Abstract
Natural populations worldwide are increasingly fragmented by habitat loss. Isolation at small population size is thought to reduce individual and population fitness via inbreeding depression. However, little is known about the time-scale over which adverse genetic effects may develop in natural populations or the number and types of traits likely to be affected. The benefits of restoring gene flow to isolates are therefore also largely unknown. In contrast, the potential costs of migration (e.g. disease spread) are readily apparent. Management for ecological connectivity has therefore been controversial and sometimes avoided. Using pedigree and life-history data collected during 25 years of study, we evaluated genetic decline and rescue in a population of bighorn sheep founded by 12 individuals in 1922 and isolated at an average size of 42 animals for 10–12 generations. Immigration was restored experimentally, beginning in 1985. We detected marked improvements in reproduction, survival and five fitness-related traits among descendants of the 15 recent migrants. Trait values were increased by 23–257% in maximally outbred individuals. This is the first demonstration, to our knowledge, of increased male and female fitness attributable to outbreeding realized in a fully competitive natural setting. Our findings suggest that genetic principles deserve broader recognition as practical management tools with near-term consequences for large-mammal conservation.
Keywords: mammals, immigration, genetic rescue, conservation genetics
1. Introduction
A prominent paradigm in conservation science predicts that populations isolated at small size will experience reductions in individual fitness due to inbreeding depression. Loss of individual adaptation may adversely affect population demography and increase the risk of population extinction (Gilpin & Soulé 1986). Given widespread and continuing habitat fragmentation (Jenkins 2003), this implies the need for management that maintains, restores or substitutes for historical patterns of between-population gene flow. Habitat fragmentation is of particular concern in large-bodied wildlife species having low natural population density because even large fragments may only support small populations. However, preventative and corrective measures (e.g. corridor protection or animal translocation) can be costly, and limiting the spread of disease in the present is often a greater concern than genetic viability over the longer term (Simberloff et al. 1992). Disease is a particular worry for game mammals because large economic costs may follow disease-mediated die-offs.
Studies of genetic decline and rescue in natural populations could help decide whether these costs and risks should be assumed. Examples of natural rescue to date have relied upon relatively indirect evidence, such as a positive change in population trend coinciding with renewed immigration (Westemeier et al. 1998; Madsen et al. 1999; Vilà et al. 2003). Associations of this kind could have environmental as well as genetic causes. Experimental studies can control for environmental effects and provide replication (reviewed in Tallmon et al. 2004). However, experimental designs may sacrifice realism for control and will usually be limited to species whose life histories differ substantially from those of large mammals, which, for better or worse, garner the lion's share of management attention. Individual, pedigree-based studies of natural populations offer an attractive alternative approach (Marr et al. 2002), which we follow here.
We studied Rocky Mountain bighorn sheep (Ovis canadensis) residing on the National Bison Range (NBR; Montana, USA). The population was established by transplant in 1922. It remained isolated by distance and human development until 1985, when bighorn derived from two outbred herds were introduced. We use historical demographic data and individual genetic and life-history data collected during 1979–2003 to describe the severity of the population size bottleneck, to quantify changes in genetic variation during isolation and to compare the fitness of individuals coexisting in time and space, but differing in level of recent outbreeding. Fitness losses during isolation should be reflected in fitness increases among offspring of the recent immigrants. Finally, we consider whether the effects of admixture were evident at the population level. Our results are most relevant to population structures in which gene flow ceased recently, population isolates are relatively small and, hence, loss of fitness during isolation is a greater concern than is loss of local adaptation due to renewed migration (Hedrick 2005).
2. Material and methods
(a) Study site and population
The NBR study site is an 8000 ha National Wildlife Refuge located in northwestern Montana (47° N, 114° W). The study population was established in February 1922 by transplant of four males and eight females from Banff National Park (Alberta, Canada). Two of the males were apparently offspring of transplanted ewes and all or most ewes should have been carrying fetuses conceived in Banff during the autumn of 1921. There is no further record of natural or artificial immigration until 1985, when five rams were introduced. An additional 10 sheep (three males and seven females) were successfully introduced during 1990–1994. Fourteen of the immigrants originated in herds established by recent transplant from the Sun River (MT) complex of native populations. One female immigrant derived from the Whiskey Basin (WY) complex of native populations.
(b) Field data
All bighorn were individually recognizable from 1979 forward. Tissue collections for genetic analysis were initiated in 1988. Tissue was collected following hand-capture of neonates, by biopsy dart or after chemical immobilization. With the exception of one 2-year-old ram in 1988, all adults living during 1988–2002 were sampled. Population size estimates prior to 1979 were taken from end-of-year totals reported in refuge records. Thereafter, we used exact end-of-year counts made during this study, except in 1986 when we used an exact spring count and 1987 when we averaged the 1986 spring and 1988 end-of-year counts. Annual male and female reproductive success were estimated as the number of paternities assigned to ram i in year j on genetic evidence (below) and the number (0, 1) of ewe i's lambs surviving to age six months in year j, respectively. Survival to half-year intervals was determined by repeated spring and autumn censuses. We estimated female breeding date as the last day of the first observed oestrus, birth weight as the capture weight (kg) of lambs handled within 3 days of birth and gestation as the interval (days) between conception breeding date and birth date. Finally, male consort success, copulatory success and dominance interactions were recorded in autumn behavioural observations as described previously (Hogg 1987). Male dominance rank was assigned using MatMan software (v. 1.0; http://www.noldus.com).
(c) DNA extraction, amplification and typing
Protocols for DNA extraction and PCR reactions are described in Forbes et al. (1995) and Maudet et al. (2004). We typed 16 (AC)n repeat microsatellite loci cloned and characterized in domestic sheep (Ovis aries). The loci and annealing temperatures in degrees Celsius were as follows: OarFCB11 (63), OarFCB266 (63), OarFCB304 (63), MAF33 (54), MAF36 (63), MAF48 (63), MAF65 (63), MAF209 (63), OarFCB128 (63), ADCYAP1 (54), OarAE16 (54), OarCP20 (63), OarFCB20 (54), OarFCB226 (63), OarHH47 (63) and OarHH62 (63) (http://rubens.its.unimelb.edu.au/~jillm/jill.htm). The first nine loci were selected for typing independently of their level of genetic variation. These loci were therefore used to characterize genetic diversity in the 1985 founder descendants.
(d) Demography, population genetics and paternity assignment
Generation time was estimated using data on age-specific female fecundity and survival from 1979 to 2003. We estimated effective population size for the bottleneck period from the reduction in heterozygosity using the relation (Hartl & Clark 1989), where Ho and Ht are observed heterozygosity at founding and just before admixture t generations later, respectively. As a surrogate for Ho, which is unknown, we substituted observed heterozygosity for the 1993 Sheep River (Alberta) population, a native herd within 100 km of Banff (Forbes & Hogg 1999).
We used Cervus (v. 2.0; http://www.helios.bto.ed.ac.uk/evolgen/cervus) to assign paternity to a total of 165 lambs. Likelihoods and critical values for the delta test statistic were computed from year-specific data on allele frequencies, number of candidate fathers (14–24), proportion of candidate fathers genotyped (0.89–1.00) and average fraction of loci genotyped (0.69–0.86). Allele frequencies were calculated using all adults surviving to the breeding season. Rams aged 1.5 years and older were considered potential fathers. We assumed a 0.01 genotype error rate and used 10 000 simulated paternity tests to establish critical values for the delta statistic. Fathers for 148 of the 165 tested lambs were assigned with 95% or higher confidence. Lambs for which paternity was not established with 95% confidence were nonetheless assigned to the male with the greatest likelihood of paternity. These 17 lambs had more candidate fathers (those with positive log-likelihood ratios) that were less outbred (expected fraction of introduced alleles≤0.50). Their inclusion avoids underestimation of paternity by less outbred rams and is conservative with respect to the hypothesis that outbreeding increased male annual reproductive success (ARS).
(e) Crossbreeding analysis
We evaluated the effect of recent outbreeding on components of fitness and fitness-related traits using Lynch's (1991) model of expected phenotype under crossbreeding. This model provides a formal framework for evaluating the phenotypic effects of population admixture and the genetic bases of differentiation between admixed populations. The Lynch model is completely general with respect to the causes of population differentiation. Here, we outline the method in the specific context of two populations (termed migrant and founder), differing as a result of genetic drift occurring primarily in one population (founder). A more technical introduction is provided in the electronic supplementary material. Full details can be found in Lynch (1991) and Lynch & Walsh (1998).
In our application of the model, founder individuals and their pure descendants comprised one source (parental) population (PF). Recent migrants and their pure descendants comprised a second source population (PM). Degree of outbreeding in the Lynch model is measured on two dimensions by the source and hybridity indices. The source index (θS) is a simple transformation of the expected fraction (S) of migrant versus founder alleles present in an individual (θS=2S−1). S varied from 0.0 for an individual carrying only founder-derived alleles to 1.0 for an individual carrying only migrant-derived alleles. S and θS measure the ‘dosage’ of migrant versus founder alleles in an individual. The hybridity index (θH) is a simple transformation of the probability (H) that an individual carried one founder and one migrant allele at a given locus (θH=2H−1). H varied from 0.0 for individuals carrying only founder-derived or only migrant-derived alleles to 1.0 in the case of individuals with one PF and one PM parent (the F1 crossbreeding class). H and θH measure within-locus ‘mixing’ of migrant versus founder alleles and hence the potential for beneficial, dominant alleles originating in one population to mask the deleterious effects of recessive alleles originating in the other (heterosis). Additional genetic variables in the Lynch model measure the potential for individuals in different crossbreeding classes to suffer outbreeding depression from disruption of coadapted gene complexes. These are quadratic terms derived from the source and hybridity indices. We tested for outbreeding depression by including three such variables (designated θS2, θSθH and θH2). These terms comprehensively model the potential in each crossbreeding class for loss (or gain) in favourable coadaptation between pairs of loci. Values for S and H (hence θS, θH and the three quadratic terms) were calculated on Mendelian principles from maternal and paternal pedigree.
We regressed individual trait values on these five genetic variables and a set of non-genetic predictor variables (age, gender, etc.) appropriate to the trait in question. We also tested a ‘persistent environmental effects’ variable indicating presence/absence (1, 0) of potential for trait values to be influenced by more (or less) favourable conditions obtaining in the migrant source population. Such influences could be direct, as in the case of migrants, or indirect, in the form of maternal effects transmitted to the descendants of female migrants. Thus, individuals scored ‘1’ for this variable included all migrants and all descendants of migrants connected to the migrant environment by one or more generation links unbroken on the maternal side (i.e. all offspring of female migrants, all offspring of daughters of female migrants, and so on). Thirty-six percent of measured bighorn (48 of 135) were connected to the migrant environment in this way. Individuals were separated from the migrant environment by an average of 1.02 generations and a range of zero (for migrants) to three generations. We tested genetic terms in the sequence described by Lynch & Walsh (1998), after all significant non-genetic variables were identified and included. The persistent environmental effects variable was tested after all significant genetic variables were identified and included.
For repeated measurement traits (all traits but adult survival), we used generalized linear mixed model regression (GLMM; Breslow & Clayton 1993) to estimate the effect of each predictor variable. Year and individual identity were included as random factors in these models. To test for the effects of outbreeding on age-specific adult survival, we used Cox proportional hazards regression (Therneau & Grambsch 2000). Additional information on regression models is provided in the electronic supplementary material.
(f) Male dyadic dominance status
In addition to a crossbreeding analysis of male dominance status measured as a population rank, we tested for effects of outbreeding on male status measured as a pairwise (dyadic) dominance rank. We extracted all pairwise combinations of rams (dyads) over all years in which one ram (the dominant) had a majority of ‘wins’. One ram in each dyad was selected at random to be the focal (versus reference) ram. Focal rams were assigned a score of 1 if dominant to the reference ram and zero if subordinate. From this list, we sub-sampled all records in which members of the dyad were of the same age or the dominant was younger and neither ram was older than 8 years. We did this to control for age-related advantages in body size and any effect of senescence, respectively. Most ram pairs were represented in this sub-sample by two or more records reflecting their status in different years. Therefore, we randomly selected one record per dyad for a final sample of 126 unique dyads. Finally, we regressed dyad dominance score (0, 1) on the difference between focal and reference rams in both the source (ΔθS) and hybridity (ΔθH) indices. Identity of the focal ram was modelled as a random factor. We repeated the regression analysis 11 times using different random sub-samples (n=126) of one record per dyad.
(g) Recovery of individual fitness
We measured recovery of individual fitness relative to a PM standard. For individual i, recovery was estimated as , where is the expected lifetime reproductive success (LRS) of individual i and , are that of a maximally outbred (PM) and minimally outbred (PF) same-sex individual, respectively. We calculated expected LRS using age-specific values of reproductive success and survival probability as predicted by crossbreeding analysis for that individual's level of outbreeding and sex (table A1 of electronic supplementary material). This index ranged from 0 (no recovery of individual fitness) to 1 (complete recovery of individual fitness).
3. Results
(a) Pre-admixture demography and genetics
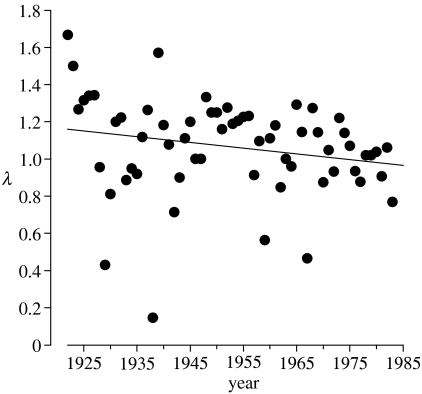
The demographic profile of the pre-admixture NBR population was fairly representative of contemporary bighorn populations in terms of both size and variation in size. Within 8 years of founding, the study population increased to a 1922–2003 maximum of 90 individuals. Population size over the entire pre-admixture period (1922–1985) averaged 41.7 individuals. The standard deviation of log-population size (SDLN) for this period was 0.28. Bighorn population size estimates are often less than 100 individuals (Toweill & Geist 1999), whereas estimates of SDLN compiled for 10 populations of mammals having comparable body mass (50–175 kg) to bighorn averaged 0.25 (Pimm 1991). The average annual population growth rate during the pre-admixture period was slightly positive (geometric mean λ=1.02). However, there was a fairly strong negative trend in population growth rate over this time (figure 1). The estimated annual change in population size near the time of founding was large and positive (), whereas that immediately prior to admixture was slightly negative ().
Figure 1.
Population growth rate (λ) followed a negative trend during the pre-admixture period (1923–1984) (; n=62 years; p=0.10).
Genetic variability prior to admixture was extremely low relative to that reported for other Ovis populations (Özüt 2001). For 20 founder descendants alive in 1985, observed heterozygosity and allelic diversity averaged only 0.44 (±0.07 s.e.) and 2.2 alleles per locus, respectively. More importantly, the distribution of alleles across allele frequency classes matched that expected for a population subjected to strong genetic drift during a recent bottleneck; rare alleles were far less common than would be expected in the absence of a recent bottleneck (figure A1 of electronic supplementary material). Such ‘allele frequency distortion’ occurs because under strong drift most rare alleles are lost altogether, while others drift into higher frequency classes (Cornuet & Luikart 1996).
The demographic and genetic data in combination provide further evidence of a prolonged and severe bottleneck. We estimated a generation time equal to 5.7 years and a bottleneck duration of 11.1 generations (63 years÷5.7 years per generation). Assuming a loss of heterozygosity during 1922–1985 equal to the difference in observed heterozygosity between that reported for the Sheep River surrogate population and the 1985 founder descendants (0.59 versus 0.44), this implies an effective population size of only 18.6 individuals. The reduction in heterozygosity corresponds to an increase of 0.25 in the average inbreeding coefficient.
(b) Individual-level effects of admixture
The beneficial effects of outbreeding on individual phenotype were striking and broadly consistent across traits. Tested traits included male and female ARS, adult survival and seven traits potentially related to ARS. The 135 bighorn contributing measurements represented 18 distinct levels of outbreeding (figure A2 of electronic supplementary material).
(i) Male annual reproductive success
Outbred rams were markedly more successful in reproductive competition than their more inbred contemporaries (table 1; figure 2a). Maximally outbred (PM) rams could expect a 2.6-fold increase in ARS relative to minimally outbred (PF) rams of the same age. In bighorn, male ARS is positively correlated with horn and body size (Coltman et al. 2002), dominance status and time in consort with oestrous females (Hogg & Forbes 1997). However, controlling for the effect of age, greater outbreeding was not associated with higher dominance rank or increased consort success (table 1). Dominance status in the preceding analysis was a population rank inferred from pairwise (dyadic) ranks. Direct analysis of the dyadic ranks (see §2) showed, in contrast, strong positive effects of outbreeding on the probability of dominating same- or older-aged rams. Focal rams in dyads were significantly more likely to be dominant when they carried more migrant alleles (ΔθS>0) and/or had greater hybridity (ΔθH>0) than reference rams (effect of ΔθS: median p=0.001; effect of ΔθH: median p=0.0002; n=11 replicate tests using different random samples of 126 dyads). When rams in a dyad had the same level of outbreeding (ΔθS and ΔθH=0), focal rams were dominant in about one-half the cases as expected (mean predicted probability of dominance=0.54±0.09 s.d.; n=11 replicate tests). However, when the focal ram had, for example, a fraction of migrant alleles that was 0.50 greater than the reference ram (but equal hybridity), the probability of being dominant was much better than even (mean predicted probability of dominance=0.74±0.06 s.d.; n=11 replicate tests). We attribute the failure to detect outbreeding effects on population dominance rank to variation in cohort size. Rams having fewer same-age or older contemporaries may attain relatively high population rank (and consort success), despite below average competence in pairwise contests. The analysis of dyadic rank is the stronger test, because it is not sensitive to variation in the overall strength of the competition.
Table 1.
The estimated effect of outbreeding on annual reproductive success (ARS), adult survival and seven fitness-related traits in males and females. (Coefficients for the source (θS) and hybridity (θH) index give the estimated change in trait value per unit increase in the index. A unit increase in θS corresponds to an increase of 0.50 in the fraction of migrant versus founder alleles. A unit increase in θH corresponds to an increase of 0.50 in the probability that a given locus carried one migrant and one founder allele. p-values≤0.05 and 0.01 are denoted by * and **, respectively. θH was not tested when θS was not significant. See electronic supplementary material for additional detail.)
| trait | θS | θH | na | periodb | modelc | units | increased |
|---|---|---|---|---|---|---|---|
| male | |||||||
| ARS | 0.64** | 0.20 | 327 (57) | 1987 (16) | P | ln(no. lambs conceived) | 2.57 |
| dominance rank | 2.43 | 254 (56) | 1988 (14) | N | %-tiles of rank | — | |
| consort success | 0.46 | 275 (57) | 1988 (13) | P | ln(no. annual consorts) | — | |
| defensive success | 0.92* | 0.54* | 175 (32) | 1988 (13) | B | ln(odds of paternity) | 1.62 |
| coursing success | 0.53* | 0.19 | 275 (57) | 1988 (13) | P | ln(no. annual matings) | 1.86 |
| female | |||||||
| ARS | 0.83** | 0.57** | 254 (61) | 1989 (15) | B | ln(odds of weaning lamb) | 2.17 |
| lamb birth weight | 0.49** | 0.22* | 118 (44) | 1989 (14) | N | kg | 0.23 |
| breeding date | −4.53** | −0.99 | 243 (59) | 1989 (14) | N | days | — |
| gestation | −0.12 | 92 (39) | 1989 (13) | N | days | — | |
| adult survival | 0.69* | 1.03 | 135 (135) | 1988 (16) | Cox | relative risk of death | 0.28 |
Number of measurements (number of individuals).
First year of measurement (number of years with data).
GLMM regression assuming Poisson (P), normal (N) or binomial (B) error structure or Cox model (Cox).
Increase in trait value for PM individuals relative to a PF standard. Estimated as , where and are predicted trait means for these classes. The entry for survival is the fractional increase in lifespan estimated assuming the annual reduction in risk of death per unit increase in θS given in column 1.
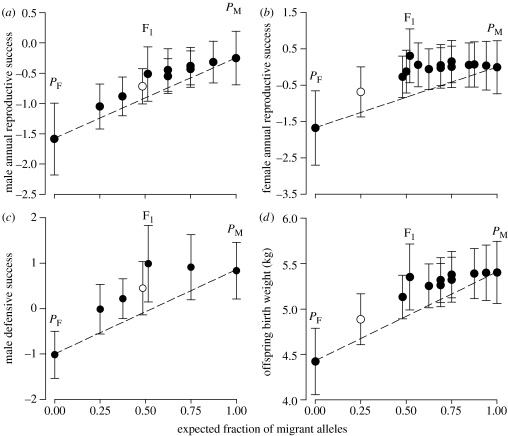
Figure 2.
The estimated effect of outbreeding on annual reproductive success and two fitness-related traits. (a) Male annual reproductive success (ARS) expressed as the natural logarithm of the number of lambs fathered per annum. (b) Female ARS expressed as the log odds of weaning a lamb. (c) Male defensive success expressed as the log odds of paternity given defensive consort with an oestrous ewe. (d) Offspring birth weight relative to the maternal level of outbreeding. Symbols (circles) give mean trait values predicted by crossbreeding analysis for each combination of the source (θS) and hybridity (θH) index observed in the study at fixed values for all non-genetic covariates. Open circles indicate a crossbreeding class not measured for a particular trait. Vertical brackets are 95% confidence intervals for the predicted mean. Outbreeding is measured on the horizontal axis in terms of the expected fraction (S) of migrant alleles carried by an individual. The dashed lines indicate the estimated independent effect of S on the trait value. Values of S=0.0, 0.50 and 1.0 correspond to a source index (θS) of −1.0, 0.0 and 1.0, respectively. Deviations of the predicted means (symbols) from the dotted line reflect the estimated effect of the hybridity index (θH). Positive deviations (symbols above the dashed lines) indicate that migrant alleles tended to be dominant to founder alleles and masked their deleterious effects. Differences in the magnitude of these deviations reflect variation in hybridity among crossbreeding classes. The estimated effect of hybridity was not significant in the case of male ARS (table 1).
More outbred rams also had greater likelihoods of paternity given defensive consort with an oestrous ewe (table 1; figure 2c). The estimated probability of paternity for maximally outbred (PM) rams that defended a female during her entire oestrus (duration of male consort=100%) was 0.70 versus 0.27 for minimally outbred (PF) defending rams. Subordinate ‘coursing’ rams routinely fight defending rams to force breedings with oestrous females (Hogg 1984). Approximately 40% of all lambs may be fathered via the coursing mating tactic (Hogg & Forbes 1997). Thus, it is significant that more outbred rams were also markedly more successful in obtaining copulations via the coursing mating tactic (table 1).
(ii) Female annual reproductive success
Outbreeding was associated with greater annual female reproductive success (table 1; figure 2b). Maximally outbred (PM) females could expect a 2.2-fold average increase in ARS relative to minimally outbred (PF) ewes of the same age. For ewes 5 years of age in an average recruitment year, this represents an increase in the estimated probability of successful reproduction from 0.16 to 0.50. More outbred females also produced markedly larger lambs and had much earlier breeding (hence birthing) dates (table 1; figure 2d). For example, lambs born to PM versus PF mothers were 0.98 kg heavier at birth and PM females entered oestrus 9.1 days before PF females. These differences represent a 23% increase in birth weight and an advance in breeding date equal to ca 30% of the typical range in annual breeding dates. Because outbred ewes tended to have shorter gestations (table 1), increased lamb birth weight meant similarly large increases in prenatal growth rate. Both early birth and increased offspring size are associated with increased female reproductive success in bighorn (Festa-Bianchet 1988; Hogg et al. 1992; Festa-Bianchet et al. 1997).
(iii) Survival
Finally, adult survival increased substantially with outbreeding. We detected no effect of hybridity on survival. However, a unit increase in θS (corresponding to an increase of 0.50 in the fraction of migrant alleles) reduced the age-specific probability of death by a factor of 0.69 (table 1). Although age-specific probabilities of mortality were generally small to begin with, such reduction in risk applied annually with significant cumulative effect. Thus, for example, the average predicted lifespan (given survival to 1 year) was fully 2.1 years longer for maximally outbred (PM) versus minimally outbred (PF) individuals of either sex.
There was no evidence for any trait of outbreeding depression. Coefficients for genetic terms designed to test for outbreeding depression were never significantly different from zero (mean and median p-values across all traits were 0.50 and 0.55, respectively). Similarly, we found no evidence of persistent environmental effects attributable to more (or less) favourable conditions obtaining in the migrant source population. Coefficients for the persistent environmental effects variable were never significantly different from zero (mean and median p-values across all traits were 0.65 and 0.69, respectively).
(c) Genetic and demographic recovery
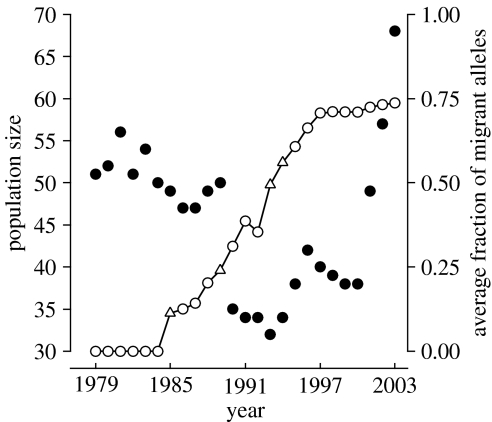
Genetic variation at marker loci was rapidly restored by admixture. Observed heterozygosity increased from 0.50 (±0.05 s.e.) in 1984 just prior to admixture to 0.67 (±0.06 s.e.) in 2003 (n=8 loci typed in all individuals). Allelic diversity increased stepwise upon arrival of each migrant group from 2.4 alleles per locus in 1984 to 5.4 alleles per locus by 2003. These values are comparable to those reported for large, outbred bighorn populations (Forbes & Hogg 1999). Average outbreeding, as measured by the expected fraction of migrant alleles (S), increased from zero in the immediate pre-admixture population (1984) to 0.74 in 2003 (figure 3). Thus, approximately 74% of the gene pool in 2003 derived from migrants; 26% derived from the 1922 founders and presumably still contributed elevated frequencies of disadvantageous alleles.
Figure 3.
Total number of native bighorn (residents minus migrants; filled symbols) in relation to the average fraction of migrant alleles carried by resident bighorn (natives plus migrants; open symbols) during 1979–2003. Reversal of a multi-year decline in population size coincided with the year (1993) in which the average resident carried a majority of migrant alleles. Because migrants were excluded from annual population number (filled symbols, left axis), population changes are entirely due to reproduction and mortality within the NBR population. Years in which migrants arrived are indicated by open triangles (left to right, migrants were five males, one female, three males/three females and three females). Ten rams removed at various ages for transplant prior to 1985 were included in the population count by (conservatively) assigning each a lifespan of 10.5 years.
Restoration of genetic variation was associated with demographic recovery at both the individual and population levels. For the 58 adult members of the 2003 population, the average estimated rebound in individual fitness (relative to a PM standard) was 81%. At the population level, increases in average outbreeding were associated with reversal of a multi-year decline in population size. Onset of the reversal coincided with the year in which the average individual in the NBR population carried a majority of introduced alleles (figure 3). In contrast to the pre-admixture period when growth rate followed a negative trend (figure 1), annual population growth rate in the post-admixture period (1985–2003) followed a fairly strong positive trend (; n=18 years; p=0.11).
4. Discussion
(a) Genetic interpretation of outbreeding effects
The principal result of this study was the large net positive effect of outbreeding on total fitness realized in a fully competitive natural setting. We detected favourable effects of outbreeding in both sexes and for nearly all tested characters. These included major components of fitness (ARS and adult survival) and five functionally distinct morphological, behavioural and life-history traits (dyadic dominance rank, defensive success, coursing success, offspring birth weight and breeding date).
There was substantial between-trait consistency in the outbreeding effects estimated by crossbreeding analysis. The effect of the source index (dosage of migrant alleles) was invariably in the direction of improved performance with increased dosage, tended to be large and was generally significant. These results suggest that disadvantageous alleles were more common at differentiated loci in the founder (NBR) population. The effect of the hybridity index was typically substantial, statistically significant in key cases (e.g. female ARS and offspring birth weight) and in the direction of improved performance for individuals with greater within-locus mixing of migrant versus founder alleles (greater hybridity). These results suggest that recessive or partially recessive deleterious alleles were more frequent in the founder population, but their effects were more often masked by beneficial dominant alleles from the migrant population. We found no evidence of outbreeding depression for any trait.
These patterns in the crossbreeding results, the marked reduction in allelic diversity and strong allele frequency distortion among founder descendants are all consistent with founder versus migrant population differentiation due primarily to increased frequencies of recessive deleterious alleles in the NBR population caused by genetic drift during isolation. The crossbreeding results cannot be explained as environmental change that differentially increased resource availability for more outbred individuals. All analyses were restricted to post admixture periods when individuals with very different levels of outbreeding coexisted in identical environments. Moreover, individual values for repeated measures traits were always evaluated with respect to the annual mean trait value (via the year random factor). This increased power to control for unmeasured environmental change is one advantage of an individual pedigree-based evaluation of genetic rescue.
An alternative genetic explanation is that local adaptation in the founder (Banff) and migrant (Sun River) source herds produced Banff genotypes that were less well adapted to the NBR environment. Evidence against this explanation includes: the proximity of Banff and Sun River in the same biogeographical region; lack of genetic evidence in bighorn for isolation by distance within geographical regions over distances up to about 1000 km (Forbes & Hogg 1999); demographic evidence of high levels of regional connectivity (rapid spread of epizootics) (Andryk & Irby 1986); the absence in this study of evidence for outbreeding depression, a crossbreeding effect typically associated with adaptive divergence (Lynch 1995). Although a role for local adaptation cannot be ruled out on this evidence (Lynch 1995), the genetic data and crossbreeding results are more consistently and simply explained as consequences of genetic drift. Divergence of source populations by local adaptation could be applied in the same way as an alternative explanation for other examples of genetic rescue of natural populations (Westemeier et al. 1998; Madsen et al. 1999; Vilà et al. 2003).
(b) Outbreeding effects and inference of fitness loss
An under-emphasized and perhaps under-appreciated limitation of admixture studies is that outbreeding effects measured on coexisting individuals do not necessarily estimate the magnitude of fitness loss during isolation or, conversely, fitness gain in a fully recovered population. This is most easily appreciated for male ARS. Maximally outbred (PM) rams had 2.6 times the success of minimally outbred (PF) rams in the admixed NBR population. However, even this large outbreeding effect will not increase the population mean male ARS provided that two conditions hold. First, prior inbreeding was not severe enough to cause dysfunction in non-competitive aspects of male reproduction (such that some females did not conceive because of male reproductive failure). Second, success in male–male reproductive competition depends upon relative rather than absolute values of the physical traits influencing ARS. Under these conditions, more versus less outbred males will simply be sorted into opposite ends of a roughly constant range of variation in ARS centred about a roughly constant mean. Outbreeding effects will then be closely related to the normal variance in male ARS (large) and not at all to expected gain in the population mean trait value (zero). Both conditions probably applied in our study. This does not mean that more versus less outbred NBR males were similar in fitness-related phenotype, but only that such differences would not be reflected in ARS except in a mixed-pedigree competition. Moreover, outbreeding effects on male ARS were still fundamentally important in accelerating the spread of immigrant alleles.
Female reproduction in bighorn does not obviously involve strong competition of the type experienced by males. Moreover, phenotypic differences associated with the increased ARS of outbred females (earlier breeding date, larger offspring birth weight and greater offspring growth rate) should confer some advantage independently of any such competition. Nonetheless, outbreeding effects on female ARS were surprisingly large. Earlier breeding (and birthing) by more outbred ewes and increased offspring growth rate could introduce an element of competition based on relative ability if, for example, predators more frequently attack less developmentally advanced lambs in a group because they are easier to capture.
In general, strong competition (e.g. for mates) tends to increase the variance of downstream traits (e.g. ARS). When success in competition is determined by relative ability, outbreeding effects measured on coexisting individuals will be scaled by the trait variance rather than by change in the population mean trait value. Even so, large outbreeding effects imply strong sorting according to competitive ability, which in turn implies significant phenotypic differences attributable to prior inbreeding. Outbreeding effects on traits more probably shaped by absolute values of underlying characteristics (adult survival and offspring birth weight) suggest that post-admixture improvements in population mean trait values at the NBR were substantial and of the order of 20–30%.
(c) Population rescue
Tallmon et al. (2004) suggest that genetic rescue occurs when population fitness increases more than can be explained by the direct demographic contributions of migrants. They further suggest that the ideal criterion for rescue is an increased population growth rate sustained over many generations. But natural populations typically inhabit variable environments and experience large variation in size (Pimm 1991). Population growth coincident with immigration could be due to improving ecological conditions. Conversely, an immediate or sustained response could be prevented by deteriorating conditions. Separating genetic and environmental effects on population response will usually be difficult in non-experimental settings. In this study, for example, we documented trends in population growth rate consistent with genetic decline and rescue, but cannot exclude environmental causes for these population-level patterns.
The individual-based crossbreeding results were more decisive. Here, substantial separation of genetic from environmental effects was possible because more versus less outbred individuals coexisted in time and space. Repeated measurements of individuals provided additional leverage in controlling for non-genetic sources of individual variation and unmeasured annual variation in environmental conditions. Genetic rescue defined by recovery of individual adaptation may be a more practical management objective than genetic rescue defined by sustained population growth. Although monitoring trends in individual adaptation will rarely be feasible, it is still important to make a clear distinction between the unambiguous management objective (recovery of individual adaptation) and an indirect measure of that recovery (population response). Otherwise, management for gene flow may, in critical cases, be falsely judged ineffective and prematurely abandoned.
The large average rebound to date in relative individual fitness (81%) combined with evidence for pre-admixture reductions in absolute trait values on the order of 20% suggest that the probability of NBR population persistence has been significantly increased by migration. Whether post-admixture environmental trends will permit that improvement to be manifested in sustained population growth is an open question.
(d) Management implications
Our findings provide strong empirical support for the view that substantial loss of fitness can develop rapidly in the absence of gene flow at population sizes currently typical of many large mammal species (Lynch et al. 1995). Such loss could be accelerated in population isolates also subject to trophy harvest. In extreme cases, selective harvest of large males can actually favour alleles with deleterious effects on male size (Coltman et al. 2003) and correlated traits (Coltman et al. 2005). More subtly, less intense selective harvest might increase fixation of such alleles by making selection coefficients less negative (smaller absolute value). Genetic drift becomes relatively more influential in determining allele frequencies when selection coefficients decline in absolute value (Lynch et al. 1995).
In agreement with other studies of renewed migration (reviewed in Tallmon et al. 2004), our results suggest that genetic rescue can be surprisingly rapid. We have extended previous work on genetic rescue of natural populations by documenting large outbreeding effects on total individual fitness in both sexes and by using variation in individual pedigree measured over multiple generations to: (i) control for unmeasured environmental influences, and (ii) test for the possibility of compensatory outbreeding depression arising with increasing genetic mixture. Compelling evidence of significant risk to insular populations and of rapid recovery following renewed migration combine to provide managers with a strong practical justification for applying genetic as well as demographic, epidemiological and economic principles in actions affecting the ecological connectivity of large-mammal populations.
Acknowledgments
We are indebted to Don Jenni and Bart O'Gara for initiating study of the NBR bighorn population. We thank the US Fish and Wildlife Service for cooperation and assistance at the NBR study site. Fred Allendorf, David Coltman, Marco Festa-Bianchet and David Tallmon provided helpful comments on the manuscript. Funding was provided by The Charles Engelhard Foundation, Eppley Foundation for Research, Juniper Hill, Inc., National Geographic Society, Tim and Karen Hixon Foundation, US Forest Service, Bureau of Land Management, Turner Foundation, Boone and Crockett Club, Foundation for North American Wild Sheep, Campfire Conservation Fund, James H. Woods Foundation and Max McGraw Wildlife Foundation. Special thanks go to the many field assistants who worked on the study.
Footnotes
Present address: Division of Medical Genetics, University of Washington, Seattle, WA 98195, USA.
Supplementary Material
Demonstration of allele frequency distortion and additional detail on regression model structure and interpretation.
References
- Andryk T.A, Irby L.R. Population characteristics and habitat use by mountain sheep prior to a pneumonia die-off. Bienn. Symp. North. Wild Sheep Goat Counc. 1986;5:272–289. [Google Scholar]
- Breslow N.E, Clayton D.G. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993;88:9–22. [Google Scholar]
- Coltman D.W, Festa-Bianchet M, Jorgenson J.T, Strobeck C. Age-dependent sexual selection in bighorn rams. Proc. R. Soc. B. 2002;269:165–172. doi: 10.1098/rspb.2001.1851. doi:10.1098/rspb.2001.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman D.W, O'Donoghue P.O, Jorgenson J.T, Hogg J.T, Strobeck C. Undesirable evolutionary consequences of trophy hunting. Nature. 2003;426:655–658. doi: 10.1038/nature02177. doi:10.1038/nature02177 [DOI] [PubMed] [Google Scholar]
- Coltman D.W, O'Donoghue P.O, Jorgenson J.T, Hogg J.T, Festa-Bianchet M. Selection and genetic (co)variance in bighorn sheep. Evolution. 2005;59:1372–1382. [PubMed] [Google Scholar]
- Cornuet J.M, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa-Bianchet M. Birthdate and survival in bighorn lambs (Ovis canadensis) J. Zool. 1988;214:653–661. [Google Scholar]
- Festa-Bianchet M, Jorgenson J.T, Berube C.H, Portier C, Wishart W.D. Body mass and survival of bighorn sheep. Can. J. Zool. 1997;75:1372–1379. [Google Scholar]
- Forbes S.H, Hogg J.T. Assessing population structure at high levels of differentiation: microsatellite comparisons of bighorn sheep and large carnivores. Anim. Conserv. 1999;2:223–233. doi:10.1017/S1367943099000554 [Google Scholar]
- Forbes S.H, Hogg J.T, Buchanan F.C, Crawford A.M, Allendorf F.W. Microsatellite evolution in congeneric mammals: domestic and bighorn sheep. Mol. Biol. Evol. 1995;12:1106–1113. doi: 10.1093/oxfordjournals.molbev.a040284. [DOI] [PubMed] [Google Scholar]
- Gilpin M.E, Soulé M.E. Minimum viable populations: processes of species extinction. In: Soulé M.E, editor. Conservation biology: the science of scarcity and diversity. Sinauer Associates; Sunderland, MA: 1986. pp. 19–34. [Google Scholar]
- Hartl D.L, Clark A.G. Sinauer Associates; Sunderland, MA: 1989. Principles of population genetics. [Google Scholar]
- Hedrick P. ‘Genetic restoration’: a more comprehensive perspective than ‘genetic rescue’. Trends Ecol. Evol. 2005;20:109. doi: 10.1016/j.tree.2005.01.006. doi:10.1016/j.tree.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Hogg J.T. Mating in bighorn sheep: multiple creative male strategies. Science. 1984;225:526–529. doi: 10.1126/science.6539948. [DOI] [PubMed] [Google Scholar]
- Hogg J.T. Intrasexual competition and mate choice in Rocky Mountain bighorn sheep. Ethology. 1987;75:119–144. [Google Scholar]
- Hogg J.T, Forbes S.H. Mating in bighorn sheep: frequent male reproduction via a high-risk ‘unconventional’ tactic. Behav. Ecol. Sociobiol. 1997;41:33–48. doi:10.1007/s002650050361 [Google Scholar]
- Hogg J.T, Hass C.C, Jenni D.A. Sex-biased maternal expenditure in Rocky Mountain bighorn sheep. Behav. Ecol. Sociobiol. 1992;31:243–251. doi:10.1007/BF00171679 [Google Scholar]
- Jenkins M. Prospects for biodiversity. Science. 2003;302:1175–1177. doi: 10.1126/science.1088666. doi:10.1126/science.1088666 [DOI] [PubMed] [Google Scholar]
- Lynch M. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution. 1991;45:622–629. doi: 10.1111/j.1558-5646.1991.tb04333.x. [DOI] [PubMed] [Google Scholar]
- Lynch M. A quantitative-genetic perspective on conservation issues. In: Avise J.C, editor. Conservation genetics: case histories from nature. Chapman & Hall; New York, NY: 1995. pp. 471–501. [Google Scholar]
- Lynch M, Walsh B. Sinauer Associates; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am. Nat. 1995;146:489–518. doi:10.1086/285812 [Google Scholar]
- Madsen T, Shine R, Olsson M, Wittzell H. Restoration of an inbred adder population. Nature. 1999;402:34–35. doi:10.1038/46941 [Google Scholar]
- Marr A.B, Keller L.F, Arcese P. Heterosis and outbreeding depression in descendants of natural immigrants to an inbred population of song sparrows (Melospiza melodia) Evolution. 2002;56:131–142. doi: 10.1111/j.0014-3820.2002.tb00855.x. [DOI] [PubMed] [Google Scholar]
- Maudet C, et al. A standard set of polymorphic microsatellites for threatened mountain ungulates (Caprini, Artiodactyla) Mol. Ecol. 2004;4:49–55. doi:10.1046/j.1471-8286.2003.00563.x [Google Scholar]
- Özüt, D. 2001 Conservation genetics of Anatolian mouflon (Ovis gmelinii anatolica, Valenciennes, 1858). M.Sc. Middle East Technical University, Ankara, Turkey.
- Pimm S. University of Chicago Press; Chicago, IL: 1991. The balance of nature? Ecological issues in the conservation of species and communities. [Google Scholar]
- Simberloff D, Farr J.A, Cox J, Mehlman D.W. Movement corridors—conservation bargains or poor investments. Conserv. Biol. 1992;6:493–504. doi:10.1046/j.1523-1739.1992.06040493.x [Google Scholar]
- Tallmon D.A, Luikart G, Waples R.S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. doi:10.1016/j.tree.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Therneau T, Grambsch P. Springer; New York, NY: 2000. Modeling survival data. [Google Scholar]
- Toweill D, Geist V. Boone and Crockett Club and Foundation for North American Wild Sheep; Missoula, MT: 1999. Return of royalty: wild sheep of North America. [Google Scholar]
- Vilà C, et al. Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc. R. Soc. B. 2003;270:91–97. doi: 10.1098/rspb.2002.2184. doi:10.1098/rspb.2002.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westemeier R, Brawn J, Simpson S, Esker T, Jansen R, Walk J, Kershner E, Bouzat J, Paige K. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. doi:10.1126/science.282.5394.1695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demonstration of allele frequency distortion and additional detail on regression model structure and interpretation.