Abstract
Sexual reproduction, typically conceived of as a puzzling feature of eukaryotes, has posed an extraordinary evolutionary challenge in terms of the twofold replicative advantage of asexual over sexual organisms. Here we show mathematically that a greater than twofold cost is paid by retroviruses such as HIV during reverse transcription. For a retrovirus, replication is achieved through RNA reverse transcription and the effectively linear growth processes of DNA transcription during gene expression. Retroviruses are unique among viruses in that they show an alternation of generations between a diploid free living phase and a haploid integrated phase. Retroviruses engage in extensive recombination during the synthesis of the haploid DNA provirus. Whereas reverse transcription generates large amounts of sequence variation, DNA transcription is a high-fidelity process. Retroviruses come under strong selection pressures from immune systems to generate escape mutants, and reverse transcription into the haploid DNA phase serves to generate diversity followed by a phase of transcriptional clonal expansion during the restoration of diploidy from a stable, long lived, DNA encoded provirus.
Keywords: evolution of sex, virus evolution, retrovirus, HIV, twofold cost, mutation accumulation
1. Introduction
The Darwinian theory of evolution makes the average rate of replication of an organism a measure of competitive status. The greater the rate of replication, the greater the frequency of genes placed back in the population gene pool. Endogenous mechanisms that increase this frequency are typically deemed adaptive, whereas those that decrease this frequency are deemed maladaptive. Sexual reproduction according to this simple definition is maladaptive, as rather than allowing each genome to place two copies back into the gene pool as it could if asexual, it only allows a single copy to be placed back into the gene pool. This feature has been called the twofold cost of sex, the cost of males and the cost of meiosis (Smith 1978). The fundamental feature of sexual reproduction in contrast to asexual replication, according to measurement by gene frequencies, is the halving of the intrinsic growth rate. This preference for reduced rates of growth in a wide range of eukaryotes has been considered one of the more puzzling traits observed in nature (Smith 1978).
Here we show that this trait is not restricted to sexual reproduction or to eukaryotic organisms, but is a prominent feature of the life cycles of the retroviruses, such as human immunodeficiency virus (HIV) and human T-cell leukaemia virus (HTLV; Coffin et al. 1997). Retroviruses are RNA viruses that integrate a copy of their genome into the DNA genome of their host (Temin 1991). This is achieved through the action of an RNA-dependent DNA polymerase, called reverse transcriptase (RT; Skalka & Goff 1993). Reverse transcription proceeds when a retrovirus specific tRNA binds to a complementary region of the virus RNA called the primer-binding site (PBS). A DNA segment is extended from the bound tRNA in the 3′ to 5′ direction through the action of the polymerase. The underlying replicated genome is then removed by the RNase H activity of RT. The newly synthesized sequence, thus liberated, then binds to the complementary 3′ sequence and extends in the 5′ direction to complete synthesis of the proviral DNA genome with an accompanying breakdown of the remaining RNA genome. The virus encoded protein integrase, then inserts the virus genome into the host DNA genome.
2. Dynamics of integration
Most RNA viruses replicate their genomes using an RNA-dependent RNA polymerase in the cytoplasm. Each new genome synthesized in this way serves indirectly as a template for another round of replication. With retroviruses, replication disappears to be replaced by transcription. In other words, for a retrovirus, replication has become a modified form of host gene expression. We model the intracellular dynamics of the virus life cycle as follows.
Let p(t) be the probability that a viral genome is integrated into the host genome by a time t following infection:
The parameter λ is the rate of integration in a unit time interval. From an integrated provirus, the genomic RNA (G) and viral messenger RNAs are produced:
The parameter mH is the rate of (host-transcriptase-dependent) transcription from the integrated DNA and fG is the fraction of viral genomic RNA in the total transcripts (the remaining fraction fP=1−fG is to be translated into viral proteins). The initial conditions are p(0)=0 and G(0)=0. This gives and
For t≫1/λ,
| 1.1 |
That is, the viral genomic RNAs accumulate linearly with time after a grace period 1/λ=8∼12 h for integration.
Now we compare this with the corresponding rate of genomic RNA accumulation in a model describing a positive-strand RNA virus (e.g. Flavi- and picornaviruses; Krakauer & Komarova 2003). We focus on the rate of genomic RNA accumulation in an infected cell. Because genomic RNA G+ of positive-strand RNA virus and negative-strand RNA G− is templated from G− and G+, respectively, assisted by viral RNA replicase P,
| 1.2 |
| 1.3 |
Here mV is the rate of viral RNA-dependent transcription. As genomic RNAs (G+) also act as messenger RNAs for viral proteins, RNA polymerases (P) are translated with the rate
where k is the rate of translation and μ is the degradation rate of replicase. The initial conditions are G(0)=G0 and P(0)=0, where G0 corresponds to the concentration of a viral genome packaged inside the infected virion. Assuming quasi-equilibrium for the production and degradation of P (i.e. ), we find after some algebra that
| 1.4 |
which diverges to infinity at
where a=mVk/μ. Thus, the number G(t) of genomic RNA tends to infinity in a finite time t=tc. Hence, the RNA virus gains an infinite advantage over the retrovirus in terms of genome production. The rate of growth near tc produces a significantly greater than twofold advantage over the retrovirus life cycle. In classic evolutionary models of sex, the rate of replication of an asexual organism is held constant; hence its population growth rate is kx where k is a rate constant and x population density. With a positive-strand RNA virus, the rate is accelerating since the replication rate is proportional to the product GP. This can be thought of as a simple form of niche construction, whereby the virus synthesizes components of its environment (in this case P) that feed back positively to increase its net rate of replication. With co-infection, each virus strain benefits from the polymerase synthesized by homologous strains.
(a) Constraints on replication, transcription and the role of accessory genes
The rate of production of viral genomes will eventually reduce as a result of depletion of nucleotides, energy supplies and space limitations. Hence, the magnitude of the RNA virus replicative advantage over retrovirus transcription is unlikely to be infinite in practice. However, studies on the knockout of retrovirus accessory genes demonstrates that retrovirus replication strategies are not limited by cell resources but regulatory genes; indeed, HIV may have required the subsequent evolution of these genes to remain competitive in the cell. Knockout studies of the ‘non-essential’ accessory genes vpu and vpr—associated among other things with up-regulating transcription—in HIV-1 produce a 1000-fold reduction in virion production in macrophages (Balliet et al. 1994). Further support for their role in replicative pathogenesis comes from the observation that long term non-progression to AIDS is associated with loss of accessory genes (Yamada & Iwamoto 2000). The accessory genes are also a property of the derived retroviruses (Foley 2000), which have a significant replicative edge over ancestral forms. These studies together illustrate how retroviruses have had to evolve specialized means of reducing the initial replicative cost of integration. Thus, the contemporary replicative efficiency of retroviruses obscures the greater cost that had to be paid at the time at which integration first evolved. The rate of production of virions is important for HIV because higher rates of production reduce the time required for the population to reach the cell burst size and increase the burst size per cell per unit time (Gilchrist et al. 2004). This can maximize within-host relative viral fitness.
In summary, for a retrovirus the number of genomic RNAs accumulates only linearly with time after a long grace period following integration (see equation (2.1)), whereas copy numbers tend to infinity in a finite time tc (as in equation (2.4)) for a positive-strand RNA virus. Though the initial production rate of virus genomes is small for an RNA virus as a result of a dependency on low copy numbers of viral RNA transcriptases, the integration of the retroviral genome depends in a similar way on the RTs packaged inside the infected virion. Overall, retroviruses suffer significant opportunity costs of replication by virtue of interposing a DNA phase in the positive-strand RNA life cycle. Over the course of evolution this cost was reduced through the evolution of accessory genes.
3. Evolving Integration in the ancestral retrovirus
A retrovirus genome is a diploid genome comprising two positive-sense, single-stranded RNAs. During reverse transcription of the virus genome, the DNA polymerase switches back and forth between the two RNA templates, in a process of homologous recombination, producing a recombinant provirus with sequence information derived from both parental RNAs (Hu et al. 2003). Furthermore RT has a high error rate, with approximately 1 in every 2000 bases being a misincorporation (Roberts et al. 1988). Thus, retroviruses, just like sexual eukaryotes, exploit diploidy and recombination as a means of generating genomic variation (Hughes & Otto 1999). As the fidelity of RT is low, there is a concomitant increase in the rate of mutation during the recombination process.
The question therefore arises, why not have evolved recombination with a diploid RNA genome and forgo the DNA phase in the life cycle? This strategy would serve to circumvent the greater than twofold cost and render a significant growth rate advantage. There are two possible sets of answers to this question. The first set is mechanistic and relates to recombination in RNA viruses, and the second is functional and relates to: (i) the durability of the error-correcting DNA provirus in host cells, (ii) the fidelity of replication through repeated transcription of a single DNA provirus as opposed to replication from derived transcripts and (iii) the diversity generation achieved during reverse transcription.
Consider the first reason. The retroviruses are the only diploid positive-strand RNA viruses. As a result, homologous genomes are always in close proximity and potentially physically linked. Whereas a number of RNA viruses have been observed to engage in recombination through copy choice mechanisms—including coronaviruses and picornaviruses—recombination involves collisions between free viral RNAs concentrated at membranes (Lai 1992). For a retrovirus, recombination rates are limited by mechanisms of template switching; for a positive-strand RNA virus, recombination rates are limited by the multiplicity of co-infection and template switching. Furthermore, it seems that RNA-dependent DNA polymerase is more efficient at template switching than RNA-dependent RNA polymerase based on rates of recombination in in vitro experiments. Why this should be the case remains unknown. Thus, the diploid retroviruses are better suited structurally to recombination than the non-integrating RNA viruses. The protracted selection pressure on the RNA-dependent DNA polymerase, by virtue of the persistently diploid state of retroviruses, will also have lead to more effective mechanisms of homologous recombination.
Consider the second set of reasons. Retroviruses are able to simultaneously exploit reverse integration to (i) generate high levels of diversity, (ii) as a mechanism for generating a DNA genome from which genomic transcripts are generated with high fidelity during transcription and (iii) benefit from persistent representation in host cells in the form of DNA provirus undergoing mitotic cell division.
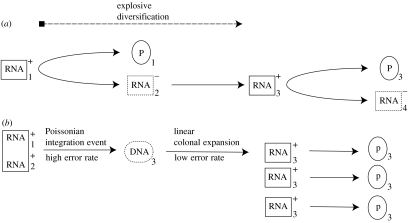
After a phase of recombination and hypermutation during the synthesis of the provirus, the virus mutation rate drops considerably and new genomes are produced through transcriptional clonal expansion (see figure 1b) using a DNA-dependent RNA polymerase II. This is not true for ordinary RNA viruses, which experience a very high rate of diversification during every round of replication using RNA-dependent RNA polymerase (see figure 1a). Thus, for the retrovirus, each new genome is derived from the same DNA provirus benefiting from host DNA repair processes; whereas, for RNA virus, new genomes are reproduced from increasingly aged templates with a non-error-correcting polymerase.
Figure 1.
Logic of virus life cycles. (a) Positive-strand RNA virus of strain type 1 infects cell. RNA is translated into polyprotein 1 and replicated with error into negative sequence strain 2. Positive genome synthesized with error into strain 3. Growth is ‘explosive’ (greater than exponential) as both new genomes and additional replicatory proteins are synthesized throughout the life cycle. (b) Diploid, heterozygous retrovirus infects cell. Proviral genome of strain type 3 is synthesized during reverse integration after a waiting time. Genomes of type 3 are transcribed at high fidelity at a linear rate and translated, producing an effectively clonal population of new retroviruses of strain type 3. Contrast this with (a), the RNA virus, where each new genome must be synthesized from an aging template. While we have not shown it, co-infection with multiple virus strains can produce heterozygous diploids at the final segregation stage of the life cycle by packaging heterologous genomes into the virion.
Hence, retroviruses have killed two birds with one stone: mitigating mutational-error accumulation during replication via transcription while simultaneously producing potentially adaptive variability during integration. A cell infected by a single retrovirus presents a very diverse ensemble of clonal populations of virus, where each population in the ensemble is the transcriptional progeny of a single integration event.
4. Retroviruses and the evolution of sex
Traditionally, three forms of explanation have been provided to account for the evolutionary persistence of sex in eukaryotes: (i) sexual recombination generates diverse progeny to occupy diverse environments (tangled bank (TB) hypothesis; Bell 1982), (ii) sex allows hosts to generate sufficient antigenic diversity to evade parasites (parasite–host coevolution (CE) hypothesis; Hamilton et al. 1990) and (iii) recombination promotes efficient purging of deleterious mutations from the population (synergistic mutation (M) hypothesis; Kondrashov 1988). Empirical evidence has been adduced in support of each of these hypotheses (West et al. 1999). Somewhat surprisingly, similar if not identical arguments can be applied to reverse integration by retroviruses. We examine the explanatory power of each of these theories.
Under the TB hypothesis retroviral diversification becomes a function of the diversity of host niches which the virus population finds itself in. Immune memory establishes a diversity of niches negatively by excluding virus epitopes for which their exists complementary T-cell receptors (Dutton et al. 1998). Furthermore, during the course of a single HIV infection following inoculation with a single strain, variants emerge that are specialists for different tissue types (Ostrowski et al. 1998). The pattern of virus evolution in different tissues can proceed at very different rates, and can favour different amino acid substitutions. Since the infection bottleneck for a retrovirus can be very small, it might be important that sufficient diversity can be generated over the course of a single infection to allow for maximum population growth. However, it is unclear whether such high rates of virus mutation are necessary given that host genomes associated with tissues are highly conserved. Furthermore, non-integrating RNA viruses generate sufficient variation during replication to exploit a diversity of host niches over the course of infection, without recourse to recombination and hypermutation during reverse integration (Evans & Almond 1998). For these comparative reasons, the TB hypothesis for retroviral-integration is somewhat weakened.
Under the CE hypothesis, pressure from the host adaptive immune system favours mechanisms by which the virus can quickly generate variable epitopes promoting immune evasion. It is well known that viruses such as HIV are under very strong selection pressures for diversification, and that reducing virus mutation rates promotes more effective clearance (McMichael & Phillips 1997). This is not unique to retroviruses: it will also be experienced by RNA viruses. Evidence for selective pressures comes from escape variants that mutate away from drug target sequences thereby restoring high rates of replication (Bonhoeffer et al. 1997). The adaptive immune system is the only antagonistic host response to a virus that can evolve on a comparable time scale to the virus and, therefore, imposes strong and variable pressures on mechanisms of diversification. This can be achieved through multiple rounds of replication such as in an RNA virus (polio for example), but excessive mutation can lead to loss of heredity (Eigen 2002) and non-functional viruses. The DNA phase of the retrovirus serves to lessen mutation and promotes a phase of transcriptional clonal expansion analogous to the clonal expansion of immune effector cells of the adaptive immune system. In this way, the retrovirus derives the benefits of recombination and hypermutation, producing diversity comparable to a non-integrating RNA virus through replication, but with the added possibility of exploiting selectively favourable genotypes repeatedly that are stored as a proviral DNA and creating clonal RNA pools through transcription of DNA. Furthermore, integrated viruses would also benefit from vertical transmission as host cells divide and survive for extended periods between host propagation opportunities.
Under the M hypothesis recombination becomes a means of parcelling groups of mutations among the members of a virus population. Recombination allows that some genomes will harbour large numbers of deleterious mutations, whereas others will have very few to none. Assuming that selection works more efficiently in genomes with larger numbers of mutations, then recombination can be favoured. Recently this has been shown to work only in the case of synergistic epistasis among mutations (Bretscher et al. 2004). Furthermore, with segmented viruses, negative complementation can overcome the recombinational advantages (Froissart et al. 2004) and lead to a net increase in mutation load. Moreover, unlike the TB and CE hypotheses, the M hypothesis for reverse transcription does not favour a DNA phase, as it could work just as well for a non-integrating diploid RNA virus capable of recombination. Indeed it would be preferable, as the additional hypermutation associated with generating the provirus could be avoided. It seems, therefore, that the M hypothesis is not a strong explanation for the greater than twofold disadvantage of reverse integration.
5. Conclusions
The twofold cost is not restricted to sexual reproduction as much as the evolutionary literature would seem to imply. The twofold or greater than twofold cost is a more fundamental property related to the tradeoff between diversity-promoting and diversity-preserving mechanisms, and those mechanisms promoting replication. Retroviruses are an ancient evolutionary lineage that have elected to solve their replication-diversity problem in much the same way as complex, multicellular eukaryotic lineages. Interestingly, the most plausible explanation for why retroviruses reverse transcribe, is a mirror image of one of the dominant theories for why sexual eukaryotes produce males. For the retrovirus, the greater than twofold cost pays for stable diversity capable of escaping immune detection; whereas, for the eukaryotes, the twofold cost pays for diversity required to clear virus infection. Once integrated, the provirus derives additional benefits from the increased life span afforded by replication and repair in host cells. Thus, the host cell could be seen as ‘refrigerating’ diversity, increasing the opportunity for transmission to new susceptible cells and hosts.
Acknowledgments
We thank the referees for their constructive suggestions and Sebastian Bonhoeffer for correspondence. D.C.K. is supported by the NIH and the Packard Foundation.
References
- Balliet J.W, Kolson D.L, Eiger G, Kim F.M, McGann K.A, Srinivisan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficieny virus type 1 accessory genes vpr, vpu and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. 10.1006/viro.1994.1225 [DOI] [PubMed] [Google Scholar]
- Bell G. Croom Helm; London: 1982. The masterpiece of nature: the evolution and genetics of sexuality. [Google Scholar]
- Bonhoeffer S, May R.M, Shaw G.M, Nowak M.A. Virus dynamics and drug therapy. Proc. Natl Acad. Sci. USA. 1997;94:6971–6976. doi: 10.1073/pnas.94.13.6971. 10.1073/pnas.94.13.6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M.T, Althaus C.L, Muller V, Bonhoeffer S. Recombination in HIV and the evolution of drug resistance: for better or for worse? Bioessays. 2004;26:180–188. doi: 10.1002/bies.10386. 10.1002/bies.10386 [DOI] [PubMed] [Google Scholar]
- Coffin J.M, Hughes S.H, Varmus H.E, Coffin J.M. Cold Spring Harbor Laboratory Press; Cold spring Harbor, NY: 1997. Retroviruses. [PubMed] [Google Scholar]
- Dutton R.W, Bradley L, Swain S.L. T cell memory. Annu. Rev. Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. 10.1146/annurev.immunol.16.1.201 [DOI] [PubMed] [Google Scholar]
- Eigen M. Error catastrophe and antiviral strategy. Proc. Natl Acad. Sci. USA. 2002;99:13 374–13 376. doi: 10.1073/pnas.212514799. 10.1073/pnas.212514799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.J, Almond J.W. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 1998;6:198–202. doi: 10.1016/s0966-842x(98)01263-3. 10.1016/S0966-842X(98)01263-3 [DOI] [PubMed] [Google Scholar]
- Foley B.T. An overview of the molecular phylogeny of lentiviruses. In: Kuiken C.L, et al., editors. HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Almos National Laboratory; Los Almos, NM: 2000. pp. 35–43. [Google Scholar]
- Froissart R, Wilke C.O, Montville R, Remold S.K, Chao L, Turner P.E. Co-infection weakens selection against epistatic mutations in RNA viruses. Genetics. 2004;168:9–19. doi: 10.1534/genetics.104.030205. 10.1534/genetics.104.030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M.A, Coombs D, Perelson A.S. Optimizing within-host viral fitness: infected cell lifespan and virion production rate. J. Theor. Biol. 2004;229:281–288. doi: 10.1016/j.jtbi.2004.04.015. 10.1016/j.jtbi.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D, Axelrod R, Tanese R. Proc. Natl Acad. Sci. USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.S, Otto S.P. Ecology and the evolution of biphasic life cycles. Am. Nat. 1999;154:306–320. doi: 10.1086/303241. 10.1086/303241 [DOI] [PubMed] [Google Scholar]
- Hu W.S, Rhodes T, Dang Q, Pathak V. Retroviral recombination: review of genetic analyses. Front Biosci. 2003;8:143–155. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. 10.1038/336435a0 [DOI] [PubMed] [Google Scholar]
- Krakauer D.C, Komarova N.L. Levels of selection in positive-strand virus dynamics. J. Evol. Biol. 2003;16:64–73. doi: 10.1046/j.1420-9101.2003.00481.x. 10.1046/j.1420-9101.2003.00481.x [DOI] [PubMed] [Google Scholar]
- Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;3:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael A.J, Phillips R.E. Escape of immunodeficiency virus from immune control. Annu. Rev. Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. 10.1146/annurev.immunol.15.1.271 [DOI] [PubMed] [Google Scholar]
- Ostrowski M, Krakauer D.C, Nowak M, Fauci A. The effect of immune activation on the dynamics of HIV replication on the distribution of viral quasispecies. J. Virol. 1998;72:7772–7784. doi: 10.1128/jvi.72.10.7772-7784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.D, Bebenek K, Kunkel T.A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Smith J.M. Cambridge University Press; Cambridge, UK: 1978. The evolution of sex. [Google Scholar]
- Temin H.M. Sex and recombination in retroviruses. Trends Genet. Mar. 1991;7:71–74. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- West S.A, Lively C.M, Read A.F. A pluralist approach to sex and recombination. J. Evol. Biol. 1999;12:1003–1012. 10.1046/j.1420-9101.1999.00119.x [Google Scholar]
- Skalka A.M, Goff S.P. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. Reverse transcriptase. [Google Scholar]
- Yamada T, Iwamoto A. Comparison of proviral accessory genes between long-term nonprogressors and progressors of human immunodeficiency virus type 1 infection. Arch. Virol. 2000;145:1021–1027. doi: 10.1007/s007050050692. 10.1007/s007050050692 [DOI] [PubMed] [Google Scholar]