Abstract
The extreme polymorphism of the vertebrate major histocompatibility complex (Mhc) is famous for protecting hosts against constantly evolving pathogens. Mate choice is often evoked as a means of maintaining Mhc variability through avoidance of partners with similar Mhc alleles or preference for heterozygotes. Evidence for these two hypotheses mostly comes from studies on humans and laboratory mice. Here, we tested these hypotheses in a wild outbred population of house sparrows (Passer domesticus). Females were not more or less closely related to the males they paired with when considering neutral genetic variation. However, males failed to form breeding pairs when they had too few Mhc alleles and when they were too dissimilar from females at Mhc loci (i.e. had no common alleles). Furthermore, pairs did not form at random as Mhc diversity positively correlated in mating pairs. These results suggest that mate choice evolves in response to (i) benefits in terms of parasite resistance acquired from allelic diversity, and (ii) costs associated with the disruption of co-adapted genes.
Keywords: Mhc, mate choice, outbreeding avoidance, inbreeding avoidance, heterozygote advantage
1. Introduction
Mate choice is often evoked as a means of maintaining major histocompatibility complex (Mhc) variability through preference for heterozygotes (Doherty & Zinkernagel 1975; Landry et al. 2001; Arkush et al. 2002; Penn et al. 2002; McClelland et al. 2003), or avoidance of partners with similar Mhc alleles (Yamazaki et al. 1976, 1988; Potts et al. 1991; Penn & Potts 1998; Freeman-Gallant et al. 2003). Such preference may be better understood in the light of two commonly cited indirect benefits of mate choice, the search for good genes for the progeny (Hamilton & Zuk 1982) and inbreeding avoidance (Yamazaki et al. 1976, 1988; Potts et al. 1991; Penn & Potts 1998; Freeman-Gallant et al. 2003). Less attention has been devoted to the risk of outbreeding depression (Thornhill 1993), despite the fact that disruption of co-adapted genes may result in a severe fitness reduction (Hendry et al. 2000; Neff 2004).
The Mhc genes are particularly relevant for assessing whether mate choice is based on good genes, inbreeding or outbreeding avoidance. This multiloci gene complex codes for highly polymorphic molecules involved in defence against invading pathogens (Bodmer 1972; Carrington et al. 1999; Klein et al. 1993). As a result, individuals with either maximal numbers of Mhc alleles (heterozygote advantage) and/or with specific Mhc alleles (frequency-dependent selection) are privileged in a host population under pathogenic pressures (Doherty & Zinkernagel 1975; Hill 1998; Landry et al. 2001; Langefors et al. 2001; Arkush et al. 2002; Penn et al. 2002; McClelland et al. 2003). A female may therefore benefit from mating with a particular partner if she acquires ‘resistance’ genes for her progeny. Mhc genes are also known to be markers of the degree of relatedness between individuals, and a few studies have shown that Mhc-based mate choice may reflect the avoidance of incestuous mating rather than the search for ‘good genes’ (Yamazaki et al. 1976, 1988; Potts et al. 1991; Brown & Eklund 1994; Penn & Potts 1998).
Each of these hypotheses leads to testable predictions: (i) if there is a selective advantage of carrying many different alleles, then we expect the most diverse individuals to be preferred over the less diverse ones during pair formation; (ii) if inbreeding avoidance is the main force driving mate preference, we expect pairs to be assembled of more dissimilar individuals than found at random; and finally, (iii) if mate choice aims at preserving the linkage of co-adapted genes, we expect mates to share more alleles than found at random.
We tested these hypotheses in a wild outbred population of house sparrows (Passer domesticus). All individuals were screened both at neutral microsatellite loci and at the peptide-binding region of the most polymorphic gene family of Mhc class I (Bonneaud et al. 2004a). Previous work on this and another population showed that (i) Mhc diversity positively correlates with reproductive success (Bonneaud et al. 2004b), and (ii) specific Mhc alleles are associated with stronger immune responses to T-dependent antigens (Bonneaud et al. 2005), and confer increased resistance to malaria parasites (Bonneaud et al. in press). These results suggest that Mhc genes may be under intense selection in the house sparrow.
2. Material and methods
(a) The population
This study was conducted during spring 2003 at the ‘Centre d'Etude Biologique de Chizé’ (France), on a nest-box house sparrow population established in 1992 (Chastel et al. 2003). Breeding males and females were captured at the nest-box when chicks were 8 days old. Breeding individuals that we failed to capture at nest, as well as non-mated individuals, were captured with mist nets permanently set up throughout the breeding season. Here we define mate choice as the selection of a social mate, and mating success as the outcome of pair formation. Non-mated males and females are therefore individuals who have failed to form a breeding pair. We measured body mass (±0.1 g), tarsus length (±0.01 mm) and male badge size (±0.01 mm, adult males only) for all captured adults and nestlings (when they were 10 days old). In addition, blood was sampled (ca 150 μl) and stored in PBS/EDTA 2 mM buffer at −20 °C.
(b) Mhc screening
We screened all individuals (adults and chicks) to assess allelic diversity at the most variable Mhc class I gene family using the PCR-based denaturing gradient gel electrophoresis (DGGE) method (Bonneaud et al. 2004a). This method allows us to examine single-nucleotide polymorphism at Mhc class I exon 3, corresponding to the highly variable peptide binding site of the protein (α2 domain). The PCR primers used were GCA21M-fA23M. Each DGGE band is considered to correspond to one allele (Bonneaud et al. 2004a).
This genotyping method does not allow us to determine the level of heterozygosity present at each individual locus. Instead, it gives us an estimate of the overall number of alleles present in the most variable lineage of Mhc class I genes. Individuals who carry the largest numbers of Mhc alleles are considered to be the most diverse. Although there seems to be a maximal number of six Mhc loci (the most diverse individuals have 11 alleles), we cannot rule out the possibility that we may be amplifying a gene family encompassing more than six loci.
(c) Microsatellite analyses
Adults were genotyped using seven microsatellite markers: Pdo3, Pdo4, Pdo5, Pdo6 (Griffith et al. 1999), Mjg1 (Shou-Hsien et al. 1997), Fhu2 (Primmer et al. 1996) and Ase18 (Richardson et al. 2000). Amplifications were run in a final volume of 10 μl, including 15–50 ng of DNA, 50–200 nM of each primer, 300 μM of dNTPs, 1 μl of 10× incubation buffer (50 mM KCl, 10 mM Tris–HCl, 1.5 mM MgCl2, 0.1% TritonX-100, pH 9.0) and 0.25 U of Taq DNA polymerase (Qbiogene). The reaction was performed in a Gene Amp PCR System 9700 thermocycler (Applied Biosystems). Samples were then run in an ABI 310 automated sequencer (Applied Biosystems). Allele sizes were determined using GENESCAN software v. 2.1 with reference to the GENESCAN ROX 500 size standard.
(d) Population structure and measures of relatedness
Standard diversity indices, molecular indices and test of Hardy–Weinberg equilibrium were performed using Arlequin v. 2.0 software (Schneider et al. 2000). In addition, we used Guo & Thompson's method (1992) to detect significant departure from Hardy–Weinberg equilibrium (Fisher's exact test with the Markov chain method (chain length=10 000 and dememorizations steps=10 000)). Coefficients of relatedness (r) based on microsatellite genotype similarity were calculated between all male and female pairs using the KINSHIP program (Goodnight & Queller 1999). Pairwise relatedness values between all individuals of the population were also estimated to determine whether the population was inbred or outbred. Finally, mean individual heterozygosity was calculated across all loci.
(e) Statistical analyses
The probability of forming a breeding pair was modelled using a generalized linear model with number of Mhc alleles, body mass and tarsus length as continuous variables and sex as a factor (SAS institute 1999). Mhc similarity between females and males were calculated as Mhc band-sharing values; the proportion of band-sharing in a pair is twice the number of the bands shared by two individuals divided by the sum of the bands of each individual [D=2Fab/(Fa+Fb)] (Wetton et al. 1987). Band-sharing and relatedness values were analysed using a randomization test (Manly 1997). The observed median value of band-sharing and relatedness in breeding pairs was compared with the value obtained by 10 000 bootstrapped band-sharing and relatedness medians.
Spatial autocorrelation of number of Mhc alleles was assessed using Moran's I autocorrelation coefficients (R Package, v. 4.0; see http://www.bio.umontreal.ca/Casgrain/en/labo/R/).
3. Results
We assessed mate choice in 45 female and 56 male adult house sparrows. Only 30 individuals of each sex successfully formed a breeding pair over the entire reproductive season (out of 37 breeding attempts, each involving different pairs of individuals).
(a) Microsatellite genotyping
The number of alleles per locus was 17 for Pdo3, 24 for Mjg1, 85 for Pdo4, 14 for Fhu2, 16 for Pdo5, 15 for Ase18 and 70 for Pdo6. The mean number of pairwise differences between all pairs of haplotypes was 6.321±3.010 alleles and the average gene diversity was 0.903±0.476. We found a significant deficiency in heterozygotes in three loci (Pdo4: p<0.0001; Fhu2: p<0.0001; Pdo6: p=0.045). Females were not more or less related to their mates than found at random (randomization test: observed median relatedness value=−0.014, median of 10 000 bootstrapped band-sharing values=−0.018, p>0.05). Negative pairwise values of relatedness indicate that the individuals were less genetically alike than found on average. In addition, relatedness values between mated pairs did not significantly differ from those obtained when pairing females and non-breeding males (randomization test: observed median band-sharing value=−0.014, median of 10 000 bootstrapped band-sharing values=−0.009, p>0.05). Furthermore, it was found that mated males were not more heterozygous across all loci than non-mated males (mated males=0.114±0.020; non-mated males=0.120±0.018; Student's t78=−0.20, p=0.841). Finally, calculation of overall pairwise population relatedness showed that this population was outbred (median=−0.017, mean=6.627×10−5±0.131).
(b) Mhc-based mate choice
We found a total of 46 Mhc alleles present in 1–50% of the population, and individuals exhibited between 1 and 11 alleles (mean±s.e., males=3.48±2.39, females=3.40±1.90, ANOVA: F1,99=0.04, p=0.852). We tested whether the occurrence of each allele was independent of others in a restricted sample of the most frequent Mhc alleles (range 20–51%, n=6). Analysis of contingency tables indicated that two pairs of Mhc alleles occurred together more frequently than expected at random (alleles a161–a165, p=0.0006; alleles a172–a163, p=0.0093).
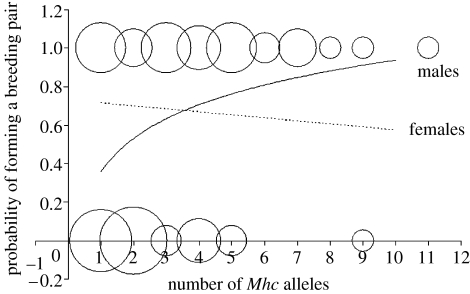
Males that succeeded in forming a breeding pair displayed higher Mhc allele diversity than non-mated ones (χ2=6.63, p=0.010; figure 1), but females did not (χ2=0.77, p=0.381). This resulted in a statistically significant sex by allelic diversity interaction (χ2=3.85, p=0.0497). However, the association between Mhc diversity and probability of forming a breeding pair was not due to the confounding effect of body condition since neither body mass nor tarsus length were retained in the model (p=0.524 and p=0.625, respectively).
Figure 1.
Probability of forming a breeding pair as a function of the number of Mhc alleles for both males (solid line) and females (dotted line). Probabilities of forming a breeding pair have been modelled using a logistic regression model. Observed values of male pairing success are represented by circles that vary in size according to the number of observations (n=1, 2, 3, 4 or 5).
Within the breeding population, pairs were not assembled of more dissimilar individuals than found at random (randomization test: observed median band-sharing value=0.2, median of 10 000 bootstrapped band-sharing values=0.182, p=0.306; figure 2). Instead, females were paired to mates with whom they shared more Mhc alleles, as shown by the significant difference in the band-sharing values between females and breeding males versus non-breeding males (randomization test: observed median band-sharing value=0.2, median of 10 000 bootstrapped band-sharing values=0, p=0.006; figure 3). Importantly, random pairs of males and females had an extremely low probability of carrying identical Mhc genotypes (0.3%), whereas they had a 54% chance of possessing completely different sets of alleles (i.e. band-sharing=0).
Figure 2.
Distribution of the 10 000 bootstrapped Mhc band-sharing values for pairs assembled at random from the whole male population (i.e. mated and non-mated males). The observed value indicates the band-sharing value observed in true breeding pairs.
Figure 3.
Distribution of the 10 000 bootstrapped Mhc band-sharing values for pairs composed solely of females and non-mated males. The observed value indicates the band-sharing value observed in true breeding pairs.
The numbers of Mhc alleles found in males and females were significantly correlated (Spearman's r=0.371, p=0.024, n=37). This non-random configuration of mating events affected the average number of Mhc alleles in chicks, so that highly diverse pairs produced highly diverse broods of chicks (slope±s.e.=0.523+0.129, p=0.0003, n=34). The mean allelic diversity did not differ between adults (mean±s.e.=3.45±2.18) and offspring (mean±s.e.=3.39±1.79, Student's t174=0.21, p=0.834).
Mhc-based mate selection implies that Mhc genotypes can be predicted by phenotypic traits used as cues by the choosing partner. Nevertheless, we failed to show any significant correlation between Mhc genotypes and body mass and tarsus length in males and females, or the expression of a male secondary sexual trait (badge size; all p>0.1). The house sparrow is a colonial nesting species with individuals aggregating at variable levels of density around nesting sites. We therefore tested whether a spatial aggregation of breeding individuals could generate the observed assortative mating. Coefficients of spatial autocorrelation did not, however, indicate any particular structuring of males, females or pairs in relation to their number of Mhc alleles (Moran's I, all p>0.1).
4. Discussion
The mating patterns observed in this study strongly promote the idea of Mhc-related mate choice in birds. Mate choice in house sparrows proved to be based not only on the partners' allelic diversity at Mhc loci, but also on the number of shared Mhc alleles. As a result, males of low Mhc diversity that were too dissimilar from females at Mhc loci (i.e. no common alleles) were excluded from reproductive events. Moreover, Mhc diversity was positively correlated in both mating partners. Importantly, variation at neutral microsatellite markers did not influence reproductive decisions, suggesting that variation at Mhc loci per se may be driving mate choice in this population.
It is currently debated whether selection favours an intermediate or a maximum number of Mhc alleles (Penn et al. 2002; McClelland et al. 2003; Hedrick 2004; Wegner et al. 2004). An optimum number of Mhc alleles could be generated by a trade-off between selection for recognition of the largest antigenic peptide repertoire (Carrington et al. 1999; Arkush et al. 2002; Penn et al. 2002; McClelland et al. 2003), which favours more alleles, and selection to minimize the loss of T cell clones due to self-tolerance induction (Nowak et al. 1992), which favours fewer. By excluding males that are too dissimilar (i.e. band-sharing=0), female house sparrows may prevent diversity from reaching detrimental levels in their offspring. Yet, by giving preference to mates with high allelic diversity, they may simultaneously endeavour to maximize the number of Mhc alleles in their progeny.
Rejection of the most dissimilar males may, however, have a larger significance than just avoiding costs of unfavourable diversity at the Mhc. Mating between highly divergent partners may disrupt local adaptations or co-adapted gene complexes (Hendry et al. 2000) and produce offspring of lower fitness. Furthermore, intermediate levels of genomic divergence have been shown to be associated with higher reproductive success than more extreme levels of divergence (Neff 2004). The optimal level of genomic divergence may hence vary according to whether it is more advantageous to produce higher-quality surviving inbreds despite the increased mortality of offspring.
The Mhc alleles a172 and a163 were found to be significantly associated with each other. In a previous study involving this population of wild house sparrows, the allele a172 was found to be associated with higher resistance to infections with the most common local Plasmodium strain, whereas none of the individuals that carried the a163 allele endured simultaneous infections with two strains of malaria parasites (Bonneaud et al. 2005). We can easily speculate that rupture of this association of alleles would be selectively detrimental to the individual in terms of increased susceptibility to malaria infections. Co-adapted genes therefore exist in this population and probably undergo both parasite-mediated selection and reproductive selection.
Analyses of relatedness at neutral microsatellite loci showed that this population was outbred. In addition, mate choice was found to be based uniquely on Mhc genotypes, as females were not more or less closely related to the subset of males they paired with. Females only had 0.03% chances of pairing with a male carrying an identical combination of microsatellite alleles and 8% chances of pairing with a completely different male, so the absence of microsatellite-based reproductive preferences may be explained by a lack of inbreeding or outbreeding risks at neutral loci.
Our study reveals that females favoured males of comparable Mhc diversity and with higher numbers of matching alleles than found at random. It is fundamental to point out that females did not select males with identical Mhc genotypes, but rather excluded males with whom they shared no alleles. Hence our findings do not contradict previous work reporting avoidance of Mhc-similar mates (Yamazaki et al. 1976, 1988; Potts et al. 1991; Freeman-Gallant et al. 2003). Most experimental assessments of mate choice have examined female preference between males of Mhc genotypes identical or different from their own. Yet, recent evidence suggests preference for mates with small or intermediate numbers of matching Mhc alleles rather than for mates with no or all matching alleles (Jacob et al. 2002). In this outbred population, the probability of pairing with a male of identical Mhc genotype was extremely low (0.3%), whereas the chance of randomly pairing with a male carrying a completely different set of Mhc alleles (i.e. band-sharing=0) was 54%. A biased decision may allow females to control the number of Mhc alleles in their progeny and their overall level of genomic divergence, while still maximizing the number of Mhc alleles by favouring the most diverse males.
An underlying assumption to Mhc-based mate choice is that Mhc genotypes can be predicted by phenotypic traits used as cues by choosy partners. The direct implications of Mhc genes in fighting off diseases make obvious the links between Mhc genotypes and condition-dependent traits (Zelano & Edwards 2002). We failed to show any significant correlation between Mhc genotypes and body mass or tarsus length in males and females, or expression of a male secondary sexual trait (badge size). In the same way, coefficients of spatial autocorrelation did not indicate any particular structuring of males, females or pairs as a function of their number of Mhc alleles. Studies on Mhc-based mate choice in mammals and fish indicate that olfactory cues are generally used to discriminate among Mhc genotypes (Brown & Eklund 1994; Reusch et al. 2001). Because birds are commonly thought to be anosmatic, targets of sexual selection usually involve visual signals. Yet recent experimental work on different bird orders has revealed unsuspected abilities to discriminate between the odours of conspecifics and between those of aromatic plants (Petit et al. 2002; Hagelin et al. 2003; Bonadonna & Nevitt 2004). Association between the Mhc, odours, and mating preferences in birds is a promising question that awaits further exploration.
Mhc-based mate choice has now been reported in a variety of species throughout the vertebrate realm. Evidence in mammals (Yamazaki et al. 1976, 1988; Potts et al. 1991; Penn & Potts 1998; Jacob et al. 2002; but see Paterson & Pemberton 1997), fish (Landry et al. 2001; Reusch et al. 2001; Aeschlimann et al. 2003), reptiles (Olsson et al. 2003) and birds (Freeman-Gallant et al. 2003; but see Westerdahl 2004; Richardson et al. 2005) point towards a preference for the most heterozygous individuals or the use of Mhc-based cues to evaluate the degree of relatedness of a potential partner in order to avoid inbreeding or outbreeding. The optimal level of genomic diverge may however lie at more intermediate levels that maximize reproductive success (Thornhill 1993; Neff 2004). In this outbred population of house sparrows, female choice probably maintains a certain level of diversity in the offspring, while avoiding the costs resulting from the disruption of local adaptations.
Acknowledgments
We are grateful to Mark Kirkpatrick and Jean Clobert for their useful comments, to Céline Chastel for wonderful help in the field, and to John Ewen and Ryan Calsbeek for valuable advice. Financial support came from the Ministère de la Recherche (bourse doctorale to C.B. and ACI Jeunes Chercheurs to G.S.).
Footnotes
Present address: BioGéoSciences, CNRS UMR 5561, Université de Bourgogne, 6 Boulevard Gabriel, 21000 Dijon, France.
References
- Aeschlimann P.B, Häberli M.A, Reusch T.B.H, Boehm T, Milinski M. Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. Behav. Ecol. Sociobiol. 2003;54:119–126. [Google Scholar]
- Arkush K.D, Giese A.R, Mendonca H.L, McBride A.M, Marty G.D, Hedrick P.W. Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawyscha): effects of inbreeding and major histocompatibility complex genotypes. Can. J. Fish. Aquat. Sci. 2002;59:966–975. 10.1139/f02-066 [Google Scholar]
- Bodmer W.F. Evolutionary significance of the HLA-system. Nature. 1972;237:139–145. doi: 10.1038/237139a0. 10.1038/237139a0 [DOI] [PubMed] [Google Scholar]
- Bonadonna F, Nevitt G.A. Partner-specific odor recognition in an Antarctic seabird. Science. 2004;306:835. doi: 10.1126/science.1103001. 10.1126/science.1103001 [DOI] [PubMed] [Google Scholar]
- Bonneaud C, Mazuc J, Chastel O, Westerdahl H, Sorci G. Terminal investment induced by immune challenge and Mhc associated fitness traits in the house sparrow. Evolution. 2004a;58:2823–2830. doi: 10.1111/j.0014-3820.2004.tb01633.x. [DOI] [PubMed] [Google Scholar]
- Bonneaud C, Sorci G, Morin V, Westerdahl H, Zoorob R, Wittzell H. Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus) Immunogenetics. 2004b;55:855–865. doi: 10.1007/s00251-004-0648-3. 10.1007/s00251-004-0648-3 [DOI] [PubMed] [Google Scholar]
- Bonneaud C, Richard M, Faivre B, Westerdahl H, Sorci G. An Mhc class I allele associated to the expression of T-dependent immune response in the house sparrow. Immunogenetics. 2005;57:782–789. doi: 10.1007/s00251-005-0046-5. [DOI] [PubMed] [Google Scholar]
- Bonneaud, C., Pérez-Tris, J., Federici, P., Chastel, O. & Sorci, G. In press. Mhc alleles associated with local resistance to malaria in a passerine. Evolution [PubMed]
- Brown J.L, Eklund A. Kin recognition and the major histocompatibility complex: an integrative review. Am. Nat. 1994;143:435–461. 10.1086/285612 [Google Scholar]
- Carrington M, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. 10.1126/science.283.5408.1748 [DOI] [PubMed] [Google Scholar]
- Chastel O, Lacroix A, Kersten M. Pre-breeding energy requirements: thyroid hormone, metabolism and the timing of reproduction in house sparrows Passer domesticus. J. Avian Biol. 2003;34:298–306. 10.1034/j.1600-048X.2003.02528.x [Google Scholar]
- Doherty P.C, Zinkernagel R.M. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. 10.1038/256050a0 [DOI] [PubMed] [Google Scholar]
- Freeman-Gallant C.R, Meguerdichian M, Wheelwright N.T, Sollecito S.V. Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Mol. Ecol. 2003;12:3077–3083. doi: 10.1046/j.1365-294x.2003.01968.x. 10.1046/j.1365-294X.2003.01968.x [DOI] [PubMed] [Google Scholar]
- Goodnight K.F, Queller D.C. Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol. Ecol. 1999;8:1231–1234. doi: 10.1046/j.1365-294x.1999.00664.x. 10.1046/j.1365-294x.1999.00664.x [DOI] [PubMed] [Google Scholar]
- Griffith S.C, Stewart I.R.K, Dawson D.A, Owens I.P.F, Burke T. Extra-pair paternity in mainland and island populations of a socially monogamous bird, the house sparrow (Passer domesticus): is there an “island effect”? Biol. J. Linn. Soc. 1999;68:303–316. 10.1006/bijl.1999.0343 [Google Scholar]
- Guo S, Thompson E. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hagelin J.C, Jones I.L, Rasmussen L.E.L. A tangerine-scented social odour in a monogamous seabird. Proc. R. Soc. B. 2003;270:1323–1329. doi: 10.1098/rspb.2003.2379. 10.1098/rspb.2003.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.D, Zuk M. Heritable true fitness and bright birds: a role for parasites? Science. 1982;218:384–387. doi: 10.1126/science.7123238. [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Comment on “Parasite selection for immunogenetic optimality”. Science. 2004;303:957a. doi: 10.1126/science.1093355. 10.1126/science.1092163 [DOI] [PubMed] [Google Scholar]
- Hendry A.P, Burg J.K, Bentzen P, Volk E.C, Quinn T.P. Rapid evolution of reproductive isolation in the wild: evidence from introduced salmon. Science. 2000;290:516–518. doi: 10.1126/science.290.5491.516. 10.1126/science.290.5491.516 [DOI] [PubMed] [Google Scholar]
- Hill A.V.S. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. 10.1146/annurev.immunol.16.1.593 [DOI] [PubMed] [Google Scholar]
- Jacob S, McClintock M.K, Zelano B, Ober C. Paternally inherited HLA alleles are associated with women's choice of male odor. Nat. Genet. 2002;30:175–179. doi: 10.1038/ng830. 10.1038/ng830 [DOI] [PubMed] [Google Scholar]
- Klein J, Satta Y, O'hUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu. Rev. Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. 10.1146/annurev.iy.11.040193.001413 [DOI] [PubMed] [Google Scholar]
- Landry C, Garant D, Duchesne P, Bernatchez L. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar) Proc. R. Soc. B. 2001;268:1279–1285. doi: 10.1098/rspb.2001.1659. 10.1098/rspb.2001.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langefors Å, Lohm J, Grahn M, Andersen Ø, von Schantz T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. B. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. 10.1098/rspb.2000.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B.F.J. 2nd edn. Chapman & Hall; London: 1997. Randomisation, bootstrap and Monte Carlo methods in biology. [Google Scholar]
- McClelland E.E, Penn D.J, Potts W.K. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 2003;71:2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. 10.1128/IAI.71.4.2079-2086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff B.D. Stabilizing selection on genomic divergence in a wild fish population. Proc. Natl Acad. Sci. USA. 2004;101:2381–2385. doi: 10.1073/pnas.0307522100. 10.1073/pnas.0307522100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A, Tarczy-Hornoch K, Austyn J.M. The optimal number of major histocompatibility complex molecules in an individual. Proc. Natl Acad. Sci. USA. 1992;89:10 896–10 899. doi: 10.1073/pnas.89.22.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Madsen T, Norby J, Wapstra E, Ujvari B, Wittzell H. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. B. 2003;270(Suppl. 2):S254–S256. doi: 10.1098/rsbl.2003.0079. 10.1098/rsbl.2003.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S, Pemberton J.M. No evidence for major histocompatibility complex-dependent matings patterns in a free-living ruminant population. Proc. R. Soc. B. 1997;264:1813–1819. doi: 10.1098/rspb.1997.0250. 10.1098/rspb.1997.0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D, Potts W. MHC-disassortative mating preferences reversed by cross-fostering. Proc. R. Soc. B. 1998;265:1299–1306. doi: 10.1098/rspb.1998.0433. 10.1098/rspb.1998.0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D.J, Damjanovich K, Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA. 2002;99:11 260–11 264. doi: 10.1073/pnas.162006499. 10.1073/pnas.162006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Hossaert-McKey M, Perret P, Blondel J, Lambrechts M.M. Blue tits use selected plants and olfaction to maintain an aromatic environment for nestlings. Ecol. Lett. 2002;5:585–589. 10.1046/j.1461-0248.2002.00361.x [Google Scholar]
- Potts W.K, Manning C.J, Wakeland E.K. Mating patterns in seminatural populations of mice influenced by MHC genotypes. Nature. 1991;352:619–621. doi: 10.1038/352619a0. 10.1038/352619a0 [DOI] [PubMed] [Google Scholar]
- Primmer C.R, Møller A.P, Ellegren H. New microsatellites from the pied flycatcher Ficedula hypoleuca and the swallow Hirundo rustica genomes. Hereditas. 1996;124:281–283. doi: 10.1111/j.1601-5223.1996.00281.x. 10.1111/j.1601-5223.1996.00281.x [DOI] [PubMed] [Google Scholar]
- Reusch T.B.H, Häberli M.A, Aeschlimann P.B, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. 10.1038/35104547 [DOI] [PubMed] [Google Scholar]
- Richardson D.S, Jury F.L, Dawson D.A, Salgueiro P, Komdeur J, Burke T. Fifty Seychelles warbler (Acrocephalus sechellensis) microsatellite loci polymorphic in Sylviidae species and their cross-species amplification in other passerine birds. Mol. Ecol. 2000;9:2155–2234. doi: 10.1046/j.1365-294x.2000.105338.x. 10.1046/j.1365-294X.2000.105338.x [DOI] [PubMed] [Google Scholar]
- Richardson D.S, Komdeur J, Burke T, von Schantz T. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B. 2005;272:759–767. doi: 10.1098/rspb.2004.3028. 10.1098/rspb.2004.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS institute 1999 SAS user's guide: statistics, v. 6.12. Cary, NC: SAS institute.
- Schneider S, Roessli D, Excoffier L. 2000. Arlequin: a software for population genetics data analysis. Version 2.0. Genetics and Biometry Lab, Department of Anthropology, University of Geneva. [Google Scholar]
- Shou-Hsien L, Yi-Jiun H, Brown J.L. Isolation of tetranucleotide microsatellites from the Mexican jay Aphelocoma ultramarina. Mol. Ecol. 1997;6:499–501. doi: 10.1046/j.1365-294x.1997.00215.x. 10.1046/j.1365-294X.1997.00215.x [DOI] [PubMed] [Google Scholar]
- Thornhill, N. W. (ed.) 1993 The natural history of inbreeding and outbreeding: theoretical and empirical perspectives Chicago, IL: University of Chicago Press.
- Wegner K, Kalbe M, Kurtz J, Reusch T.B.H, Milinski M. Response to comment on “Parasite selection for immunogenetic optimality”. Science. 2004;303:957b. doi: 10.1126/science.1088293. 10.1126/science.1093355 [DOI] [PubMed] [Google Scholar]
- Westerdahl H. No evidence of an MHC-based female mating preference in great reed warblers. Mol. Ecol. 2004;13:2465–2470. doi: 10.1111/j.1365-294X.2004.02238.x. 10.1111/j.1365-294X.2004.02238.x [DOI] [PubMed] [Google Scholar]
- Wetton J.H, Carter R.E, Parkin D.T, Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987;327:147–149. doi: 10.1038/327147a0. 10.1038/327147a0 [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Boyse E.A, Mike V, Thaler H.T, Mathieson B.J, Abbott J, Boyse J, Zayas Z. Control of mating preferences in mice by genes of the major histocompatibility complex. J. Exp. Med. 1976;144:1324–1335. doi: 10.1084/jem.144.5.1324. 10.1084/jem.144.5.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Beauchamp G.K, Kupniewski J, Bard J, Thomas L, Boyse E.A. Familial imprinting determines H-2 selective mating preferences. Science. 1988;240:1331–1332. doi: 10.1126/science.3375818. [DOI] [PubMed] [Google Scholar]
- Zelano B, Edwards S.V. An Mhc component to kin recognition and mate choice in birds: predictions, progress, and prospects. Am. Nat. 2002;160:S225–S237. doi: 10.1086/342897. 10.1086/342897 [DOI] [PubMed] [Google Scholar]