Abstract
The elemental composition of proteins influences the quantities of different elements required by organisms. Here, we considered variation in the sulphur content of whole proteomes among 19 Archaea, 122 Eubacteria and 10 eukaryotes whose genomes have been fully sequenced. We found that different species vary greatly in the sulphur content of their proteins, and that average sulphur content of proteomes and genome base composition are related. Forces contributing to variation in proteomic sulphur content appear to operate quite uniformly across the proteins of different species. In particular, the sulphur content of orthologous proteins was frequently correlated with mean proteomic sulphur contents. Among prokaryotes, proteomic sulphur content tended to be greater in anaerobes, relative to non-anaerobes. Thermophiles tended to have lower proteomic sulphur content than non-thermophiles, consistent with the thermolability of cysteine and methionine residues. This work suggests that persistent environmental growth conditions can influence the evolution of elemental composition of whole proteomes in a manner that may have important implications for the amount of sulphur used by living organisms to build proteins. It extends previous studies that demonstrated links between transient changes in environmental conditions and the elemental composition of subsets of proteins expressed under these conditions.
Keywords: cysteine, methionine, sulphur, proteome, atomic composition, thermophile
1. Introduction
The elemental composition of biopolymers influences the quantities of different elements required for their synthesis, and has an important role in determining nutrient relations between organisms and their environment (Sterner & Elser 2002). Increasingly, there is recognition that systematic biases in the elemental composition of biopolymers can be related to environmental and resource constraints. This was first demonstrated by Mazel & Marlière (1989), who found that sulphur-starved cyanobacteria express sulphur-depleted versions of light-harvesting proteins, thus reducing the quantity of sulphur required to build proteins. Later, we demonstrated that in Escherichia coli and Saccharomyces cerevisiae, proteins used for assimilating sulphur and carbon are significantly depleted of sulphur and carbon atoms, respectively. We proposed that these specific impoverishments in elemental protein components help maintain the integrity of metabolic networks during transitory shortages of these nutrients (Baudouin-Cornu et al. 2001). Similarly, yeast cells exposed to cadmium were demonstrated to reduce their use of sulphur in proteins by up to 30% through a large reprogramming of protein synthesis, thus releasing more sulphur atoms for the synthesis of glutathione, a major cadmium detoxifying agent (Fauchon et al. 2002). These studies each reveal adaptive responses by organisms to transient shortages in nutrients, through the synthesis of a subset of proteins with biases in their elemental composition. They demonstrate the importance of protein elemental composition in influencing the quantities of nutrients required by organisms for protein synthesis.
Recently, full genome sequences have become available for many different organisms, providing the opportunity to study the elemental composition of biopolymers among large numbers of organisms at the levels of whole genomes and proteomes. Such studies have revealed striking variation among species in the carbon content of whole proteomes, and strong associations between the carbon and nitrogen content of whole genomes and proteomes (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004). However, several key questions remain unanswered. For example, we do not yet know whether organisms adapted to specific environmental conditions have consistent differences in the elemental composition of their biopolymers, and hence in the quantities of different nutrients they require for growth. Biases in amino acid frequencies of proteomes have been observed for organisms living under specific environmental conditions (e.g. high temperature; see Hickey & Singer 2004), and potentially have consequences for elemental composition. Recognition of systematic variation in whole proteome elemental composition in organisms adapted to specific environmental conditions would represent a significant advance, complementing previous studies that have focused on subsets of proteins expressed during transient changes in environmental conditions.
An analysis of proteomic sulphur composition among organisms may be particularly interesting in this regard. Sulphur is contained in only two coded amino acids, cysteine and methionine, which share a number of unique properties (e.g. high susceptibility to oxidation; Berlett & Stadtman 1997). Many organisms obtain inorganic sulphur in the form of sulphate, and consume NADPH when performing assimilatory reduction of sulphate into sulphide, which is the only one-sulphur form that can be incorporated into a carbon chain for the synthesis of sulphur-containing amino acids. Moreover, among prokaryotes, diverse sulphur metabolic pathways are employed in the production of energy. For example, dissimilatory sulphate-reducing species use sulphate as a terminal respiratory electron acceptor (Barton & Tomei 1995). Full genome sequences are now available for three dissimilatory sulphate-reducing species: the eubacteria Desulfovibrio vulgaris and Desulfotalea psychrophila, and the archaeon Archaeoglobus fulgidus (Klenk et al. 1997; Heidelberg et al. 2004; Rabus et al. 2004).
In this study, we examine variation in protein sulphur content within and among 151 species. Our aims were to (i) determine whether proteomic sulphur content varies substantially among species; (ii) investigate patterns in protein sulphur content within proteomes, by considering cumulative frequency (quantile) distributions of proteomic sulphur content and the sulphur content of orthologous proteins; and (iii) investigate factors that may influence proteomic sulphur content in different organisms, including genomic base composition, and traits that reflect adaptations to persistent environmental features, anaerobiosis and growth temperature.
2. Material and methods
(a) Proteome sequences and calculations of protein sulphur content
Protein and nucleic acid sequence data for 151 species (10 eukaryotes, 19 Archaea and 122 Eubacteria) were collected from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes). Where data were available for multiple strains of a single species, strains were excluded at random, so that each species is represented only once in analyses. Data were pooled for multiple chromosomes of a species, and plasmids were excluded. Non-standard amino acids and bases (e.g. ‘X’) were ignored in our calculations. See the electronic supplementary material for a list of the species and strains that were used, and their accession numbers.
For each protein sequence of each species, we calculated protein length, and the frequency of use of each amino acid. We also counted the number of sulphur atoms per protein, and sulphur content per amino acid of each protein. For these analyses, starting methionine residues were excluded, since they are frequently removed from mature proteins (e.g. see Meinnel et al. 1993). We then calculated the mean proteomic sulphur content (per amino acid) for each species, as the average value across all the proteins.
For each species, GC content (the proportion of guanine plus cytosine bases) and AG content (the proportion of adenine plus guanine bases) were calculated for each coding sequence, and averaged.
(b) Sulphur content of orthologous proteins
Sequences for proteins in two sets of clusters of orthologous groups (hereafter COGs; Tatusov et al. 1997) were downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/pub/COG/COG). The first set consists of 63 COGs, whose proteins are represented in all species in the unicellular COGs database (widely distributed COGs; see electronic supplementary material for a list). A large proportion of these COGs (52 out of 63) are involved in translation. The second set of COGs contains proteins that were determined to be abundant in S. cerevisiae (Ghaemmaghami et al. 2003) and which we used to investigate the influence of expression on carbon use in whole proteomes (high expression COGs; Baudouin-Cornu et al. 2004). This set contains 25 COGs. We calculated the sulphur content and length of proteins in these COGs as described above. However, we did not exclude starting amino acids, as COGs do not always include a starting methionine. When a species had more than one protein in a COG, we calculated an average value of sulphur content and length, and these averages were used in subsequent analyses.
(c) Species trait information
Prokaryotic species were classified as being either obligately anaerobic, or non-anaerobes, based on sources including The Institute of Genomic Research website (http://www.tigr.org/tigr-scripts/CMR2/genome_properties), and literature sources (e.g. Holt 1984, 1986, 1989a,b and Dworkin 2004; see electronic supplementary material for more information). Non-anaerobic species include those considered obligately aerobic, facultatively anaerobic or microaerophilic. Among the 141 prokaryotes in our dataset, we identified 25 anaerobes and 106 non-anaerobes. The other species were undetermined.
Prokaryotic species were classified as thermophiles or non-thermophiles, mainly using Dworkin (2004). Among the 141 prokaryotes, we identified 21 thermophiles in our dataset, and 119 non-thermophiles (including 118 mesophiles, and D. psychrophila, which is usually considered a psychrophile). We obtained optimal growth temperature data for 92 species in our dataset, mainly from the German National Resource Centre for Biological Material website (www.dsmz.de; see the electronic supplementary material for other data sources). Where a range of optimal temperatures was given, we calculated an average.
(d) Analyses of protein sulphur content within proteomes
For each species in our dataset, we calculated quantile distributions of protein sulphur content within proteomes (as in Baudouin-Cornu et al. 2004) following the method suggested by Karlin & Brendel (1992) for representing the amino acid composition of proteins.
To test whether variation among species in mean proteomic sulphur content was reflected in the sulphur content of orthologous proteins, we considered sulphur content quantile distributions for different species based only on the widely distributed COGs, and considered the variation in protein sulphur content of orthologous proteins by drawing sulphur content quantile plots for each individual COG. We tested whether mean protein sulphur content of these COGs (n=63) was correlated with mean proteomic sulphur content, among 29 species (3 eukaryotes, 13 Archaea and 13 Eubacteria, see electronic supplementary material) that were common to our whole proteome dataset and the COGs database. Finally, we tested the correlation between the sulphur content of each of these 63 COGs with mean proteomic sulphur content, among the same species (see electronic supplementary material).
Since the number of sulphur atoms immobilized in a proteome is influenced not only by the sulphur content of each protein, but also by the relative abundance of proteins, we considered sulphur content in proteins that are likely to be highly expressed (‘high expression COGs’, see §2b) and tested whether the mean sulphur content in these COGs was correlated with mean proteomic sulphur content. This analysis was done for the same 29 species used in the analysis of widely distributed COGs.
(e) Relationships between proteomic sulphur content and other traits, among organisms
We tested the relationship among prokaryotes between the average GC content of coding sequences (hereafter genomic GC content) and proteomic sulphur content by fitting a polynomial regression, and calculated residual deviations from the resulting curve (SPSS v. 12.0). We performed simulations of this relationship, based on the genetic code. We generated three randomized proteomes corresponding to each of 11 values of genomic GC content: 20.8%, 26.0%, 31.2%, 36.2%, 41.2%, 46.2%, 51.2%, 56.1%, 60.9%, 65.8% and 70.6%. Each of these 33 proteomes consists of 2000 proteins of length 280, with each codon chosen randomly with a probability determined by the required GC content. When stop codons were encountered in sequences they were discarded, and final GC content was corrected to take into account the 2000 STOP codons. We did not enforce the use of starting methionines in these simulations, since we excluded starting methionines from our proteomic sulphur content calculations.
We compared proteomic sulphur content between anaerobes and non-anaerobes, and between thermophiles and non-thermophiles, using Mann–Whitney U-tests (hereafter, Mann–Whitney U abbreviated by U) (SPSS v. 12.0). These analyses were repeated using residuals from the relationship between GC content and proteomic sulphur content, to consider the influence of these traits on proteomic sulphur content independently of GC content. Additionally, thermophiles tend to have short proteins (Tekaia et al. 2002) and elevated transcript AG content (Schultes et al. 1997; Lambros et al. 2003) relative to mesophiles. To ensure that these features of thermophiles did not influence our analyses, we compared total mean protein sulphur content between thermophiles and non-thermophiles, and tested whether there was an association between proteomic sulphur content and the mean AG content of coding sequences.
Individual species may not represent independent data points if closely related species have similar trait values due to shared ancestry (see Harvey & Pagel 1991). Statistical methods have been developed to analyse relationships among species while accounting for phylogeny (e.g. Felsenstein 1985; Harvey & Pagel 1991). However, the application of these methods to prokaryotes may be complicated by the difficulty of resolving phylogenetic relationships among deeply diverging groups (e.g. see Creevey et al. 2004). Therefore, we have addressed this issue by repeating our analyses of proteomic sulphur content in relation to genomic base composition and environmental factors among prokaryotic orders. We obtained taxonomic information from the NCBI Taxonomy website (see http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi), and calculated average values (across species) of genomic base composition and proteomic elemental composition for each order. To test the influence of anaerobiosis and thermophily at the level of order, we classified prokaryotic orders as anaerobic (n=16) or non-anaerobic (n=33), and as thermophilic (n=14) or non-thermophilic (n=36), based on the representatives of orders in our species-level dataset. For each comparison, we excluded species that could not be classified according to the corresponding environmental factor (anaerobiosis or thermophily), as well as orders that contained representatives of both groups.
3. Results
(a) Protein sulphur content: proteome quantile distributions and orthologous proteins
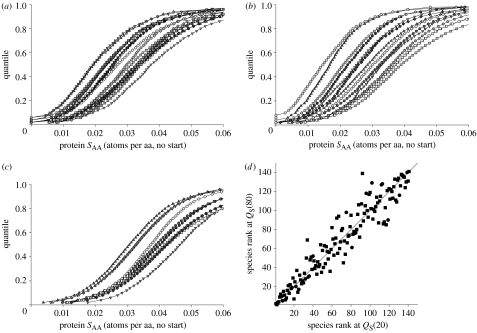
Among species, the quantile distributions of protein sulphur content (SAA) exhibited stochastic ordering (Karlin & Brendel 1992; Baudouin-Cornu et al. 2004; see figure 1a–c). That is, the quantile curves of different species intersect relatively infrequently, such that the ranks of species were quite consistent at different quantile values. To illustrate this further, the protein sulphur content of all 141 prokaryote species were ranked at quantile values of QS(20), QS(50) and QS(80). Most species had a similar rank at QS(20), QS(50) and QS(80), such that ranks at these quantiles were strongly and positively correlated among species (for each pair-wise correlation rS>0.95, one-tailed p<0.001; figure 1d). Further, median values of proteomic sulphur content were correlated very strongly with mean values of proteomic sulphur content (n=141, rS=0.992, one-tailed p<0.001). These results suggest that differences among species in mean proteomic sulphur content are typically indicative of differences across the quantile distribution of their protein sulphur content values.
Figure 1.
Quantile (cumulative frequency) distributions of protein sulphur content (atoms per amino acid, excluding start methionines; SAA) for (a) 19 Archaea, (b) 14 Eubacteria and (c) 10 eukaryotes. See electronic supplementary material for larger versions of these graphs, with legends. (d) Plot of species ranks for protein sulphur content at quantile value QS(20), versus ranks at quantile value QS(80), for 141 prokaryotes. The squares are Eubacteria, circles are Archaea. The line is 1 : 1.
Sulphur quantile distributions using only widely distributed COGs had shapes broadly similar to those of whole proteomes (figure 2a), and exhibit considerable variation among species. Further, quantile distributions of sulphur content of individual COGs showed considerable variation (figure 2b), demonstrating that proteins thought to perform the same function can vary substantially in their sulphur content. Among species, the mean sulphur content in the widely distributed COG proteins (n=63 for each species) was strongly and positively correlated with the mean sulphur content of whole proteomes (n=29, rS=0.876, one-tailed p<0.001; figure 2c). Further, when we considered the relationship between mean proteomic sulphur content and the sulphur content of the proteins of each of these 63 COGs individually, there was a positive and significant correlation for 43 out of 63 COGs (68% had n=29, rS>0.312, one-tailed p<0.05 (Zar 1999); see electronic supplementary material for more details).
Figure 2.
COG sulphur content (SAA denotes sulphur atoms per amino acid). (a) Quantile distributions for the protein sulphur content of 63 COGs, plotted for 13 Archaea. (b) Quantile distributions of sulphur content in 20 individual COGs (randomly selected from the 63 widely distributed COGs). See electronic supplementary material for larger versions of these graphs, with legends. (c) Plot of mean sulphur content of the proteins of 63 COGs as a function of mean proteomic sulphur content (including starting methionines), among 29 species. The line is 1 : 1.
Similarly, mean values of sulphur content per amino acid in high expression COGs were correlated positively with mean proteomic sulphur content (n=29, rS=0.774, one-tailed p<0.001). Taken together, our analyses of sulphur content in orthologous proteins suggest that differences in mean values of proteomic sulphur content are indicative of real differences among species in the quantities of sulphur contained in proteins.
(b) Variation among species in proteomic sulphur content
Among species, the maximum and minimum values of mean proteomic sulphur content varied by 99.5% relative to the average value. Eukaryotes tended to have higher mean proteomic sulphur content than prokaryotes (U=181, p<0.001). This was likely attributable to higher use of cysteine in eukaryotes (as observed by Karlin & Brendel 1992; U=64, p<0.001), since eukaryotes did not use significantly more methionine than prokaryotes (U=698, p=0.96). Among prokaryotes, there was no significant difference between Eubacteria and Archaea in proteomic sulphur content (U=1090, p=0.68). Across prokaryotic species, maximum and minimum species values of mean proteomic sulphur content varied by 85.6% relative to the average value.
(c) Genomic GC content and proteomic sulphur content
The three codons for cysteine and methionine (UGU, UGC, AUG) have a mean GC content of 0.44, suggesting that sulphur content might be greatest at intermediate GC content, and lower at high and low values of GC content. Among both randomized proteomes and real proteomes, mean sulphur content was related to GC content in a manner well approximated by a convex parabola (figure 3; among random proteomes, n=33, SAA=−0.215 GC2+0.184 GC+0.012, r2=0.99, F=2297.34, p<0.001; among real species, n=141, SAA=−0.171 GC2+0.164 GC−0.005, r2=0.291, F=28.36, p<0.001; among orders, n=53, SAA=−0.196 GC2+0.188 GC−0.011, r2=0.278, F=9.61, p<0.001). That is, mean proteomic sulphur content was highest at intermediate values of GC content, and lower at high and low GC content. Also, at a given GC content, real proteomes tended to contain less sulphur than expected according to the simulations, with the exception of the sulphate-reducing bacterium D. vulgaris, whose proteomic sulphur content was comparable to the simulated value, and which had the greatest value of proteomic sulphur content among prokaryotes (figure 3).
Figure 3.
Mean proteomic sulphur content (atoms per amino acid, excluding start methionines; SAA) as a function of mean GC content for randomly generated genomes (small circles), unicellular eukaryotes (filled triangles), multicellular eukaryotes (open triangles), anaerobic prokaryotes (filled diamonds), non-anaerobic prokaryotes (open diamonds) and unclassified prokaryotes (diamonds with dot). Fitted lines indicate relationships between GC content and mean proteomic sulphur content for randomly generated genomes (dashed) and real prokaryotes (unbroken) (statistics provided in the text). Arrows indicate points corresponding to the species Desulfovibrio vulgaris (Dvu), Mycobacterium avium (Mav), Rickettsia conorii (Rco) and Vibrio vulnificus (Vvu).
(d) Anaerobiosis
Obligately anaerobic prokaryotes had higher values of mean proteomic sulphur content than non-anaerobes based on raw values of proteomic sulphur content (among species, U=803.5, p=0.002, figure 3; among orders, U=150, p=0.015), and residuals from the relationship between GC content and proteomic sulphur content (among species, U=855, p=0.006; among orders, U=149, p=0.014). Both methionine and cysteine appeared to contribute to elevated proteomic sulphur content in anaerobes. Methionine was used more frequently in anaerobes than non-anaerobes (among species, U=954, p=0.030; among orders, U=146, p=0.012). Cysteine was used more frequently by anaerobic species than non-anaerobic species (U=842, p=0.005), and anaerobic orders had a marginally significant tendency to use more cysteine than non-anaerobic orders (U=183, p=0.084). We tested whether other easily oxidized residues: tyrosine, phenylalanine, tryptophan and histidine (Y+F+W+H, hereafter FYWH; Berlett & Stadtman 1997; Naya et al. 2002) were also used more frequently by anaerobic species than non-anaerobes. We found no difference in the use of FYWH between anaerobes and non-anaerobes before (among species, U=1316, p=0.958; among orders, U=236, p=0.551) or after (among species, U=1048, p=0.105; among orders, U=207, p=0.224) accounting for the influence of GC content on FYWH.
(e) Growth temperature and proteomic sulphur content
The optimal growth temperatures of the species in our dataset varied widely (from 7 to 103 °C, figure 4). There was a marginally significant tendency for sulphur content to be lower in thermophiles than non-thermophiles (among species, U=953, p=0.084; among orders, U=174, p=0.092). This was probably attributable to use of cysteine being lower in thermophiles (among species, U=870, p=0.027; among orders, U=160, p=0.047), as methionine was not significantly lower in thermophiles (among species U=1155, p=0.581; among orders, U=223, p=0.531). The tendency of thermophiles to use less sulphur than non-thermophiles was stronger when considered as residuals calculated from the relationship between GC content and mean proteomic sulphur content (among species, U=753, p=0.004; among orders, U=126, p=0.006). Moreover, among thermophiles, there was a significant negative correlation between proteomic sulphur content and optimal growth temperature among species (n=21, rS=−0.536, two-tailed p=0.012), and a marginally significant negative correlation among orders (n=14, rS=−0.494, p=0.073). This appears to be primarily due to decreasing use of methionine with increasing optimal growth temperature (among species, n=21, rS=−0.481, two-tailed p=0.027; among orders, n=14, rS=−0.627, two-tailed p=0.016), as cysteine use was not related significantly to optimal growth temperature among thermophiles (among species, n=21, rS=−0.126, two-tailed p=0.587; among orders, n=14, rS=0.011, two-tailed p=0.970).
Figure 4.
Mean proteomic sulphur content (atoms per amino acid, excluding start methionines; SAA) as a function of optimal growth temperature, among 92 prokaryotes. Plot of mean proteomic sulphur content (atoms per amino acid, excluding start methionines) as a function of optimal growth temperature. Circles are thermophiles (optimal growth temperature greater than or equal to 47 °C), non-thermophiles are squares, filled symbols are anaerobes, open symbols are non-anaerobes.
Two additional analyses suggested that previously observed traits of thermophiles, elevated AG content (Schultes et al. 1997; Lambros et al. 2003) and reduced protein length (Tekaia et al. 2002) did not account for our observations of reduced proteomic sulphur content in thermophiles. First, when we calculated mean proteomic sulphur content per protein (without dividing by length), we still found that thermophiles had lower mean proteomic sulphur content per protein than non-thermophiles (among species, U=720, p=0.002; among orders, U=133, p=0.010). Second, there was no significant relationship between proteomic AG content and proteomic sulphur content (among species, n=141, rS=−0.031, two-tailed p=0.714; among orders, n=53, rS=−0.091, two-tailed p=0.518).
4. Discussion
This study of proteomic sulphur content in 151 species extends our independent previous studies on proteomic nitrogen and carbon content (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004), and has yielded several important findings.
First, variation among species in proteomic sulphur content is reflected quite uniformly across frequency distributions of proteins in proteomes, and in proteins thought to perform common functions in different species, similar to previous observations for proteomic carbon content (Baudouin-Cornu et al. 2004). In particular, proteins that are orthologous in different organisms can vary considerably in their sulphur content, and across species, the sulphur content of these proteins tends often to be correlated with mean proteomic sulphur content. This is important in that it refutes the possibility that variation among species in proteomic sulphur content is due solely to the presence of unique proteins in different species. It also suggests a way to estimate mean proteomic sulphur content rapidly for different species, by sequencing a modest number of orthologous proteins (see electronic supplementary material for a list of COGs and their correlations with mean proteomic sulphur content). Taken together, these observations suggest that mean values of proteomic sulphur content among species ought to provide a good indication of the relative sulphur content of proteins of different organisms. Our observation that protein sulphur content in highly expressed COGs is correlated with mean proteomic sulphur content among species is particularly important in this regard, since these proteins may have an inordinate influence on the total sulphur content of proteins in organisms. Therefore these observations lend considerable weight to our analyses of mean proteomic sulphur content among prokaryotes.
Our observations that mean values of proteomic sulphur content vary widely among species (among the prokaryotes, the maximum and the minimum values vary by 85.6% relative to the mean value) imply that different species vary greatly in the quantities of sulphur required to build their proteins. This suggests interesting possibilities, such as testing whether prokaryotes with low proteomic sulphur content are competitively or adaptively favoured under conditions of sulphur scarcity. Variation among species in proteomic carbon content (12.5%, for the same 141 prokaryotes; J. G. Bragg 2005, unpublished results) and nitrogen content (8.8%, for the same 141 prokaryotes; J. G. Bragg 2005, unpublished results) is more modest. However, an important feature of variation among organisms in proteomic carbon and nitrogen content is that these properties are related negatively to one another, probably because they are both related linearly to genomic GC content, but negatively in the case of carbon and positively in the case of nitrogen (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004). This implies tradeoffs among organisms in the quantities of carbon and nitrogen atoms that are used in proteins (Bragg & Hyder 2004). Here we show that proteomic sulphur content is also related to genomic GC content, and may therefore be subject to similar tradeoffs. That is, forces influencing the frequency of use of any one of these three elements in proteomes might additionally influence the frequency with which the other two are used.
Similar to relationships among GC content and proteomic nitrogen and carbon content (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004), the relationship between GC content and proteomic sulphur content appears to be specified in the genetic code. This is demonstrated by the observation that simulations based on the genetic code suggest a convex dependence of proteomic sulphur content on GC content, as we observed across real organisms. However, the relationship between proteomic sulphur content and GC content contrasts to those of carbon and nitrogen in three important ways. First, sulphur is not a component of DNA. Genomes contain carbon and nitrogen atoms in quantities that vary deterministically with GC content (McEwan et al. 1998), such that for both carbon and nitrogen, atomic counts are positively correlated in whole genomes and proteomes (Bragg & Hyder 2004). Such a relationship does not exist for sulphur. Second, the dependence of proteomic sulphur content on GC content among real organisms is convex, rather than linear. That is, organisms with intermediate GC content tend to have the highest proteomic sulphur content. This is exemplified by the observation that Vibrio vulnificus, which has a moderate GC content (47%), has a proteomic sulphur content that is 15.3% greater than that of low GC content (32%) species Rickettsia conorii, and 34.5% greater than that of high GC content (69%) species Mycobacterium avium (figure 3). Similarly, the proteomic carbon and nitrogen contents of these species reflect the influence of GC content: R. conorii has greater proteomic carbon than V. vulnificus, which in turn has greater proteomic carbon than M. avium, while for proteomic nitrogen content, this order is reversed (data not shown). Third, the influence of GC content on proteomic sulphur content is relatively weak, particularly relative to carbon, and the variation in proteomic sulphur content at a given value of GC content is quite substantial. Here, we report a correlation coefficient (r2) of 0.29 for the polynomial relationship between GC content and proteomic sulphur content, among species. Previously it was shown that GC content is very strongly related to proteomic carbon use (r2=0.81, Baudouin-Cornu et al. 2004; rS=0.88, Bragg & Hyder 2004), while the relationship between GC content and proteomic nitrogen use is weaker (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004).
We also observed that all species, with the exception of D. vulgaris, have proteomic sulphur values lower than predicted on the basis of the simulations. Two other species of dissimilatory sulphate reducers, D. psychrophila and A. fulgidus, also have relatively high values of proteomic sulphur, ranking fifth and thirtieth, respectively, among 141 prokaryotes. Possibly this reflects the cost of biosynthesis of cysteine and methionine, especially given the energetic cost of assimilatory sulphate reduction (Akashi & Gojobori 2002; Fauchon et al. 2002). That is, these three dissimilatory sulphate-reducing species may have relatively high values of proteomic sulphur content because they produce reduced sulphur during their respiration.
Our observation that anaerobic prokaryotes tend to have higher proteomic sulphur content than non-anaerobes is consistent with the tendency of anaerobes to have elevated use of cysteine-containing motifs (Major et al. 2004). However, the sulphur in cysteine-containing motifs cannot explain elevated sulphur content in anaerobes entirely, since we find that anaerobes also have higher use of methionine than non-anaerobes. It has been reported previously that a group of six easily oxidized amino acids, including cysteine and methionine, were used less frequently by obligate anaerobic species than by anaerobes (Naya et al. 2002). It was suggested that these residues are disfavoured in aerobes due to greater exposure to reactive oxygen species. Here we did not find a significant tendency for the other four easily oxidized amino acids (tyrosine, phenylalanine, tryptophan and histidine) to be used with reduced frequencies in non-anaerobes. These results could imply that something other than susceptibility to oxidation accounts for reduced use of sulphur-containing amino acids in non-anaerobes, or it could reflect the greater susceptibility to oxidation of cysteine and methionine (Berlett & Stadtman 1997). Elucidating the biochemical basis for the higher use of sulphur-containing amino acids in anaerobes may be a profitable avenue for future investigation.
We observe a strong tendency for reduced use of sulphur-containing amino acids by prokaryotes adapted to high growth temperatures. Both cysteine and methionine contribute to this pattern, in slightly different ways. Cysteine content was lower in proteomes of thermophiles than non-thermophiles, consistent with previous observations (Tekaia et al. 2002; Singer & Hickey 2003; Beeby et al. 2005). However, we also found that methionine content decreases with optimal growth temperature among thermophiles, and that this probably causes a negative relationship between optimal growth temperature and proteomic sulphur content among thermophiles. Previously, both cysteine and methionine have been classified as thermolabile residues, due to their tendency to undergo deamidation at high temperatures (Russell et al. 1997). This potentially accounts for their reduced use in thermophiles.
This research contributes to an expanding literature suggesting that the elemental composition of biopolymers can exhibit considerable plasticity, and can be related to environmental factors. Previously, links have been demonstrated between transient changes in environmental conditions and the elemental composition of proteins that are expressed under these conditions (e.g. Mazel & Marlière 1989; Baudouin-Cornu et al. 2001; Fauchon et al. 2002). Here we extend this by demonstrating associations between traits reflecting evolutionary adaptation to persistent environmental factors (thermophily and anaerobiosis) and the elemental composition of biopolymers at the level of whole proteomes.
Acknowledgements
We thank J. Brown and M. Tyers, and their respective lab groups, for useful discussion and support. Additionally, we thank L. Barton and U. Bergthorsson for useful discussion, A. Evangelisti and A. Hurlbert for valuable advice on programming, and J. Brown for comments on a draft of this manuscript. J.G.B. was supported by an NSF Biocomplexity Fellowship. P.B.-C. was supported by a grant from the Leukemia and Lymphoma Society and the Commissariat à l'Energie Atomique.
Footnotes
Additional author for correspondence (jbragg@unm.edu).
Supplementary Material
Protein and nucleic acid sequence data for 151 species (10 eukaryotes, 19 Archaea and 122 Eubacteria) were collected from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes). This electronic supplementary material contains a list of the species and strains that were used, and their accession numbers
References
- Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilus. Proc. Natl Acad. Sci. USA. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. 10.1073/pnas.062526999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L.L, Tomei F.A. Characteristics and activities of sulfate-reducing bacteria. In: Barton L.L, editor. Sulfate-reducing bacteriavol. 8. Plenum Press; New York, NY: 1995. pp. 1–32. [Google Scholar]
- Baudouin-Cornu P, Surdin-Kerjan Y, Marlière P, Thomas D. Molecular evolution of protein atomic composition. Science. 2001;293:297–300. doi: 10.1126/science.1061052. 10.1126/science.1061052 [DOI] [PubMed] [Google Scholar]
- Baudouin-Cornu P, Schuerer K, Marlière P, Thomas D. Intimate evolution of proteins: proteome atomic content correlates with genome base composition. J. Biol. Chem. 2004;279:5421–5428. doi: 10.1074/jbc.M306415200. 10.1074/jbc.M306415200 [DOI] [PubMed] [Google Scholar]
- Beeby M, O'Connor B.D, Ryttersgaard C, Boutz D.R, Perry L.J, Yeates T.O. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:e309. doi: 10.1371/journal.pbio.0030309. 10.1371/journal.pbio.0030309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett B.S, Stadtman E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20 313–20 316. doi: 10.1074/jbc.272.33.20313. 10.1074/jbc.272.33.20313 [DOI] [PubMed] [Google Scholar]
- Bragg J.G, Hyder C.L. Nitrogen versus carbon use in prokaryotic genomes and proteomes. Proc. R. Soc. B. 2004;271(Suppl. 5):S374–S377. doi: 10.1098/rsbl.2004.0193. 10.1098/rsbl.2004.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevey C.J, Fitzpatrick D.A, Philip G.K, Kinsella R.J, O'Connell M.J, Pentony M.M, Travers S.A, Wilkinson M, McInerney J.O. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc. R. Soc. B. 2004;271:2551–2558. doi: 10.1098/rspb.2004.2864. 10.1098/rspb.2004.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M, editor. The prokaryotes: an evolving electronic resource for the microbiological community. 3rd edn. Springer-Verlag; New York: 2004. [Google Scholar]
- Fauchon M, et al. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. 10.1016/S1097-2765(02)00500-2 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. doi: 10.1086/703055. 10.1086/284325 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W.-K, Bower K, Howson R.W, Belle A, Dephoure N, O'Shea E.K, Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Harvey P.H, Pagel M.D. Oxford University Press; Oxford, UK: 1991. The comparative method in evolutionary biology. [Google Scholar]
- Heidelberg J.F, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. 10.1038/nbt959 [DOI] [PubMed] [Google Scholar]
- Hickey D.A, Singer G.A.C. Genomic and proteomic adaptations to growth at high temperature. Genome Biol. 2004;5:1171–1177. doi: 10.1186/gb-2004-5-10-117. 10.1186/gb-2004-5-10-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J.G, editor. Gram-negative bacteria of general, medical, or industrial importance. Bergey's manual of systematic bacteriology. vol. 1. Williams & Wilkins; Baltimore, MD: 1984. [Google Scholar]
- Holt J.G, editor. Gram-positive bacteria other than Actinomycetes. Bergey's manual of systematic bacteriology. vol. 2. Williams & Wilkins; Baltimore, MD: 1986. [Google Scholar]
- Holt J.G, editor. Actinomycetes. Bergey's manual of systematic bacteriology. vol. 4. Williams & Wilkins; Baltimore, MD: 1989a. [Google Scholar]
- Holt J.G, editor. Archaeobacteria, Cyanobacteria, and remaining Gram-negative bacteria. Bergey's manual of systematic bacteriology. vol. 3. Williams & Wilkins; Baltimore, MD: 1989b. [Google Scholar]
- Karlin S, Brendel V. Chance and statistical significance in protein and DNA sequence analysis. Science. 1992;257:39–49. doi: 10.1126/science.1621093. [DOI] [PubMed] [Google Scholar]
- Klenk H.P, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. 10.1038/37052 [DOI] [PubMed] [Google Scholar]
- Lambros R.J, Mortimer J.R, Forsdyke D.R. Optimum growth temperature and the base composition of open reading frames in prokaryotes. Extremophiles. 2003;7:443–450. doi: 10.1007/s00792-003-0353-4. 10.1007/s00792-003-0353-4 [DOI] [PubMed] [Google Scholar]
- Major T.A, Burd H, Whitman W.B. Abundance of 4Fe-4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiol. Lett. 2004;239:117–123. doi: 10.1016/j.femsle.2004.08.027. 10.1016/j.femsle.2004.08.027 [DOI] [PubMed] [Google Scholar]
- Mazel D, Marlière P. Adaptive eradication of methionine and cysteine from cyanobacterial light-harvesting proteins. Nature. 1989;341:245–248. doi: 10.1038/341245a0. 10.1038/341245a0 [DOI] [PubMed] [Google Scholar]
- McEwan C, Gatherer D, McEwan N. Nitrogen-fixing aerobic bacteria have higher genomic GC content than non-fixing species within the same genus. Hereditas. 1998;128:173–178. doi: 10.1111/j.1601-5223.1998.00173.x. 10.1111/j.1601-5223.1998.00173.x [DOI] [PubMed] [Google Scholar]
- Meinnel T, Mechulam Y, Blanquet S. Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-d. 10.1016/0300-9084(93)90005-D [DOI] [PubMed] [Google Scholar]
- Naya H, Romero H, Zavala A, Alvarez B, Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. 10.1007/s00239-002-2323-3 [DOI] [PubMed] [Google Scholar]
- Rabus R, et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 2004;6:887–902. doi: 10.1111/j.1462-2920.2004.00665.x. 10.1111/j.1462-2920.2004.00665.x [DOI] [PubMed] [Google Scholar]
- Russell R.J.M, Ferguson J.M.C, Hough D.W, Danson M.J, Taylor G.L. The crystal structure of citrate synthase from the hyperthermophilic Archaeon Pyrococcus furiosus at 1.9 angstrom resolution. Biochemistry. 1997;36:9983–9994. doi: 10.1021/bi9705321. 10.1021/bi9705321 [DOI] [PubMed] [Google Scholar]
- Schultes E, Hraber P.T, LaBean T.H. Global similarities in nucleotide base composition among disparate functional classes of single-stranded RNA imply adaptive evolutionary convergence. RNA. 1997;3:792–806. [PMC free article] [PubMed] [Google Scholar]
- Singer G.A.C, Hickey D.A. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene. 2003;317:39–47. doi: 10.1016/s0378-1119(03)00660-7. 10.1016/S0378-1119(03)00660-7 [DOI] [PubMed] [Google Scholar]
- Sterner R.W, Elser J.J. Princeton University Press; Princeton, NJ: 2002. Ecological stoichiometry. [Google Scholar]
- Tatusov R.L, Koonin E.V, Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- Tekaia F, Yeramian E, Dujon B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene. 2002;297:51–60. doi: 10.1016/s0378-1119(02)00871-5. 10.1016/S0378-1119(02)00871-5 [DOI] [PubMed] [Google Scholar]
- Zar J.H. Prentice Hall; Englewood Cliffs, NJ: 1999. Biostatistical analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein and nucleic acid sequence data for 151 species (10 eukaryotes, 19 Archaea and 122 Eubacteria) were collected from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes). This electronic supplementary material contains a list of the species and strains that were used, and their accession numbers