Abstract
The barn swallow (Hirundo rustica) is one of most widely distributed swallows, owing in part to its recent switch from natural nest sites to human structures. We conducted phylogenetic analysis of mitochondrial (mt) and nuclear DNA to explore the recent evolutionary history of this species. Strongly supported mtDNA clades corresponded to Europe, Asia and North America plus the Baikal region of Asia. Analysis of sequence data from a sex-linked nuclear gene was unable to recover the phylogenetic splits in the mtDNA tree, confirming that the main clades evolved recently. The phylogenetic pattern suggests that the ancestral area of the barn swallow was the holarctic; most divergence events are consistent with vicariance. Most unexpectedly, analyses show that barn swallows from North America colonized the Baikal region in the recent past (one fixed substitution). This dispersal direction is opposite of that for most nearctic–palearctic taxon exchanges. Although this invasion was envisioned to coincide with the appearance of new types of human dwelling in the Baikal region, calibration of molecular divergence suggests an older dispersal event. A recent history of gene flow within the main palearctic clades is consistent with range and population expansion owing to new nesting opportunities provided by human settlements. Contrary to expectation, populations in North America appear historically larger and more stable than those in the palearctic. The Baikal population apparently has not increased greatly since colonization.
Keywords: Holarctic, mtDNA, vicariance, phylogeography, biogeography, dispersal
1. Introduction
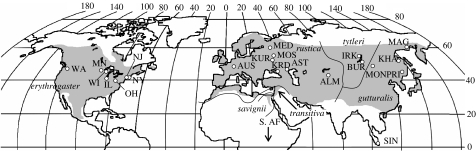
By virtue of its vernacular name, the barn swallow (Hirundo rustica) would seem to be dependent on human settlements for its existence. Indeed, barn swallows often attach their open-cup mud nests to the interior walls of barns. Of course, this begs the question of how barn swallows managed to nest before humans built barns. In North America, barn swallows historically nested in caves, but by the early 1800s had begun to affix their nests to Native American structures (Brown & Brown 1999), and subsequently to structures and bridges in European settlements; natural nest sites are rarely used today. The widespread availability of nest sites resulted in dramatic range and population expansion in North America. A similar history occurred in Europe (Møller 1994) and Asia. For example, Smirenskiy & Mishchenko (1981) studied barn swallows from the Baikal, Amur and Primor'e regions of eastern Russia. They reported that prior to the seventeenth century, native people of the Amur region lived mostly in subterranean structures, which together with a paucity of natural nest sites suggested to them that barn swallows were historically absent in this region. The arrival of people who built above-ground structures potentially led to colonization of the region by barn swallows. As a consequence of its association with human settlements, the barn swallow today is the most widespread and abundant swallow species in the world, breeding in Eurasia and North America (figure 1) and wintering south to tropical Africa, northern Australia, Central and South America (Turner & Rose 1989; Brown & Brown 1999). Despite the vagility of barn swallows, subspecific taxonomy suggests some isolation (see figure 1).
Figure 1.
World breeding range (grey, following Brown & Brown (1999) and Turner (2004)) and distribution of sampling localities for the barn swallow Hirundo rustica. From six (fig. 1; Cramp 1988; Turner & Rose 1989) to eight (Turner 2004) subspecies have been recognized, differing mainly in the coloration of the underparts and the extent of the breast band (Turner & Rose 1989). Lines indicate approximate borders of subspecies distribution (Cramp 1988). In general, western Eurasian rustica and eastern Asian gutturalis are whitish below, whereas the underparts of North American erythrogaster, south-central Siberian tytleri, Egyptian savignii and eastern Mediterranean transitiva are more rufous. The breast band varies in most populations, generally it is complete in rustica, transitiva and savignii, but only partial in erythrogaster. In tytleri and gutturalis, black patches on sides of breast can be connected by a narrow black band across lower chest (Cramp 1988). Abbreviations for localities are explained in table 1 of electronic supplementary material.
We analysed genetic variation in the mitochondrial DNA (mtDNA) and nuclear genomes of barn swallows to evaluate potential genetic consequences of its large range and recent history of range and population expansion.
2. Material and methods
We analysed breeding individuals from 13 Eurasian and six North American localities as well as wintering individuals from South Africa and Singapore, resulting in a total of 74 individuals (figure 1; table 1 of electronic supplementary material). A study skin and a spread wing were preserved for the majority of individuals deposited at the Burke Museum, University of Washington, Seattle and the Bell Museum, University of Minnesota, St Paul (table 1 of electronic supplementary material). Tissue samples were preserved in 95% ethanol or frozen in liquid nitrogen. Genomic DNA was extracted using DNeasy Tissue Kit (Qiagen, Valencia, CA). The complete mitochondrial NADH subunit 2 (ND2) gene was amplified with primers metL (5′-AAGCTATCGGGCCCATACCCG-3′) and ASN (5′-GATCRAGGCCCATCTGTCTAG-3′, both designed by O. Haddrath 2004, unpublished). The following thermocycling parameters were used: 2.5 min of initial denaturation at 95 °C, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 1 min, concluded with an extension of 10 min at 72 °C.
To compare evolutionary hypotheses derived from mtDNA phylogenetic tree, nine individuals were sequenced for a nuclear, Z-linked intron of the MUSK gene with the primers MUSK-I3F (5′-CTTCCATGCACTACAATGGGAAA-3′) and MUSK-I3R (5′-CTCTGAACATTGTGGATCCTCAA-3′), both designed by F. K. Barker (2004, personal communication). PCR for MUSK started with 15 min of initial denaturation at 95 °C and followed by five cycles of 20 s at 95 °C, 20 s at 58 °C and 75 s at 72 °C, then by five cycles of 20 s at 95 °C, 20 s at 56 °C and 75 s at 72 °C, then by five cycles of 20 s at 95 °C, 20 s at 54 °C and 75 s at 72 °C, then by 20 cycles of 20 s at 95 °C, 20 s at 52 °C and 75 s at 72 °C and concluded with 3 min at 72 °C.
PCR products were cleaned with a QIAquick PCR Purification Kit (Qiagen) and sequenced with amplification primers and, for ND2, with primers L5215 (Hackett 1996) and H1064 (Drovetski et al. 2004a) on an ABI 3700 automated sequencer using BigDye kit v. 3.0 according to recommended protocols (Applied Biosystems). Complete ND2 (1041 base pairs (bp)) and MUSK intron (609 bp) sequences were aligned and edited in Sequencher 3.1.1 (Gene Codes, Ann Arbor, MI) and deposited in GenBank (DQ176512–176594).
We used Hirundo aethiopica (GenBank number AY826023) as an outgroup for the ND2 tree of H. rustica, because this species was shown to be one of the closest relatives of H. rustica (Sheldon et al. 2005). For the nuclear MUSK intron, Riparia riparia was used as an outgroup.
We employed Modeltest 3.06 (Posada & Crandall 1998) and the Akaike Information Criterion to find the model of sequence evolution which best fit our data. The TrN+I model (Tamura & Nei 1993) was selected for ND2, with the proportion of invariant sites set to 0.8172. This model was used to perform a maximum-likelihood (ML) tree search with 10 replicates in PAUP* (Swofford 2000). We also performed a maximum-parsimony (MP) search with all characters weighted equally. Support for the branches was assessed by the ML and MP bootstrap analyses with 100 and 1000 replicates, respectively. Finally, Bayesian analysis was performed in MrBayes 3.1 (Ronquist & Huelsenbeck 2003) with general time reversible model and estimated proportion of invariable sites. Two independent runs with four chains each were run for 2 000 000 generations and sampled every 100 generations. The first 5000 trees from each run were discarded as a burn-in and the remaining 30 000 trees were used to construct a majority-rule consensus. Posterior probabilities (proportion of the trees with particular node) were used to assess the reliability of the nodes.
A likelihood ratio (LR) test (Felsenstein 1981) was performed to test clock-like molecular evolution. Likelihood scores of ML trees with and without a molecular clock enforced were compared and the LR was computed as 2 (ln Lclock−ln Lno clock) under the assumption that LR was χ2 distributed with the number of degrees of freedom (d.f.) equal to the number of taxa minus two (Nei & Kumar 2000).
We used DnaSP v. 4.00 (Rosas et al. 2003) to calculate the number of polymorphic sites, number of haplotypes in a population, haplotype diversity (h), nucleotide diversity (π), the R2 test for detecting population growth and McDonald & Kreitman (M–K; 1991) tests for natural selection. We used Arlequin v. 2.000 (Schneider et al. 2000) to perform analysis of molecular variance (AMOVA, Excoffier et al. 1992), calculate pairwise population Φst-values, mismatch distributions, time since population expansion (τ), relative population sizes before (θ0) and after (θ1) expansion, Tajima's D (1996) and Fu's Fs; (1997) test of selective neutrality. To test the empirical mismatch distribution against a model of sudden expansion, we used the generalized nonlinear least-squares approach (Schneider & Excoffier 1999) implemented in Arlequin. Sites with ambiguous characters were omitted.
We used Mdiv (Nielsen & Wakeley 2001) to compute θ and population divergence time (tpop). We then used the formula in Brito (2005) to convert these values to time since divergence, assuming a generation length of 1 year and a mutation rate of 5.4×10−8 (Drovetski et al. 2004b). We used DIVA (Ronquist 1996, 1997) with its default settings to reconstruct biogeographic history.
3. Results
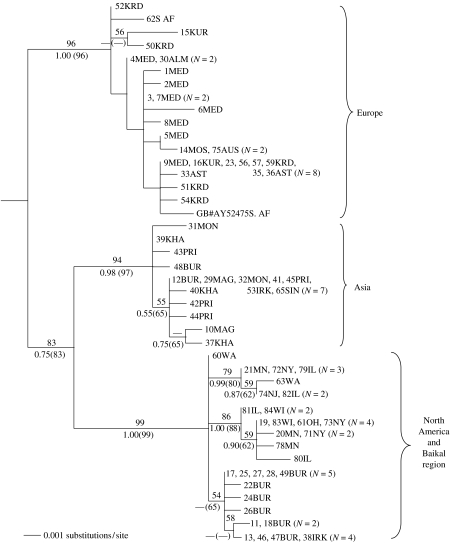
We found 42 mtDNA haplotypes among 74 individuals. Average pairwise uncorrected sequence divergence was 1.14%. Maximum-likelihood phylogenetic analysis yielded three trees, all of which shared three major clades, each with bootstrap support ≥94% (figure 2); the trees differed only in rearrangements of terminal taxa within one of the clades (Europe). Of the three clades, one included primarily European individuals (from Austria, Medvedevo, Moscow, Kursk, Krasnodar, Astrakhan' and Almaty) and two migrants collected in South Africa. The second clade comprised samples from eastern Asia (Mongolia, Khabarovsk, Magadan, Primor'ye, Irkutsk and Buryatiya) and a migrant from Singapore. The third clade, sister to the second, included samples from Baikal region (Buryatiya and Irkutsk) and North America, with samples from Baikal comprising a monophyletic group (figure 2). A close relationship among individuals from the Baikal clade indicates a recent origin of this population.
Figure 2.
One of the three maximum-likelihood trees of Hirundo rustica mtDNA haplotypes, rooted with H. aethiopica (average uncorrected distance between these two species was 3.1%). Numbers above branches are maximum-likelihood bootstrap values (100 replicates); values below branches are Bayesian posterior probabilities, followed by maximum-parsimony bootstrap values (1000 replicates, in parentheses). Lab numbers precede locality abbreviations (see table 1 of electronic supplementary material); total number of individuals sharing the same haplotype is noted in parentheses.
Strict consensus of 2124 equally parsimonious trees (not shown) and majority rule consensus of all Bayesian trees (not shown) identified the same three major clades as did the ML tree (figure 2) and recovered most of the nodes found on consensus ML tree (not shown); however, on the Bayesian consensus tree individuals from Baikal were intermixed with those from North America. A model of clock-like evolution was not rejected for the data set (−ln L without clock enforced=2004.2038, −ln L with clock enforced=2026.6461, d.f.=41, p>0.05). AMOVA indicated that 85.1% of the variation was distributed among regions (p<0.05) and pairwise Φst-values were significant and ranged from 0.48 (Baikal—North America) to 0.93 (Baikal—Asia).
The nine MUSK sequences resolved only three haplotypes (π=0.0011, h=0.417), with seven birds, whose mtDNA belonged to three different clades (18BUR, 60WA, 38IRK, 33AST, 50KRD, 20MN and 32MON), sharing the same sequence. Haplotypes 1MED and 40KHA possessed one and two unique mutations, respectively. There were 20 mutations, a 22 bp deletion and three single base gaps in the outgroup sequence compared to the ingroup. Uncorrected genetic distance between R. riparia and H. rustica was 3.4–3.6%, whereas distances within H. rustica were 0–0.4%. For ND2, uncorrected genetic distance between R. riparia (GenBank sequence AY826015) and H. rustica (our data) was 16.5–18.6%.
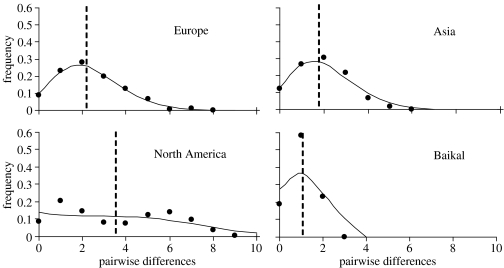
Mismatch distributions (figure 3) did not differ from the model of sudden population expansion for European, Asian and North American samples; however, a significant difference was detected for the Baikal sample (there are too few pairs of individuals with zero base-pair differences and too many with one). Population growth was also indicated for Europe and Asia by Tajima's D, Fu's Fs and R2 tests (table 1). None of these tests detected growth for the North American sample and Tajima's D and the R2 tests suggested that Baikal was from a stable distribution. McDonald & Kreitman (1991) tests were not significant (p>0.05) for any of the pairwise comparisons between Europe, Asia, North America or Baikal (haplotypes from North American clade only), with the exception that Asia versus Baikal was marginally significant (p=0.047).
Figure 3.
Mismatch distributions for four groupings of Hirundo rustica. Dots indicate observed values, solid lines show the distribution according to a model of sudden population expansion, dashed lines indicate the mean number of pairwise differences. The Baikal population is significantly different from the model of sudden population expansion and some analyses (table 1) suggest that the North American sample has also been stable.
Table 1.
Genetic characteristics of Hirundo rustica populations. (Asterisks indicate significance (p<0.05), n/s indicates p>0.05.)
| sample size | no. of haplotypes | no. of polymorphic sites | π | h | θ(S)×102 | no. of synonymous substitutions | no. of non-synonymous substitutions | Tajima's D | Fu's Fs | R2 | mismatch tests | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Europe | 27 | 17 | 19 | 0.0022 | 0.912 | 0.478 | 14 | 5 | −1.89* | −13.6* | 0.055* | n/s |
| Asia | 16 | 11 | 12 | 0.0018 | 0.875 | 0.347 | 8 | 4 | −1.83* | −8.0* | 0.057* | n/s |
| Baikal | 14 | 6 | 4 | 0.0010 | 0.813 | 0.121 | 4 | 1 | n/s | −2.8* | n/s | p=0.01 |
| North Americaa | 17 | 9 | 11 | 0.0034 | 0.912 | 0.317 | 11 | 0 | n/s | n/s | n/s | n/s |
| pooled Baikal and North America | 31 | 15 | 16 | 0.0034 | 0.938 | 0.392 | 16 | 1 | n/s | −5.28* | n/s | n/s |
| all samples pooled | 74 | 42 | 57 | 0.0114 | 0.970 | 1.180 | 50 | 9 | n/s | −13.48* | n/s | n/s |
Nucleotide and haplotype diversity for the small and large clades (figure 2) are 0.0009 and 0.73, and 0.0013 and 0.82, respectively (see text).
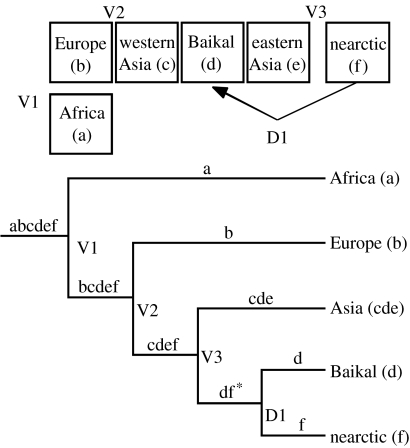
DIVA indicated that vicariance alone cannot explain the current distribution of barn swallow mtDNA lineages, and that a single dispersal event is required. According to the DIVA reconstruction (figure 4), divergence at the first three nodes resulted from vicariance. The first divergence resulted from a separation of African lineage from a widespread holarctic ancestor (figure 3). The second vicariant event separated European and Asian/nearctic clades; Mdiv results placed this split at 106 000 yBP. The third event was the split of Asian and nearctic lineages, and according to Mdiv results this occurred at 102 000 yBP. The most recent divergence resulted from dispersal of nearctic birds to the Baikal region of Central Asia (area d; figure 3). Although DIVA does not state explicitly the direction of the dispersal, dispersal of nearctic birds is supported by several independent lines of evidence. The Baikal area (Buryatiya) clade consists of very recent haplotypes embedded within the nearctic clade (figure 2). Second, birds from the Baikal area are phenotypically much more similar to nearctic birds then they are to other palearctic barn swallows. The Mdiv results suggested this split to be 27 000 yBP.
Figure 4.
DIVA reconstruction of the biogeographic history of the four mtDNA lineages of barn swallow. Small letters identify six holarctic areas. Combination of these areas identifies the geographical range of each mtDNA lineage. ‘*’ identifies a disjunct range that cannot be achieved through a vicariant event and requires a dispersal. ‘V’ with a number identifies a vicariant event, and ‘D’ identifies a dispersal.
4. Discussion
(a) Phylogenetic and biogeographic history
Phylogenetic analysis (figure 2) of mtDNA haplotypes revealed three major clades, European, Asian and North America plus the Baikal region. These clades are approximately equidistant genetically (ca 1.5% uncorrected sequence divergence). Although genetic distance should have no bearing on decisions about species limits, these clades are less differentiated than most avian species (Avise & Walker 1998). Thus, the tree topology indicates that the major clades might correspond to phylogenetic or evolutionary species although denser sampling is needed (Zink et al. 1995). The occurrence of individuals in Buryatiya from two major haplotype clades (figure 2) opens the possibility of interbreeding. Nevertheless, it would be prudent to recognize that significant historical diversity exists in the barn swallow.
Biogeographic analysis (figure 4) suggests that after separation from an African sister-taxon, the barn swallow achieved a holarctic distribution. The next historical event was vicariant separation of the western palearctic (European) clade, which was followed by the vicariant divergence of the Asian and North American clades. These population divergences appeared to be very close in time. Assuming a sequence divergence rate of 5.4% Myr−1, the splits were on the order of 100 000 years; if ND2 evolves more slowly, these estimates could double. The two clades correspond to the western and eastern palearctic regions, which mirrors a relatively common pattern (Pavlova et al. 2003; Drovetski et al. 2004b). This reconstruction is the most parsimonious assuming that vicariance is the preferred explanation. The most unexpected finding was a secondary invasion of the Baikal region from North America. Only a single synonymous substitution separates North American and Baikal groups, suggesting that dispersal and differentiation of the Baikal population was recent (and that dispersal is not ongoing). This direction of colonization runs counter to most nearctic–palearctic taxon exchanges (Cook et al. 2004). Although Smirenskiy & Mishchenko (1981) believed that barn swallows arrived recently in the Baikal region concomitant with the arrival of humans who constructed dwellings suitable for nesting sites, results from MDIV suggested that the Baikal and North American populations diverged ca 27 000 yBP (note that the coalescence point of the haplotype tree would be older). Even considering the confidence intervals around θ and tpop (not shown), molecular data suggest a divergence of the Baikal population well before the time envisioned by these authors. Perhaps, the Baikal clade persisted at low enough density to escape detection by early naturalists.
The biogeographic history of the clades in H. rustica is more complex than typically encountered (Cook et al. 2004). That is, not only was there a reinvasion of the palearctic, but also the new clade is sandwiched between two older clades. How it happened to arrive and persist in this region is unresolved. This odd biogeographic history is consistent with two observations. First, both Baikal and North American birds share a reddish-bellied plumage, whereas birds from other sampled regions have whitish underparts (see figure 1). The sharing of phenotypes by North American and Baikal birds suggests stabilizing selection or an insufficient time for divergence. Second, mtDNA analyses (Zink et al. 2006, unpublished data) of the bank swallow (R. riparia) reveal a unique clade in the Baikal region. Thus, the pattern in the barn swallow is not unique, but nonetheless reveals an intriguing biogeographic history.
(b) Population history
The spread of human settlements has increased the range and population size of barn swallows (Moller 1994; Brown & Brown 1999). In agreement with this observation, both the European and Asian clades have mismatch distributions (figure 3) and associated tests (table 1) consistent with population growth beginning approximately at roughly the same time (mean number of base-pair differences of 2.3 and 1.8, respectively). The history of the nearctic population is less clear. The mismatch distribution is bimodal, with a mean number of differences of 3.5, suggesting an older and more stable history. The bootstrap test of the bimodal mismatch distribution was consistent with population growth, whereas Tajima's D, Fu's Fs and the R2 tests indicated that the North American sample has not undergone major growth (table 1). Ramos-Onsins & Rozas (2002) suggested that the R2 test was a better test of population growth, at least under some circumstances. Thus, it appears that despite the documented history of population growth and expansion (Brown & Brown 1999), the nearctic population was historically larger and more stable than those in the palearctic, or that population expansion was not as great. However, the two clades of North American haplotypes (individuals 21, 63, 72, 74, 79, 82 and the rest excluding individual 60; see figure 2), which are not geographically structured (figure 2), are consistent with two histories: a single population that shows the expected deep coalescence structure (Hudson 1990), or prior geographical isolation followed by subsequent intermixing (see also Zink et al. 2005). If the two clades were part of the same large population, each should have the same genetic signature. We found that each clade has similar nucleotide and haplotype diversity (table 1). Although our samples of each clade are small, R2 values for each clade are not significant (p≥0.10), suggesting population stability, and both mismatch distributions (not shown) exhibit too few zero and too many one base-pair values. Combination of the two clades creates an overall bimodal mismatch (figure 3). Thus, the data are consistent with a single large population of barn swallows in the nearctic, which has not undergone dramatic growth. Potentially, the growth of barn swallow populations has been too recent to be detectable in our data. Alternatively, perhaps range expansion has not been accompanied by dramatic increases in population size. Additional study of nearctic populations is warranted.
The history of the recently established Baikal population is more clear. Like the two clades of North American haplotypes, the mismatch distribution has a dearth of zero values and an excess of one base-pair differences (figure 3), which differs from the sudden expansion model according to the bootstrap and R2 tests. The striking similarity of mismatch distributions in the two North American and Baikal clades is suggestive of common population histories and ancestry. Although populations with unimodal mismatch distributions are typically considered to be expanding, our analyses suggest otherwise. Thus, it appears that the Baikal population has remained relatively small post-colonization, as suggested by the lack of historical sightings. Relatively low levels of haplotype and nucleotide diversity, and theta in this population (table 1) are consistent with this view. Lastly, M–K tests do not implicate natural selection because the only fixed difference between North American and Baikal is a synonymous substitution. Therefore, the incipient divergence of the Baikal haplotypes from the North American clade appears to have been driven by genetic drift and not natural selection.
(c) Mitochondrial and nuclear gene trees
MtDNA gene trees represent one of many possible gene trees. Unfortunately with small mtDNA distances among clades (especially relative to differences within clades), the likelihood of differences at nuclear genes is remote, given the larger effective population size and concomitantly slower coalescence time for nuclear loci (Palumbi et al. 2001). That is, the lack of nuclear gene differences is expected given the shallow divisions in the mtDNA tree. Nonetheless, it is valuable to note that our MUSK sequences did not reveal a pattern that differed significantly from the mtDNA gene tree.
Acknowledgements
We thank the American Museum of Natural History, and the Field Museum of Natural History for tissue samples. Financial support was received from the late G. Eddy and the National Science Foundation (DEB 9707496 and DEB 0212832). A. Jones, B. Barber, Yu. Lohman, A. Tsvetkov, D. Banin, A. Andreev, V. Masterov, R. Faucett, D. Froilich, V. Rohwer, I. Fadeev, E. Nesterov, I. Karagodin, E. Koblik and Ya. Red'kin assisted with field work. R. Blackwell-Rago, S. Farrell and M. Westberg performed laboratory work. F. K. Barker assisted with the nuclear gene analyses. P. Brito, G. Barrowclough and A. Jones provided useful advice.
Footnotes
Present address: School of Biological Sciences, Monash University, Clayton, Victoria 3800, Australia.
Supplementary Material
Sampling localities and voucher information for mitochondrial groupings of Hirundo rustica
References
- Avise J.C, Walker D. Pleistocene phylogeographic effects on avian populations and the speciation process. Proc. R. Soc. B. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. 10.1098/rspb.1998.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito P.H. The influence of Pleistocene glacial refugia on tawny owl genetic diversity and phylogeography in western Europe. Mol. Ecol. 2005;14:3077–3094. doi: 10.1111/j.1365-294X.2005.02663.x. 10.1111/j.1365-294X.2005.02663.x [DOI] [PubMed] [Google Scholar]
- Brown C.R, Brown M.B. Barn swallow (Hirundo rustica) In: Poole A, Gill F, editors. The birds of North America, no. 452. The Birds of North America; Philadelphia, PA: 1999. pp. 1–32. [Google Scholar]
- Cook J.A, Runk A.M, Conroy C.J. Historical biogeography at the crossroads of the northern continents: molecular phylogenetics of red-backed voles (Rodentia: Arvicolinae) Mol. Phylogenet. Evol. 2004;30:767–777. doi: 10.1016/S1055-7903(03)00248-3. 10.1016/S1055-7903(03)00248-3 [DOI] [PubMed] [Google Scholar]
- Cramp S. Handbook of the birds of Europe, the Middle East and North Africa. vol. 5. Oxford University Press; Oxford, UK: 1988. [Google Scholar]
- Drovetski S.V, Zink R.M, Fadeev I.V, Nesterov Y.V, Koblik Y.A, Red'kin Ya.A, Rohwer S. Mitochondrial phylogeny of Locustella and related genera. J. Avian Biol. 2004a;35:105–110. 10.1111/j.0908-8857.2004.03217.x [Google Scholar]
- Drovetski S.V, Zink R.M, Rohwer S, Fadeev I.V, Nesterov Y.V, Karagodin I, Red'kin Y.A. Complex biogeographic history of a holarctic passerine. Proc. R. Soc. B. 2004b;271:545–551. doi: 10.1098/rspb.2003.2638. 10.1098/rspb.2003.2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse P.E, Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Fu Y.-X. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett S.J. Molecular phylogenetics and biogeography of tanagers in the genus Ramphocelus (Aves) Mol. Phylogenet. Evol. 1996;5:368–382. doi: 10.1006/mpev.1996.0032. 10.1006/mpev.1996.0032 [DOI] [PubMed] [Google Scholar]
- Hudson R.R. Gene genealogies and the coalescent process. In: Futuyma D.J, Antonovics J.D, editors. Oxford surveys in evolutionary biology. Oxford University Press; New York, NY: 1990. pp. 1–44. [Google Scholar]
- McDonald J.H, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Møller A.P. Oxford University Press; Oxford, UK: 1994. Sexual selection and the barn swallow. [Google Scholar]
- Nei M, Kumar S. Oxford University Press; Oxford, UK: 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Nielsen R, Wakeley J.W. Distinguishing migration from isolation: an MCMC approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi S.R, Cipriano F, Hare M.P. Predicting nuclear gene coalescence from mitochondrial data: the three-times rule. Evolution. 2001;55:859–868. doi: 10.1554/0014-3820(2001)055[0859:pngcfm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Pavlova A, Zink R.M, Drovetski S.V, Red'kin Ya, Rohwer S. Phylogeographic patterns in Motacilla flava and Motacilla citreola: species limits and population history. Auk. 2003;120:744–758. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S.E, Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Ronquist F. Uppsala University; Uppsala, Sweden: 1996. DIVA, ver. 1.1. Computer program available by anonymous ftp. [Google Scholar]
- Ronquist F. Dispersal–vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 1997;46:193–201. [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rosas J, Sanchez-DelBarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1070–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Dueffer J.-M, Roessli D, Excoffier L. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 2000. Arlequin ver. 2.0: a software for population genetic data analysis. URL: anthropologie.unige.ch/arlequin/ [Google Scholar]
- Sheldon F.H, Whittingham L.A, Moyle R.G, Slikas B, Winkler D.W. Phylogeny of swallows (Aves: Hirundinidae) estimated from nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005;35:254–270. doi: 10.1016/j.ympev.2004.11.008. 10.1016/j.ympev.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Smirenskiy S.M, Mishchenko A.L. Taxonomical status and history of formation of the range of Hirundo rustica in the Amur territory. Zool. Zh. 1981;60:1533–1541. [in Russian.] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b2. [Google Scholar]
- Tajima F. The amount of DNA polymorphism maintained in finite population when the neutral mutation rate varies among sites. Genetics. 1996;143:1457–1465. doi: 10.1093/genetics/143.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Turner A. Family Hirundinidae (swallows and martins) In: Del Hoyo J, Elliott A, Christie D, editors. The birds of the World. vol. 9. Lynx Edicions; Barcelona, Spain: 2004. pp. 602–685. [Google Scholar]
- Turner A, Rose C. Swallows and martins. An identification guide and handbook. Houghton Mifflin; Boston, MA: 1989. [Google Scholar]
- Zink R.M, Rohwer S, Andreev A.V, Dittmann D.L. Trans-Beringia comparisons of mitochondrial DNA differentiation in birds. Condor. 1995;97:639–649. [Google Scholar]
- Zink R.M, Rising J.D, Mockford S, Horn A.G, Wright J.M, Leonard M, Westberg M.C. Mitochondrial DNA variation, species limits and rapid evolution of plumage coloration and size in the Savannah Sparrow. Condor. 2005;107:21–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling localities and voucher information for mitochondrial groupings of Hirundo rustica