinus)? Can. J. Zool. 2002;80:2150–2155. 10.1139/z02-202 [Google Scholar]
- Wolff J.O. Parents suppress reproduction and stimulate dispersal in opposite-sex juvenile white-footed mice. Nature. 1992;359:409–410. doi: 10.1038/359409a0. 10.1038/359409a0 [DOI] [PubMed] [Google Scholar]
- Wolff J.O, Lundy K.I, Baccus R. Dispersal, inbreeding avoidance and reproductive success in white-footed mice. Anim. Behav. 1988;36:456–465. [Google Scholar]
- Yen J.H, Barr A.R. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature. 1971;232:657–658. doi: 10.1038/232657a0. 10.1038/232657a0 [DOI] [PubMed] [Google Scholar]