Abstract
The extent of dispersal by pelagic larvae in marine environments, including coral reefs, is central for understanding local population dynamics and designing sustainable marine reserves. We present here the first example of a clear stepping-stone genetic structure throughout the Caribbean basin for a common coral reef species, the French grunt (Haemulon flavolineatum). Analysis of microsatellite DNA markers indicated that French grunt population structure may be characterized by overlapping populations throughout the Caribbean, influenced by independent population dynamics but with no fixed geographical boundaries. In addition, different spatial genetic patterns were found in different oceanographic regions. A second species, the bluehead wrasse (Thalassoma bifasciatum), has a much longer pelagic larval duration than French grunts and showed no explicit spatial pattern of genetic variation. This finding is concordant with the hypothesis of a positive relationship between larval dispersal and duration in the plankton. While the magnitude of the genetic signal of population structure in French grunts was very low (FST≈0.003), the pattern of isolation-by-distance throughout the Caribbean indicated considerable population structure with important ecological and conservation significance.
Keywords: genetic population structure, microsatellites, coral reef fish dispersal, marine reserves, Haemulon flavolineatum, Thalassoma bifasciatum
1. Introduction
Most coral reef fish and invertebrates spend several weeks as pelagic larvae during which time they may disperse and then metamorphose into sedentary adults. This process, which largely determines the spatial scale of population structure and is central to coral reef fish population dynamics, has been the ‘Holy Grail’ of reef fish ecology for decades. An understanding of larval dispersal patterns also helps determine whether recruits to marine protected areas are local or originate from distant locations, and at what scale they may reseed the surrounding area. For this reason, knowledge of dispersal is central to the sustainability of marine reserves (Botsford et al. 2001), yet many questions remain largely unanswered (Mora & Sale 2002; Sale et al. 2005).
Mark–recapture studies, including artificial and natural otolith tagging (Jones et al. 1999, 2005; Swearer et al. 1999) as well as analyses, which model fine-scale currents combined with larval behaviour (Cowen et al. 2000, 2006), have been used to study the movement of individuals between populations of reef organisms. Only genetic methods, however, can measure effective dispersal between populations (i.e. those individuals that survive and breed in their new population). Genetic differentiation among populations also integrates dispersal or retention patterns over generations rather than seasons or years and so provides longer-term estimates of these processes.
Earlier genetic studies of coral reef fish population structure using allozymes and mitochondrial DNA have had mixed success in resolving genetic structure. Several studies showed that marine populations were more structured than was predicted based on dispersal capabilities (Palumbi 1994). In the Pacific, a few studies found significant genetic differentiation over thousands of kilometres (Planes 2002; Ovenden et al. 2004). In contrast, several studies of fish populations in the Caribbean, where populations are separated by less than 3000 km, found no clear patterns of genetic differentiation (Lacson 1992; Shulman & Bermingham 1995).
Isolation-by-distance, based on a stepping-stone model of dispersal (Wright 1943; Kimura & Weiss 1964; Slatkin 1993), is the most realistic pattern to test when populations are distributed along a coastline or chain of islands (Palumbi 2003; Hellberg 2006). A significant pattern of isolation-by-distance indicates that sampling error is not larger than the signal of population differentiation and boosts confidence in inferences drawn from even a very weak signal of genetic differentiation (Slatkin 1993; Palumbi 2003). In some cases, the observation of isolation-by-distance may be one of the only ways to establish the existence of genetic structure in large marine populations with considerable gene flow among populations.
To estimate scales of population connectivity for Caribbean reef fish, we compared two species, the French grunt (Haemulon flavolineatum) and the bluehead wrasse (Thalassoma bifasciatum) that exhibit extremes of pelagic larval duration (PLD)—very short for French grunts and very long for bluehead wrasse. Both species have been previously studied using mitochondrial DNA markers and neither showed geographically based population structure (Shulman & Bermingham 1995). We used highly polymorphic microsatellite DNA markers coupled with a sampling protocol to test for isolation-by-distance in order to resolve the weak signal of population differentiation expected for these species.
Both species are among the most widely distributed and common reef fish in the Caribbean. For French grunts and bluehead wrasse in the Florida Keys alone, fish density counts carried out over a 20-year period imply a census population over 9 and 20 million, respectively (Bohnsack et al. 1999). Our observations in the other 13 study sites throughout the Caribbean indicated only slightly lower densities, suggesting total census populations in the sampled areas of between 50 and 100 million for French grunts. For the Caribbean as a whole, census numbers approaching a billion or more adults for each species would not be unreasonable. Even if effective population sizes were orders of magnitude smaller than census populations, they would still be large. Also, French grunts and bluehead wrasse, frequent and dispersed spawners, fit the profile of species with large effective population sizes. French grunt spawning is pelagic and occurs throughout the year in a dispersed fashion on approximately a bi-monthly basis (McFarland et al. 1985). Bluehead wrasse spawn on a daily basis at the edge of the reef (Warner et al. 1975). French grunts are under a small degree of artisanal fishing pressure on some islands (Claro et al. 2001), whereas bluehead wrasse are not harvested for food.
2. Material and methods
(a) Sampling protocol
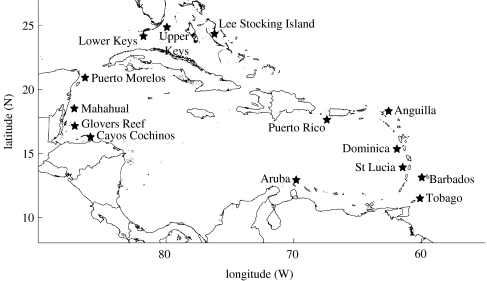
The sampling protocol, with 14 sites ringing the Caribbean basin (figure 1), was designed to detect isolation-by-distance at spatial scales ranging from basin-wide (thousands of kilometres) to intra-reef (kilometres). At each site, we sampled two sub-sites located 4 and 6 km apart to test for micro-spatial structure within the reef system. Fish were captured with hand nets, fin clipped and then released. While not always possible, an effort was made to capture 100 adults and 100 juveniles of each species at each site. A total of 1554 adult French grunts and 1498 adult bluehead wrasse were sampled from the 14 sites (an average of over 100 adults). One hundred and fifty three juvenile French grunts were sampled from two sites and 770 juvenile bluehead wrasse were sampled from eight sites. For French grunts, between 67 and 191 adults were analysed per site except for Barbados, which had 31 individuals. For bluehead wrasse, between 88 and 173 adults were sampled per site, except for Aruba, which had 57 individuals.
Figure 1.
The 14 sampling sites are indicated by stars (precise locations available from Purcell).
Two methods were used to test for the existence of temporal genetic stability. First, five sites were re-sampled for French grunts and three sites for bluehead wrasse 2 years apart, during the period 2001–2003 and genetic patterns were compared. Second, generational stability was tested by collecting equivalent numbers of juveniles at the same time and on the same reefs as for adults. This was possible at two sites for French grunts and at nine sites for bluehead wrasse. The combined temporal and generational sampling resulted in temporal stability being tested for 43% of French grunt populations and 64% of bluehead wrasse populations.
(b) Genetic analysis
Samples from each species were genotyped at nine species-specific microsatellite loci (Williams et al. 2004a,b), and fragments were separated on an ABI 3730 XL (Applied Biosystems). Alleles were scored using GENEMAPPER v. 3.0 (Applied Biosystems).
(c) Statistical analysis
Single locus estimates of expected and observed heterozygosity and number of alleles were made using the software package Microsatellite Analyzer (Dieringer & Schlotterer 2003), available at http://i122server.vu-wien.ac.at. Loci were tested for genotypic linkage equilibrium using GENEPOP (Raymond & Rousset 1995). Hardy–Weinberg equilibrium (HWE) was tested for each locus and population by determining if the inbreeding coefficient (FIS) was significantly different from zero using the program FSTAT (Goudet 2001).
Departures from HWE can be caused by biological processes such as inbreeding or population substructure (i.e. the Wahlund effect) or by technical issues such as null alleles. Deviations from HWE due to inbreeding or population substructure should result in heterozygote deficits across most or all loci, whereas technical causes such as null alleles should result in heterozygote deficits that are variable across loci and populations. The software MICROCHECKER (Van Oosterhout et al. 2004) was used to infer the most probable technical cause of HWE departures, including null alleles, mis-scoring due to stuttering, and allelic dropout due to short allele dominance (e.g. Wattier et al. 1998). When null alleles were inferred as the most likely cause of heterozygote deficits, their frequencies were estimated in MICROCHECKER using the methods of Brookfield (1996). One method (Brookfield 1) treats non-amplifying individuals as artefacts and discounts them when calculating null allele frequencies, whereas the second method (Brookfield 2) treats non-amplifications as data and regards them as null homozygotes when calculating null allele frequencies (Brookfield 1996).
Three tests were carried out to determine whether null alleles were a significant factor in determining either the presence or absence of genetic patterns in the two species. First, in both species, each locus was removed from the analysis to see whether patterns of genetic distance among populations changed significantly as a result. Second, the proportion of inferred nulls was calculated for each population and these proportions were correlated with the average genetic distance of each population to all other populations. And third, allele frequencies were re-adjusted within populations to account for null alleles and tests for isolation-by-distance were repeated using the adjusted data.
Genetic differentiation among spatially separated samples was estimated, first using Fisher's exact tests of significant pairwise allelic (genic) and genotype (genotypic) frequency differences in the program FSTAT, followed by sequential Bonferroni correction for multiple tests. Genetic differentiation was also estimated using the FST estimator, Theta (Weir & Cockerham 1984) and the FST analogue, RST, which assumes a stepwise mutation model (Slatkin 1995). Estimates of RST were made using an ANOVA approach following Michalakis & Excoffier (1996) in the program SPAGeDi (Hardy & Vekemans 2002; Hardy 2003), available at http://www.ulb.ac.be/sciences/ecoevol/spagedi.html.
Three methods were used to test for a positive relationship between pairwise population genetic differentiation (FST/(1−FST)) and geographical distance (measured as the shortest distance by sea). First, a Mantel test was used to test for a positive correlation between genetic and geographical distances with 10 000 permutations using the software IBD (Bohonak 2003), available at http://www.bio.sdsu.edu/pub/andy/IBD.html. Second, regional comparisons of genetic heterogeneity between the eastern and western Caribbean were conducted using analysis of molecular variance (AMOVA) with the program ARLEQUIN v. 2.000 (Schneider et al. 2000), available at http://anthro.unige.ch/arlequin. And third, genetic spatial autocorrelation analysis was performed using the program GenA1Ex 6 (Peakall & Smouse 2005). The spatial autocorrelation analysis implemented in GenA1Ex calculates an autocorrelation coefficient (r) for genetic distances (FST in this case) between populations for categories of geographical distance. One thousand random permutations were used to generate the 95% confidence intervals around the expectation of no spatial genetic structure. The geographical distance at which the mean r value drops below zero has been called the ‘patch size’ (Peakall et al. 2003) or the ‘neighbourhood size’ (Gold & Turner 2002). It represents the largest spatial scale at which genetic similarity is non-random.
3. Results
(a) Genetic variation and Hardy–Weinberg equilibrium
French grunts had an average of 26 alleles per locus (range, 5–44 alleles) and an average expected heterozygosity of 0.78 (range, 0.33–0.92; table 2 of electronic supplementary material). Bluehead wrasse had an average of 42 alleles per locus (range, 15–60) and average expected heterozygosity of 0.94 (range, 0.79–0.98; table 3 of electronic supplementary material). We found no evidence of genotypic disequilibrium between any of the nine loci within populations for either species (p>0.1 in all cases).
In French grunts, significant departures from HWE were observed in 21 of 126 tests (table 2 of electronic supplementary material). The locus AAT15 exhibited heterozygote deficits in six of 14 populations. No other locus had significant heterozygote deficits in more than three populations. French grunt populations only had between zero and three loci that exhibited heterozygote deficits. In bluehead wrasse, significant heterozygote deficits occurred in 44 of 126 tests (table 3 of electronic supplementary material). Three loci (AAC50, AAT41 and GTp) had heterozygote deficits in 13, 10 and 9 populations, respectively. These three loci accounted for 73% of the total cases of heterozygote deficits for the nine loci. The number of bluehead wrasse loci with significant heterozygote deficits per population was relatively evenly divided among the 14 populations. Inbreeding and population substructure seem very unlikely to have caused the observed heterozygote deficits since the patterns of deficits across loci and populations were highly variable. MICROCHECKER indicated that the most likely technical cause of these heterozygote deficits were null alleles.
(b) Population structure
There was significant genotypic differentiation for 47% and 4% of the 91 pairwise population comparisons for French grunts and bluehead wrasse, respectively, after sequential Bonferroni correction (tables 4 and 5 of electronic supplementary material). Genic differentiation showed a slightly stronger level of differentiation for both species (tables 4 and 5 of electronic supplementary material).
Global FST was 0.003 for the nine loci in French grunts (p<0.0001) and pairwise FST values among populations ranged from −0.002 to 0.009 (table 6 of electronic supplementary material). For French grunts, 26% of the 91 pairwise FST estimates (but none of the RST estimates) were significant after Bonferroni correction (table 6 of electronic supplementary material). Multilocus global FST (0.0002) for bluehead wrasse were not different from zero and no FST or RST pairwise estimates (range: −0.001 to 0.002) were significant (table 7 of electronic supplementary material). AMOVA results for grunts showed that east–west differences explained 0.29% of variance (p<0.001) while among populations within each region explained 0.00% of variance. None of the variance was explained by either between-region or among-population differences for wrasse (p>0.8). For both species, no comparisons among sub-sites or temporal samples from the same locality were significantly differentiated (French grunts FST range, −0.0001 to 0.001; bluehead wrasse FST range, −0.001 to 0.001).
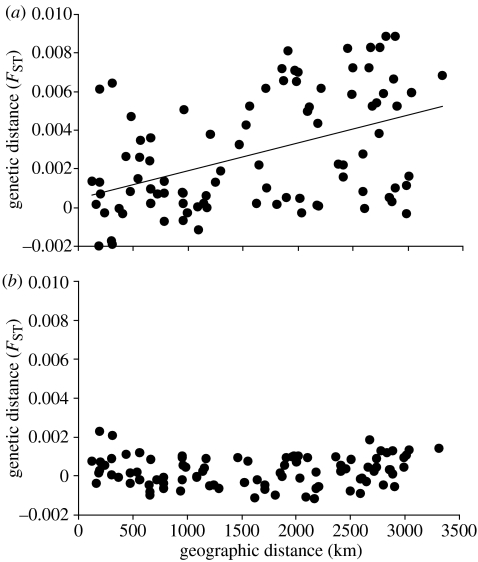
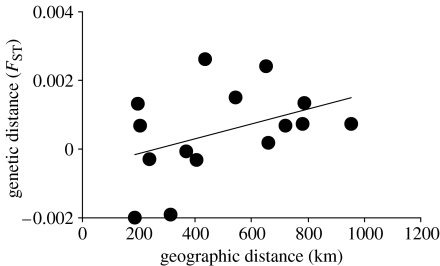
There was a significant isolation-by-distance pattern in French grunts, at both a Caribbean basin-wide scale (figure 2a) as well as at the shorter scale (approx. 900 km) of the eastern Caribbean (figure 3). Among the seven populations of the western Caribbean, however, no spatial pattern is evident (data not shown). A positive and statistically significant relationship between genetic and geographical distance remains for French grunts as each locus is removed from the dataset, showing that no single locus determined the pattern (table 1). Similarly, removing a single population at a time revealed that no one population was driving the relationship (data not presented). The consistent isolation-by-distance pattern in French grunts contrasts with the lack of any spatial pattern in bluehead wrasse at either the Caribbean basin scale (figure 2b) or any regional scale (data not shown).
Figure 2.
The relationship between geographic (km) and genetic distance (FST) among the 14 sites shown in figure 1. Geographic distance is measured as the shortest distance in kilometres by sea. (a) French grunts at a Caribbean basin-wide scale (regression analysis: y=0.000001x+0.0005; R2=0.21; n=91; p=0.001, Mantel test). (b) Bluehead wrasse at a Caribbean basin-wide scale (n=91; p=0.97).
Figure 3.
Positive relationship between geographic (km) and genetic distance (FST) for French grunts sampled at six sites in the eastern Caribbean: Puerto Rico, Anguilla, Dominica, St Lucia, Barbados and Tobago (regression analysis: y=0.000002x−0.0006; R2=0.17; p=0.04, Mantel test).
Table 1.
The relationship (using regression analyses) between geographical (km) and genetic distance (FST) at the Caribbean basin scale for French grunts. (A jack-knife procedure was performed by removing one locus at a time and the significance of the subsequent relationship was determined using a Mantel test.)
| locus excluded | R2 | slope | p |
|---|---|---|---|
| AAC3 | 0.19 | 1×10−6 | 0.002 |
| AAT15 | 0.07 | 4×10−7 | 0.002 |
| AAC41 | 0.19 | 1×10−6 | 0.002 |
| AAC10 | 0.19 | 2×10−6 | 0.002 |
| AAC37 | 0.20 | 2×10−6 | 0.002 |
| AAT3 | 0.23 | 2×10−6 | 0.002 |
| AAC54 | 0.20 | 2×10−6 | 0.001 |
| AAC43 | 0.19 | 2×10−6 | 0.001 |
| AAC46 | 0.21 | 2×10−6 | 0.001 |
| mean | 0.19 | ||
| all loci | 0.21 | 1×10−6 | 0.001 |
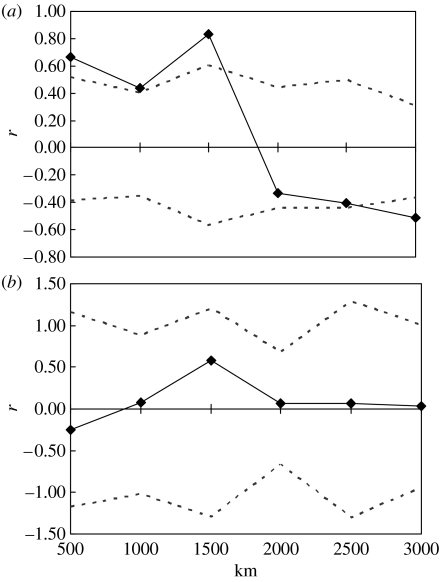
The isolation-by-distance pattern in grunts was also detected using spatial autocorrelation analysis, while wrasse showed no spatial pattern (figure 4a,b). In grunts, mean autocorrelation coefficient values (r) for distance classes of 0–500, 500–1000 and 1000–1500 km are significant (p=0.007, 0.021 and 0.006, respectively). The mean r value intercepts zero at 1857 km suggesting a patch size for French grunts between 1500 and 2000 km. In bluehead wrasse, no spatial autocorrelation distance class had a significance value less than p=0.59.
Figure 4.
Multilocus spatial autocorrelation analyses for: (a) French grunts and (b) bluehead wrasse at a Caribbean basin-wide scale. Data points (diamonds) are r (correlation coefficient) values of genetic distance between populations separated by distances in the preceding 500 km distance class. Dashed lines are upper and lower 95% confidence intervals for all data in the distance class. When an r value lies above the upper confidence interval, populations in that class are genetically more similar than would be expected by chance.
The removal of individual loci with the highest proportion of null alleles did not eliminate the significant pattern of isolation-by-distance in French grunts nor did it produce any spatial pattern in bluehead wrasse. There was also no significant relationship between the proportion of nulls in a population and either particularly high or low pairwise FST scores for that population. Furthermore, in either species, when all loci with a significant proportion of null alleles were adjusted using either the Brookfield 1 or 2 algorithms, a significant pattern of isolation-by-distance remained for French grunts and no pattern was evident for bluehead wrasse.
4. Discussion
(a) Population structure
There was a clear distinction between the genetic population structure patterns of French grunts and bluehead wrasse. Pairwise FST values were low for both species and genetic polymorphism was high: a finding common in many species of marine fish, especially for microsatellite markers (O'Reilly et al. 2004). High genetic polymorphism in turn is probably related to high mutation rates and large effective population sizes (DeWoody & Avise 2000; O'Reilly et al. 2004). However, the greater number of significant pairwise differences in French grunts than in bluehead wrasse, and the significant and robust pattern of isolation-by-distance in French grunts illustrate a well-defined population structure in French grunts in contrast to the lack of any obvious structure in bluehead wrasse.
We conclude that these findings are based on a true signal. First, there are few factors that can cause a false signal of isolation-by-distance. It is unlikely that our findings in the Caribbean represent an example of recolonization and secondary contact. There is no latitudinal component to the pattern as there is for north–south clines involving recolonization (Arnaud-Haond et al. 2003), indeed the pattern is not clinal in any direction since the sites are spaced in a circular array around the Caribbean. Further evidence for the robustness of the isolation-by-distance pattern comes from jack-knifing over the nine loci (table 1) and among the 14 populations (data not shown) suggesting the pattern is not due to selection acting through gene linkage or by geographical outliers.
In spite of the fact that heterozygote deficits for both species appear to be due to the presence of null alleles, we conclude that the pattern of population structure in French grunts and the lack of it in bluehead wrasse are unlikely to be artefacts of null alleles: the patterns persist after adjusting frequencies to take into account null alleles; patterns do not rely on loci with particularly large or small null allele proportions; and spatial patterns of null alleles do not correlate with pairwise genetic differentiation among populations. It has been suggested by O'Reilly et al. (2004), in their study of walleye pollock (Theragra chalcogramma), that null alleles may be more common in large marine populations because of large effective population sizes (which could characterize both our target species) and reduced loss of variation due to genetic drift.
(b) Effects of life-history traits on larval dispersal
The strong genetic differences between grunts and wrasse should not be over-emphasized. A study using otolith microchemistry tags indicated that between 50 and 70% of bluehead wrasse recruits to the island of St Croix carried otolith signatures of local origin (Swearer et al. 1999). Such levels of larval retention, while fairly high from a demographical perspective, are probably impossible for our genetic markers to detect. Nevertheless, the absence of spatially related genetic structure in bluehead wrasse sampled from the same reefs at the same times as grunts is probably indicative of actual differences in dispersal patterns of the two species, due to differences in life-history traits.
In particular, the difference is congruent with differences in PLD. French grunts have the shortest PLD known among Caribbean reef fish (about 15 days) and larvae are seldom caught during offshore plankton tows, suggesting they may largely be retained on their natal reefs (Lindeman et al. 2001). French grunt spawning is pelagic and occurs throughout the year in a dispersed fashion on approximately a bi-monthly basis (McFarland et al. 1985). In contrast, bluehead wrasses spawn daily and have among the longest PLD of any Caribbean reef fish (45 days or more). Larvae are caught frequently during offshore sampling.
(c) Evolutionary versus demographical significance of genetic signals
Our findings constitute an interesting contrast to the only other published study that has shown genetic structure among reef fish in the Caribbean, that of the cleaner goby, Elacatinus evelynae (Taylor & Hellberg 2003). While results from French grunts demonstrate genetic evidence for a stepping-stone model of restricted gene flow in the Caribbean, those on E. evelynae indicate virtually no gene flow (and thus no isolation-by-distance) over distances as short as 23 km and over time periods as long as 100 000 years. The contrast between these findings and our own raises a more general contrast: between genetic differences with evolutionary significance and genetic differences with demographical or ecological significance, which are nonetheless biologically important.
It is clear from the low levels of global and pairwise FST values detected for French grunts that from an evolutionary perspective these populations are highly connected, with well over 10 migrants per generation among populations. This is far above the number viewed as necessary for independent evolutionary development (Allendorf & Phelps 1981). Several authors (Palumbi 2003; Hellberg 2006) have pointed out that since total gene flow is measured as the product of effective population size and migration rate (NEm), very low FST values, while indicating an absolutely large number of migrants, may imply a very small proportion of migrants in the recipient population. Thus, when FST≈0.003 and populations are large, migration rates may be only a few percentage points or even fractions of a percentage point. When the proportion of migrants is so low, populations are likely to be self-seeding and influenced by independent population dynamics.
(d) Oceanographic factors
The two scales of isolation-by-distance observed in French grunts suggest an important role for oceanographic features in addition to geographical distance per se, for larval dispersal. Considerable differences between the oceanographic regimes in the western and the eastern Caribbean probably help to explain the genetic differences. Most notably, the western Caribbean is subjected to a fast western boundary current that may be responsible for rapid advection of larvae between some of our sampling locations, thus homogenizing the genetic signal in this region. The eastern region is dominated by the slower moving and much less clearly advective North Brazil current rings, which could gradually move a number of offshore larvae northward along the semicircle of islands (Cowen et al. 2003, 2006).
The low (less than 1%), but significant genetic differentiation for French grunts explained by the east–west division could be interpreted as a gradual build-up of genetic differentiation between overlapping populations that are themselves not different enough to achieve significance except at the extreme ends of the species' range. However, it may indicate a more substantial oceanographic break as suggested by Taylor & Hellberg (2003) for cleaner goby or by Baums et al. (2005) for elkhorn coral (Acropora palmata). Evidence for an oceanographic break was also apparent in the modelling analysis by Cowen et al. (2006).
(e) Conservation implications of overlapping neighbourhoods
The pattern of population structure of French grunts in the Caribbean basin is similar to that observed by Gold & Turner (2002) for red drum located in estuaries along the northern Gulf of Mexico: a series of overlapping populations among which gene flow is sufficient to prevent any fixed geographical boundaries but that probably have independent population dynamics. Since gene flow shown in the isolation-by-distance pattern builds up in a stepwise fashion over a number of generations, the single generation dispersal distance of larval French grunts is likely to be considerably less than the 1900 km estimated by spatial autocorrelation analysis. The existence of restricted gene flow within the eastern Caribbean also suggests that, in some areas, dispersal distances are in fact below the 900 km distance along this string of islands.
In spite of the wide range of uncertainty embedded in our estimates of larval dispersal and the size of genetically defined neighbourhoods, a number of useful insights with conservation implications are gained from the data. First, French grunts should not be managed on a Caribbean basin-wide scale since different regions would be unlikely to subsidize one another. At the very least, the eastern and the western Caribbean probably represent demographically distinct regions, though it is unlikely to be possible to define clear boundaries between the regions for species like French grunts. The spatial scale of demographically defined neighbourhoods is undoubtedly less than 2000 km and more likely in the range of hundreds of kilometres. However, neighbourhoods appear to encompass groups of islands rather than single islands, so a regional, but not a basin-wide scale appears to be appropriate for conservation purposes. Finally, while geographical distance by itself may sometimes be a convenient metric to characterize French grunt population structure, there is evidence that the scale and patterns of ecologically relevant population structure vary with large-scale differences in physical oceanographic patterns like those in the eastern versus the western Caribbean.
(f) Conclusions
Our findings represent the first genetic evidence for a stepping-stone model of restricted larval dispersal in a Caribbean coral reef fish. Previous studies have shown either no clear evidence of restricted gene flow or very strong isolation. A pattern of isolation-by-distance allowed us to conclude that larval dispersal between populations was probably demographically insignificant, and also to make a rough estimate both of the scale of the overlapping populations or perhaps more accurately ‘neighbourhoods’. Even such rough conclusions are important for marine reserve design (Palumbi 2004). These findings also indicate that combining a highly variable molecular marker with a sampling protocol to detect possible isolation-by-distance can resolve a pattern of population structure with FST values at least an order of magnitude weaker than those commonly reported (Kinlan & Gaines 2003). This ability can help to greatly expand the number of species for which genetic dispersal estimates can be made, to include some of the most common and widely dispersed coral reef taxa.
Acknowledgements
We thank P. Bentzen, S. Sponaugle, J. Trexler, A. Hale and K. Lindeman for advice, discussion and help. We thank members of the Cowen and Sponaugle labs for help in sample collection, in particular M. Paddack, M. Sullivan, E. D'Alessandro, D. Richardson, T. Smith, J. Fortuna, K. Denit, C. Paris and K. Grorud. We thank A. Miyake for help in the lab. We thank the people in the governments, universities and research stations of our Caribbean sampling sites for making this study possible, particularly C. Garcia Saez, A. Potts, H. Oxenford, J. Gonzalez Cano, F. Pagan, P. Butcher, B. Boekhoudt, S. Steiner, D. Wesby, E. Bartels, K. Wulf, A. Cubas and B. Marshall. Supported by grants from The Wildlife Conservation Society to J.F.H.P. and NSF (OCE-095955) to R.K.C., C.R.H. and J.F.H.P. Thanks also to two anonymous reviewers for helpful comments.
Supplementary Material
Tables 2–7 and figure 1 as referenced in the article.
References
- Allendorf F.W, Phelps S.R. Use of allelic frequencies to describe population structure. Can. J. Fish. Aquat. Sci. 1981;38:1507–1514. [Google Scholar]
- Arnaud-Haond S, Monteforte M, Blanc F, Bonhomme F. Evidence for male-biased effective sex ratio and recent step-by-step colonization in the bivalve Pinctada mazatlanica. J. Evol. Biol. 2003;16:790–796. doi: 10.1046/j.1420-9101.2003.00603.x. doi:10.1046/j.1420-9101.2003.00603.x [DOI] [PubMed] [Google Scholar]
- Baums I.B, Miller M.W, Hellberg M.E. Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol. Ecol. 2005;14:1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x. doi:10.1111/j.1365-294X.2005.02489.x [DOI] [PubMed] [Google Scholar]
- Bohnsack, J. A. et al 1999 Baseline data for evaluating reef fish populations in the Florida Keys, 1979–1998. NMFS Tech Memo NMFS-SEFSC-427.
- Bohonak A.J. IBD (isolation by distance): a program for analyses of isolation by distance. J. Hered. 2003;93:153–154. doi: 10.1093/jhered/93.2.153. doi:10.1093/jhered/93.2.153 [DOI] [PubMed] [Google Scholar]
- Botsford L.W, Hastings A, Gaines S.D. Dependence of sustainability on the configuration of marine reserves and larval dispersal distance. Ecol. Lett. 2001;4:144–150. doi:10.1046/j.1461-0248.2001.00208.x [Google Scholar]
- Brookfield J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996;5:453–455. doi: 10.1111/j.1365-294x.1996.tb00336.x. doi:10.1046/j.1365-294X.1996.00098.x [DOI] [PubMed] [Google Scholar]
- Claro R, Baisre J, Lindeman K, Garcia J. Cuban fisheries: historical trends and current status. In: Claro R, Lindeman K, Parenti L, editors. Ecology of the marine fishes of Cuba. Smithsonian Institution Press; Washington, DC: 2001. pp. 194–218. [Google Scholar]
- Cowen R.K, Lwiza K.M.M, Sponaugle S, Paris C.B, Olson D.B. Connectivity of marine populations: open or closed? Science. 2000;287:857–859. doi: 10.1126/science.287.5454.857. doi:10.1126/science.287.5454.857 [DOI] [PubMed] [Google Scholar]
- Cowen R.K, Paris C.B, Olson D.B, Fortuna J.L. The role of long distance dispersal versus local retention in replenishing marine populations. Gulf Carib. Res. 2003;14:129–137. [Google Scholar]
- Cowen R.K, Paris C.B, Srinivasan A. Scaling of connectivity in marine populations. Science. 2006;311:522–527. doi: 10.1126/science.1122039. doi:10.1126/science.1122039 [DOI] [PubMed] [Google Scholar]
- DeWoody J.A, Avise J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol. 2000;56:461–473. doi:10.1111/j.1095-8649.2000.tb00748.x [Google Scholar]
- Dieringer D, Schlotterer C. MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes. 2003;3:167–169. doi:10.1046/j.1471-8286.2003.00351.x [Google Scholar]
- Gold J.R, Turner T.F. Population structure of red drum (Sciaenops ocellatus) in the northern Gulf of Mexico, as inferred from variation in nuclear-encoded microsatellites. Mar. Biol. 2002;140:249–265. doi:10.1007/s002270100692 [Google Scholar]
- Goudet J. 2001 FSTAT, a program to estimate and test gene diversities and fixation indices.
- Hardy O.J. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol. Ecol. 2003;12:1577–1588. doi: 10.1046/j.1365-294x.2003.01835.x. doi:10.1046/j.1365-294X.2003.01835.x [DOI] [PubMed] [Google Scholar]
- Hardy O.J, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes. 2002;2:618–620. doi:10.1046/j.1471-8286.2002.00305.x [Google Scholar]
- Hellberg, M. E. 2006 Genetic approaches to understanding marine metapopulation dynamics. In Marine metapopulations (ed. J. P. Kritzer & P. F. Sale). San Diego, CA: Academic Press.
- Jones G.P, Milicich M.J, Emslie M.J, Lunow C. Self-recruitment in a coral reef fish population. Nature. 1999;402:802–804. doi:10.1038/45538 [Google Scholar]
- Jones G.P, Planes S, Thorrold S.R. Coral reef fish larvae settle close to home. Curr. Biol. 2005;15:1314–1318. doi: 10.1016/j.cub.2005.06.061. doi:10.1016/j.cub.2005.06.061 [DOI] [PubMed] [Google Scholar]
- Kimura M, Weiss G.H. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinlan B.P, Gaines S.D. Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology. 2003;84:2007–2020. [Google Scholar]
- Lacson J.M. Minimal genetic variation among samples of six species of coral reef fishes collected at La Parguera, Puerto Rico, and Discovery Bay, Jamaica. Mar. Biol. 1992;112:327–331. doi:10.1007/BF00702479 [Google Scholar]
- Lindeman K, Lee T, Wilson W, Claro R, Ault J. Transport of larvae originating in southwest Cuba and the Dry Tortugas: evidence for partial retention in grunts and snappers. Proc. Gulf Carib. Fish. Inst. 2001;52:732–747. [Google Scholar]
- McFarland W.N, Brothers E.B, Ogden J.C, Shulman M.J, Bermingham E.L, Kotchian-Prentiss N.M, McFarland W. Recruitment patterns in young French grunts, Haemulon flavolineatum (Family Haemulidae), at St Croix, Virgin Islands. Fish. Bull. US. 1985;83:413–426. [Google Scholar]
- Michalakis Y, Excoffier L. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996;142:1061–1064. doi: 10.1093/genetics/142.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Sale P.F. Are populations of coral reef fish open or closed? Trends Ecol. Evol. 2002;17:422–428. doi:10.1016/S0169-5347(02)02584-3 [Google Scholar]
- O'Reilly P.T, Canino M.F, Bailey K.M, Bentzen P. Inverse relationship between FST and microsatellite polymorphism in the marine fish, walleye pollock (Theragra chalcogramma): implications for resolving weak population structure. Mol. Ecol. 2004;13:1799–1814. doi: 10.1111/j.1365-294X.2004.02214.x. doi:10.1111/j.1365-294X.2004.02214.x [DOI] [PubMed] [Google Scholar]
- Ovenden J.R, Salini J, O'Connor S, Street R. Pronounced genetic population structure in a potentially vagile fish species (Pristipomoides multidens, Teleostei: Perciformes: Lutjanidae) from the East Indies triangle. Mol. Ecol. 2004;13:1991–1999. doi: 10.1111/j.1365-294X.2004.02210.x. doi:10.1111/j.1365-294X.2004.02210.x [DOI] [PubMed] [Google Scholar]
- Palumbi S.R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 1994;25:547–572. doi:10.1146/annurev.es.25.110194.002555 [Google Scholar]
- Palumbi S.R. Population genetics, demographic connectivity, and the design of marine reserves. Ecol. Appl. 2003;13:S146–S158. [Google Scholar]
- Palumbi S.R. Marine reserves and ocean neighborhoods: the spatial scale of marine populations and their management. Annu. Rev. Environ. Resour. 2004;29:31–68. doi:10.1146/annurev.energy.29.062403.102254 [Google Scholar]
- Peakall R, Smouse P.E. Australian National University; Canberra: 2005. GenA1Ex 6: Genetic analysis in Excel. Population genetic software for teaching and research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Ruibal M, Lindenmayer D.B. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution. 2003;57:1182–1195. doi: 10.1111/j.0014-3820.2003.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Planes S. Biogeography and larval dispersal inferred from population genetic analysis. In: Sale P.F, editor. Coral reef fishes: dynamics and diversity in a complex ecosystem. Academic Press; San Diego, CA: 2002. pp. 201–220. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Sale P.F, et al. Critical science gaps impede use of no-take fishery reserves. Trends Ecol. Evol. 2005;20:74–80. doi: 10.1016/j.tree.2004.11.007. doi:10.1016/j.tree.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. Arlequin version 2000: a software for population genetics data analysis. [Google Scholar]
- Shulman M.J, Bermingham E. Early life histories, ocean currents and the population genetics of Caribbean reef fishes. Evolution. 1995;49:897–910. doi: 10.1111/j.1558-5646.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swearer S.E, Caselle J.E, Lea D.W, Warner R.R. Larval retention and recruitment in an island population of a coral-reef fish. Nature. 1999;402:799–802. doi:10.1038/45533 [Google Scholar]
- Taylor M.S, Hellberg M.E. Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Science. 2003;299:107–109. doi: 10.1126/science.1079365. doi:10.1126/science.1079365 [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson W.F, Wills D.P.M, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi:10.1111/j.1471-8286.2004.00684.x [Google Scholar]
- Warner R.R, Robertson D.R, Leigh E.G. Sex change and sexual selection. Science. 1975;190:633–638. doi: 10.1126/science.1188360. [DOI] [PubMed] [Google Scholar]
- Wattier R, Engel C.R, Saumitou-Laprade P, Valero M. Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus Gv1CT in Gracilaria gracilis (Rhodophyta) Mol. Ecol. 1998;7:1569–1573. doi:10.1046/j.1365-294x.1998.00477.x [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams D.A, Purcell J, Cowen R.K, Hughes C.R. Microsatellite multiplexes for high-throughput genotyping of French grunts (Haemulon flavolineatum, Pisces: Haemulidae) and their utility in other grunt species. Mol. Ecol. Notes. 2004a;4:46–48. doi:10.1046/j.1471-8286.2003.00568.x [Google Scholar]
- Williams D.A, Purcell J, Cowen R.K, Hughes C.R. Characterization of microsatellite multiplexes for population genetic studies of bluehead wrasse (Thalassoma bifasciatum, Pisces: Labridae) Mol. Ecol. Notes. 2004b;4:525–527. doi:10.1111/j.1471-8286.2004.00713.x [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables 2–7 and figure 1 as referenced in the article.