Abstract
Recent genome analyses revealed intriguing correlations between variables characterizing the functioning of a gene, such as expression level (EL), connectivity of genetic and protein–protein interaction networks, and knockout effect, and variables describing gene evolution, such as sequence evolution rate (ER) and propensity for gene loss. Typically, variables within each of these classes are positively correlated, e.g. products of highly expressed genes also have a propensity to be involved in many protein–protein interactions, whereas variables between classes are negatively correlated, e.g. highly expressed genes, on average, evolve slower than weakly expressed genes. Here, we describe principal component (PC) analysis of seven genome-related variables and propose biological interpretations for the first three PCs. The first PC reflects a gene's ‘importance’, or the ‘status’ of a gene in the genomic community, with positive contributions from knockout lethality, EL, number of protein–protein interaction partners and the number of paralogues, and negative contributions from sequence ER and gene loss propensity. The next two PCs define a plane that seems to reflect the functional and evolutionary plasticity of a gene. Specifically, PC2 can be interpreted as a gene's ‘adaptability’ whereby genes with high adaptability readily duplicate, have many genetic interaction partners and tend to be non-essential. PC3 also might reflect the role of a gene in organismal adaptation albeit with a negative rather than a positive contribution of genetic interactions; we provisionally designate this PC ‘reactivity’. The interpretation of PC2 and PC3 as measures of a gene's plasticity is compatible with the observation that genes with high values of these PCs tend to be expressed in a condition- or tissue-specific manner. Functional classes of genes substantially vary in status, adaptability and reactivity, with the highest status characteristic of the translation system and cytoskeletal proteins, highest adaptability seen in cellular processes and signalling genes, and top reactivity characteristic of metabolic enzymes.
Keywords: gene expression, gene dispensability, protein–protein interaction, sequence evolution rate, gene loss, principal component analysis
1. Introduction
The age of genomics is, arguably, succeeded by an era of systems biology. Although systems biology defies exact definitions, it is all about connections between different parts and characteristics of biological systems at all levels (Ge et al. 2003; Provart & McCourt 2004; Herbeck & Wall 2005; Medina 2005; Pennisi 2005). The main types of data produced by genomics are nucleotide and inferred protein sequences. Comparative analysis of these sequences yields the values of variables characterizing genome evolution, such as sequence evolution rate (ER) and propensity for gene loss (PGL). In the age of systems biology, a different kind of genome-wide information is becoming increasingly available, such as gene expression level (EL), protein–protein interactions, regulatory network structure and the effect of gene knockout on the organism's fitness. Collectively, these may be considered phenotypic variables.
Many large-scale studies examined the connections between evolutionary and phenotypic variables on the premise that phenotypic characteristics of a gene determine the selective constraints, and forces acting on it during evolution and, accordingly, affect evolutionary variables. Nearly 30 years ago, Wilson et al. (1977) proposed, on general, theoretical grounds, the ‘knockout-rate hypothesis’, i.e. that a negative correlation should exist between the severity of a gene knockout effect and sequence ER such that essential genes are predicted to evolve slowly. Once genome sequences and genome-wide gene knockout data became available, this conjecture was tested in numerous empirical studies, some of which reported the predicted connection whereas other failed to do so; the outcome apparently depended on methods of measuring a gene's dispensability and the organisms involved (Hurst & Smith 1999; Hirsh & Fraser 2001; Jordan et al. 2002; Pal et al. 2003). Two recent studies that examined close-range evolutionary rates approximating the instantaneous rate and employed advanced statistical techniques seem to settle the issue, at least for yeasts of the genus Saccharomyces, by convincingly demonstrating the reality of the negative correlation between rate and dispensability, although the magnitude of the effect is not overwhelming (Wall et al. 2005; Zhang & He 2005).
The majority of these and similar studies examined pairwise relationships among genome-related variables. This way, several highly reliable connections were revealed, such as the negative correlation between a gene's EL and sequence ER (Pal et al. 2001; Krylov et al. 2003), the perhaps related negative correlation between a gene's connectivity in co-expression networks and ER (Jordan et al. 2004), and the positive correlation between sequence ER and PGL (Krylov et al. 2003). Also, a provocative link has been reported to exist between a gene's centrality in protein–protein interaction networks and the knockout effect: the hubs of the network are significantly enriched for essential genes (Jeong et al. 2001). However, some, if not most, of these and other connections between genome-related variables remain controversial. In particular, the relevance of the link between centrality and knockout effect have been suggested as being explained by hidden biases in the analysed data; examination of a supposedly unbiased dataset yielded a marginal correlation at best (Coulomb et al. 2005; Koonin 2005). Similarly, it remains unclear whether or not a significant link exists between a gene's connectivity in protein–protein interaction networks and its evolutionary rate. Although the predictable negative correlation between these variables has been reported (Fraser et al. 2002, 2003), subsequent re-analysis suggested that it held only for the most highly connected proteins, the network hubs (Jordan et al. 2003), whereas another study maintained that the connection was an artefact caused by the effect of protein abundance (Bloom & Adami 2003). More subtle and potentially interesting effects also have been reported, such as the apparent dramatic difference in the strength of the connection with evolutionary rate for the intramodule and intermodule hubs of the yeast protein–protein interaction network (Fraser 2005). The work of Fraser demonstrated that, while intramodule hubs were, on average, substantially more conserved in evolution than non-hubs, intermodule hubs showed only very modest deceleration of evolution. As a generalization, a direct dependence between a gene's ‘complexity’ and evolutionary conservation has been proposed: genes involved in complex processes and numerous interactions seem to be more conserved in evolution than less connected genes (Aris-Brosou 2005).
Most of the correlations revealed in the emerging web of links between genome-related variables are relatively weak, even if statistically significant. This suggests that these variables encompass non-overlapping information; indeed, the independence of the contributions of gene dispensability and EL to the evolutionary rate of yeast genes has been recently demonstrated (Ge et al. 2003; Provart & McCourt 2004; Herbeck & Wall 2005; Medina 2005; Pennisi 2005; Wall et al. 2005). It appears, therefore, that combined analysis of all these variables (and others, still undefined ones) is required for better understanding of the behaviour of the ‘gene community’. Several studies attempted to examine more than one pair of variables at a time by using partial correlations (Bloom & Adami 2003; Rocha & Danchin 2004). However, to uncover patterns in the web of links, true multivariate analysis seems to be required.
Here, we present principal component (PC) analysis of seven genome-related variables and propose biological interpretations for the first three PCs. The first PC seems to reflect different aspects of the intuitive notion of a gene's ‘importance’, or the ‘status’ of a gene in the genomic community. The second and third PCs may be interpreted as reflecting different aspects of a gene's evolutionary and functional plasticity.
2. Material and methods
(a) The dataset
Families of orthologues were from the dataset of clusters of eukaryotic orthologous groups of genes (KOGs; March 2003) including seven species (Arabidopsis thaliana, Encephalitozoon cuniculi, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens; Tatusov et al. 2003; Koonin et al. 2004). Additionally, proteins from eight species (Oryza sativa, Dictyostelium discoideum, Neurospora crassa, Magnaporthe grisea, Candida albicans, Caenorhabditis briggsae, Ciona intestinalis and Mus musculus) were added to existing KOGs using the Kognitor method (Tatusov et al. 2003). Index orthologues (i.e. one representative protein per organism, with the greatest similarity to the orthologues from other organisms) were identified in each KOG (Krylov et al. 2003).
On many occasions, some of the analysed data were missing for a given KOG, first, because of the lack of the relevant experimental data and, second, due to the patchy distribution of genes from different species among KOGs. Of the 10 058 KOGs, the full complement of the 38 original variables was available for only 23 KOGs. We employed two complementary strategies to expand the set of KOGs available for analysis. First, we combined data of the same nature into aggregate variables as described later, ending up with seven variables. Second, we allowed for KOGs that had, at most, a certain number of missing values. The missing values were then filled-in by the mean values of the corresponding variables. With these seven variables, 1482 KOGs had complete data and 4124 KOGs had at most one missing value. The results in this work were all obtained using the 4124-KOGs set, but we obtained qualitatively very similar results for the 1482-KOGs dataset (supplement 5 of the electronic supplementary material).
(b) Analysis of evolutionary and phenotypic variables
The PGL during evolution (Krylov et al. 2003) was attributed to each KOG on the basis of the phyletic pattern and the presumed species tree of the original seven eukaryotic species of the KOG database (figure 4aS of the electronic supplementary material). Here, we defined the PGL using a probabilistic evolutionary model based on the Dollo principle (allows for a single origin and multiple losses during evolution of any KOG) and accounting for branch-specific variability. The probability of KOG k to be lost along a branch of length Δt is assumed to be , where ϕt is the branch-specific gene loss propensity and θk is the PGL of KOG k. In order to estimate the values of θk and ϕt, we employed an expectation–maximization algorithm (see electronic supplementary material for details). The average number of paralogs (NP) in each KOG was computed from the original seven-species KOGs dataset.
For evolutionary rate estimation, alignments of index orthologues within each KOG were obtained using the MUSCLE program (Edgar 2004); evolutionary distances between all proteins were computed using PAML program (Yang 1997), with the Jones–Thornton–Taylor (JTT) substitution model adjusted to observed frequencies and the α parameter of the Γ-distribution set equal to 1.0. The minimum distance between proteins from two basal subclades in each set (figure 4bS of the electronic supplementary material) was taken to represent the divergence within the clade; the distance between orthologues was used as a measure of evolutionary rate. The distances within each clade were normalized by the median distance for this clade, resulting in a relative rate estimate; relative rates for a given KOG were averaged between different clades to provide a KOG-specific estimate of evolutionary rate.
The gene expression data for yeast, Drosophila and human were downloaded from the UCSC table browser (http://mgc.ucsc.edu/cgi-bin/hgTables?command=start; table 4S of the electronic supplementary material). Expression scores for specific probes were matched with genes using the tables available at USCS; gene sequences were identified with KOG proteins using BLAST (Altschul et al. 1997). The highest detected level of expression (among all available experiments) was taken for each gene; a KOG was represented by the median expression of its paralogues in each organism. The logarithms of the ELs for each organism were standardized (brought to zero mean and unit variance), and the maximal value among the three species was taken to yield a single EL per KOG. Skewness coefficients (Ehrenfeld & Littauer 1964) were computed for the expression profiles.
The protein and gene interaction data for yeast, Drosophila and C. elegans were downloaded from the GRID web site (http://biodata.mshri.on.ca/yeast_grid/files/Full_Data_Files/interactions.txt, http://biodata.mshri.on.ca/fly_grid/files/Full_Data_Files/interactions.txt and http://biodata.mshri.on.ca/worm_grid/files/Full_Data_Files/interactions.txt). The number of genetic and physical interaction partners was retrieved for each protein; each KOG was represented by the logarithm of the median value among all paralogues. The logarithms of the genetic and physical interaction partners for each organism were standardized, and the maximum value among the species was taken to yield a single number per KOG.
Gene disruption data for yeast were downloaded from the MIPS FTP site (ftp://ftpmips.gsf.de/yeast/catalogues/gene_disruption/gene_disruption_data_06102004); the list contained 1016 genes with a lethal knockout effect. If disruption of any of the paralogues within a KOG was lethal, the KOG was assigned a value of 1, otherwise it was assigned the value of 0. RNAi gene knockout data for C. elegans were taken from Kamath et al. (2003). Each gene was assigned a value of 1 if it had any level of embryonic lethality effect, and a value of 0 otherwise. The knockout lethality data for yeast and worm orthologues were combined by considering a KOG essential if at least one of its members was lethal upon knockout in either species.
(c) Robustness of aggregate variables
In order to assess the robustness of the results obtained with the aggregate variables, we examined the effects of several modifications to the procedures used to derive these variables. These include replacing PGL by the raw number of loss events, taking the KOG's EL as the maximum among paralogues (instead of the median), taking the KOG's number of interaction partners (both physical and genetic) as the maximum among paralogues (instead of median), using a refined (non-binary) version of knockout lethality and excluding from the analysis genes that were used as baits in yeast synthetic lethality experiments and therefore might bias the correlation structure. Neither of these modifications had any qualitative effect on the results of principal component analysis (PCA; supplement 6 of the electronic supplementary material).
(d) Dimensionality reduction by PCA
PCA is heavily affected by outliers (Koren & Carmel 2004). Thus, after standardizing all seven variables, extreme data points with expectation less than 1 (under the assumption of normality) were removed. This procedure reduced the original dataset of 4124 KOGs to 3912 KOGs. The data were re-standardized after the removal of outliers, and PCA was performed. The fractional eigenvalues of the correlation matrix, as well as the first eigenvectors (PCs) are given in figures 1 and 2, and in supplement 1 of the electronic supplementary material.
Figure 1.
Fraction of the total variance captured by each of the seven PCs.
Figure 2.
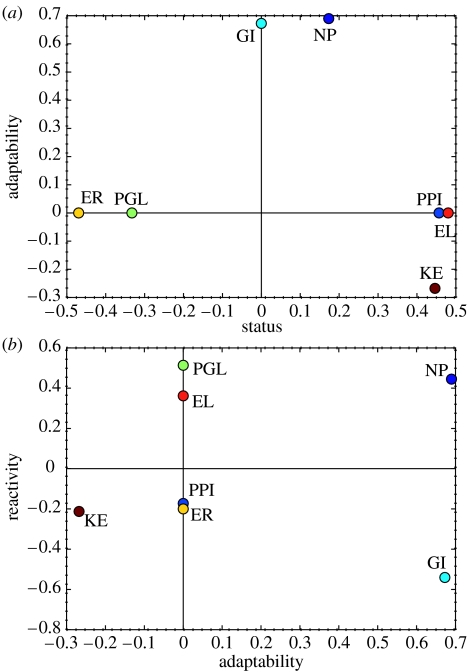
Contributions of the quantitative genome-related measures to the first three principal components (loadings plot). (a) First (PC1, horizontal axis) and second (PC2, vertical axis) principal components. (b) Second (PC2, horizontal axis) and third (PC3, vertical axis) principal components. Designation of the variables: EL, expression level; ER, evolutionary rate; GI, number of genetic interactions; KE, lethal effect of gene knockout; NP, number of paralogs; PGL, propensity for gene loss; PPI, number of physical protein–protein interaction partners.
3. Results and discussion
(a) The correlation structure
We collected or calculated the values of seven genome-related variables, namely, fitness effect of gene knockout experiments (KE), EL, number of genetic interactions between genes (GI), number of physical interactions between gene products (PPI), NP, sequence ER and PGL for 3912 clusters of orthologues eukaryotic genes (KOGs; Tatusov et al. 2003). The KOGs are a natural framework for this type of analysis because they readily allow for estimation of ER and PGL, and also because data obtained on different model systems can be combined through the knowledge of orthologues relationships, thus increasing the number of genes amenable to analysis (see §2 and electronic supplementary material for details; the values of the seven variables for each KOG are given in the electronic supplementary material).
Of the analysed variables, five characterize an organism's phenotype (KE, EL, GI, PPI, NP) and two reflect aspects of evolution (ER, PGL). Examination of the pairwise correlations between these variables reveals a clear-cut pattern: the phenotypic and evolutionary variables form two distinct classes such that variables within a class tend to be positively correlated whereas variables of different classes are negatively correlated, with most of the correlations being statistically significant (table 1). Inverse trends are detected only for GI, namely, a weak positive GI–ER correlation and weak negative GI–EL and GI–KE correlations. Since the majority of the GI data comes from synthetic lethal studies, genes with a non-zero number of GI are likely to be non-essential, which seems to explain the deviation from the general pattern. It is also of note that NP behaves in full concordance with the rest of the phenotypic variables although, a priori, there might be some ambiguity as to whether NP is to be classified as phenotypic or evolutionary.
Table 1.
The correlations between the seven genomic variables. (Asterisks denote the correlations that are significantly different from zero, p<0.05.)
| NP | PPI | GI | PGL | ER | EL | KE | |
|---|---|---|---|---|---|---|---|
| NP | — | ||||||
| PPI | 0.057* | — | |||||
| GI | 0.060* | 0.034* | — | ||||
| PGL | 0.000 | −0.125* | −0.019 | — | |||
| ER | −0.070* | −0.200* | 0.034* | 0.141* | — | ||
| EL | 0.129* | 0.199* | −0.050* | −0.099* | −0.277* | — | |
| KE | 0.027 | 0.234* | −0.048* | −0.181* | −0.155* | 0.188* | — |
Importantly, all the previously established positive correlations within the phenotypic and evolutionary classes of variables and the negative correlations between the variables of different classes held true for the analysed dataset (table 1). The only significant difference from the previous results, apart from the addition of new variables, was that, in the earlier work (Krylov et al. 2003, p. 2671), we failed to detect a significant negative correlation between ER and KE (although a marginal trend in this direction has been seen), whereas in the present work, such a correlation, weak but significant, has been detected (table 1). It appears most likely that the difference is due to the greater sensitivity of the present analysis, thanks to an expanded dataset included in the analysis and the more sophisticated procedure employed for the estimation of ER. Also, similarly to the previous studies, all observed correlations were weak to moderate although most of them reached statistical significance, thanks to the large number of data points analysed (table 1). This persistent pattern of weak (even if significant) correlations emphasizes the need for multivariate analysis in order to elucidate the actual nature of the interplay between the phenotypic and evolutionary variables.
An issue of potential concern with respect to the relatively weak correlations described here and elsewhere is the potential effect of the procedures used to derive the aggregate variables (see §2). To eliminate potential artefacts caused by the specifics of these procedures, we investigated separately the effects of several alternative approaches, e.g. transforming KE from a binary to a continuous variable or replacing the median over paralogues with the maximum as the measure of EL, PPI and GI (see §2 and supplement 6 of the electronic supplementary material for details), on the structure of the correlation matrix. None of these modifications changed the sign of any of the significant correlations, and in most cases, the changes in the magnitude of the correlations were relatively small (supplement 6 of the electronic supplementary material).
Thus, to succinctly summarize the current results of pairwise correlation analysis, genes whose knockout has a severe effect on fitness, that are highly expressed, have many protein–protein interaction partners, and many paralogues have a propensity to evolve slowly, in terms of both ER and PGL. Conceptually, one may think of these as ‘important’ genes that are subject to strong functional constraints and, as a result, refractory to evolutionary change.
(b) Principal component analysis of the genomic variables: a gene's status, adaptability and reactivity
To investigate the relationships between all the analysed genome-related variables simultaneously, we performed PCA of the 3912 KOGs in the seven-dimensional space. Each of the seven PCs accounted for a significant fraction of the variance in the data, i.e. the contribution of each PC was non-negligible (figure 1 and figure 1S of the electronic supplementary material). This shows that none of the original variables can be represented as a linear combination of other variables. Furthermore, the PCA results were found to be highly robust to various modifications of the data analysis procedures, e.g. replacing PGL with the raw number of gene losses or using the maximum among paralogues, instead of the median, to assign a KOG's EL, PPI and GI values, as described in detail in §2 and in supplement 6 of the electronic supplementary material.
The first three PCs captured over one-half (54.8%) of the total variance in the data (figure 1; table 1S and figure 1S of the electronic supplementary material). The first PC (PC1), which accounts for 25% of the overall variance, is comprised of strong positive contributions from EL, NP, KE and PPI, large negative contributions from ER and PGL, and effectively no contribution from GI (figure 2a; tables 1S and 2S of the electronic supplementary material).
PC1 appears to correspond to what may be viewed as a gene's status in the genome-wide community of genes. Indeed, the genes with the high values of PC1 are the ‘high-status’ (most ‘important’) genes—those that cannot be knocked out without a major effect on fitness, are highly expressed, occupy a prominent position in the PPI network, have many paralogues and are evolutionarily conserved. By contrast, genes with low values of PC1 can be knocked out at little cost, evolve fast, are, typically, expressed at a low level and have few (if any) paralogues and protein–protein interactions, i.e. have a low status.
The next two PCs are associated with statistically identical eigenvalues, a property known as sphericity (the p-value of the sphericity test is 0.255; figure 1 and figure 1S of the electronic supplementary material), and therefore are defined only up to a rotation in the plane PC2–PC3. Nevertheless, it seems possible to interpret this plane as a two-dimensional measure of a gene's functional and evolutionary plasticity, and the two PCs (figure 2; table 2S of the electronic supplementary material) as capturing two different facets of this plasticity.
The second PC (PC2), which accounted for 15.3% of the variance, is comprised of positive contributions from NP and GI, negative contributions from KE and effectively no contribution from ER, PGL, EL and PPI (figure 2a; tables 1S and 2S of the electronic supplementary material). Thus, PC2 gives high rank to genes that have many paralogues and often are functionally backed-up by other genes (high GI) but are non-essential (non-lethal upon knockout). We speculated that these features are associated with genes whose activity is highly malleable in response to changes in the cellular and extracellular environments. Under this interpretation, one would predict that genes with high PC2 values have highly skewed distributions of ELs under different experimental conditions, life cycle stages or different tissues of complex organisms. We tested this prediction by computing the skewness indices for expression scores obtained at different stages of the yeast cell cycle, various developmental stages of Drosophila and different human tissues, and comparing them with the PC2 values (table 2a). Indeed, the genes with high PC2 values tend to have more strongly skewed distributions of the ELs, especially those with high status (high values of PC1), i.e. with the most important biological roles (Fisher Omnibus test p-values of 0.01 and much less than 10−20 for low- and high-status KOGs, respectively, when the combined data for three organisms were analysed; Bailey & Noble 2003). Thus, we denoted PC2 gene's adaptability.
Table 2.
Median skewness of expression score distributions in relation to (a) PC1 and PC2 or (b) PC1 and PC3. (Species abbreviations: Dme, Drosophila melanogaster; Hsa, Homo sapiens; Sce, Saccharomyces cerevisiae.)
| (a) | ||||
|---|---|---|---|---|
| adaptability—bottom 50% | adaptability—top 50% | p (Mann–Whitney test) | ||
| Sce | status—bottom 50% | 0.29 | 0.29 | 0.9486 |
| status—top 50% | 0.32 | 0.44 | 0.0031 | |
| Dme | status—bottom 50% | 1.82 | 1.84 | 0.4072 |
| status—top 50% | 1.82 | 1.90 | 0.0660 | |
| Hsa | status—bottom 50% | 1.75 | 1.94 | 0.0007 |
| status—top 50% | 1.87 | 2.12 | 0.0000 | |
| (b) | ||||
|---|---|---|---|---|
| reactivity—bottom 50% | reactivity—top 50% | p (Mann–Whitney test) | ||
| Sce | status—bottom 50% | 0.26 | 0.31 | 0.2635 |
| status—top 50% | 0.22 | 0.50 | 0.0000 | |
| Dme | status—bottom 50% | 1.77 | 1.88 | 0.0631 |
| status—top 50% | 1.86 | 1.84 | 0.8857 | |
| Has | status—bottom 50% | 1.80 | 1.94 | 0.0003 |
| status—top 50% | 1.86 | 2.13 | 0.0000 | |
The third principal component (PC3), which accounts for another 14.5% of the variance (figure 1), is similar to PC2 in that it favours non-essential genes with many paralogues. In contrast to adaptability, however, the contribution of GI to PC3 is strongly negative, the contributions of ER and PPI are weakly negative, whereas PGL and EL make substantial positive contributions (figure 2b; tables 1S and 2S of the electronic supplementary material). Given that high PC3 values are also associated with significantly increased skewness of expression profiles (table 2b; Fisher Omnibus test p-values of 4×10−4 and much less than 1×10−20 for low- and high-status KOGs, respectively), we consider PC3 to be another manifestation of a gene's ability to adjust to different functional modes at different life cycle stages or in different tissues. Thus, we denoted PC3 gene's ‘reactivity’.
(c) Status, adaptability and reactivity of different functional classes of genes and multisubunit complexes
Different functional classes of genes show contrasting trends in status, adaptability and reactivity distributions. Although the individual KOGs in each class span a wide range of values, the group centroids often significantly differ from zero (table 3 and table 3S of the electronic supplementary material; see supplementary data of the electronic supplementary material for the values of status, adaptability and reactivity for all analysed KOGs). Information storage and processing systems, as a whole, are significantly biased toward high status and low adaptability and reactivity; genes involved in cellular processes show, on average, relatively high status and the highest adaptability but low reactivity; genes for metabolic enzymes and transporters are characterized by moderate status and adaptability but the highest reactivity; finally, poorly characterized genes typically fall into the low-status division, and also show low adaptability and reactivity. These trends in status, adaptability and reactivity appear biologically plausible. Thus, the high status of information processing system components is compatible with the fact that many of these are central to genome replication and expression; the characteristic high adaptability of genes involved in cellular processes, particularly, signal transduction, might reflect the involvement of these genes in complex networks of partially redundant pathways; and, the exceptionally high reactivity of metabolic genes corresponds to the notion that changes in the levels of the respective proteins in response to changes in the availability of metabolites are functionally important and do not necessarily involve much back-up. Finally, a curious observation is the distinctly low status of uncharacterized genes; it seems that the functions of the ‘most important’ eukaryotic genes are already known, at least in general terms.
Table 3.
Status, adaptability and reactivity of selected multisubunit complexes and functional classes of proteins. (*Significantly different from zero (p<0.05), using t-test with Bonferroni correction.)
| no. of KOGs | average status | average adaptability | average reactivity | |
|---|---|---|---|---|
| major functional categories | ||||
| information storage and processing | 951 | 0.553* | −0.164* | −0.146* |
| cellular processes and signalling | 1216 | 0.179* | 0.201* | −0.080* |
| metabolism | 692 | −0.057 | 0.075 | 0.494* |
| poorly characterized | 1053 | −0.669* | −0.134* | −0.100* |
| complexes | ||||
| cytoplasmic ribosome | 76 | 2.679* | 0.203 | 1.226* |
| mitochondrial ribosome | 40 | −0.004 | −0.527* | −0.089 |
| chaperonin complex TCP-1 | 8 | 2.237* | −0.291 | −0.299 |
| spliceosome | 50 | 1.234* | −0.511* | −0.393* |
| mRNA cleavage and polyadenylation | 10 | 0.968* | −0.609 | −0.705 |
| proteasome | 33 | 2.158* | −0.547* | −0.329* |
| exosome | 12 | 0.967* | −0.660 | −0.419 |
| nucleosome | 6 | 1.933 | 1.875 | 1.727 |
| vesicle coat complex | 19 | 1.360* | −0.496* | −0.049 |
| vacuolar H+-ATPase | 13 | 1.696* | −0.449 | 0.345 |
| mitochondrial F0F1-ATP synthase | 13 | 1.110* | −0.427 | 0.083 |
| replication licensing complex | 6 | 1.475* | −1.154 | −0.046 |
| aminoacyl-tRNA synthetases | 33 | 0.425 | −0.478* | −0.131 |
Genes whose protein products form multisubunit molecular complexes usually show strong coherence in status, adaptability and reactivity, whereas different complexes, even those with generally similar functions, may differ dramatically (table 3 and figure 3). Thus, comparison of cytosolic and mitochondrial ribosomal proteins shows a clean separation, with the former having a much higher status than the latter (figure 3a). Indeed, mitochondrial ribosomes are extremely diverse in different taxa, and genes coding for mitochondrial ribosomal proteins evolve fast and are often lost during evolution (Mears et al. 2002; Koonin et al. 2004; Mushegian 2005).
Figure 3.
Status and adaptability of multisubunit complex subunits and functional system components. (a) Cytoplasmic (red) and mitochondrial (green) ribosomal proteins. (b) Vacuolar ATPase (red) and vacuolar sorting complex (green). (c) Replication licensing complex (red) and histones (green). (d) RNA processing and modification. The core cluster, which was delineated by iterative removal of outliers with Mahalanobis distance exceeding the cut-off corresponding to a p-value of 0.03, is shown by red circles. The rest of the RNA processing and modification genes are shown by green circles. The blue dots show the rest of the 3912 analysed KOGs.
Complexes with the same intracellular location but distinct functions often show different, characteristic status-adaptability patterns. Thus, the vacuolar ATPase subunits are well separated from those of the vacuolar sorting complex, the former having a much higher status and somewhat lower adaptability (figure 3b). A similar pattern is seen in a comparison of histones and the replication licensing complex, two chromatin-associated complexes. The histones have a significantly higher status and greater adaptability than the licensing complex subunits (figure 3c), which presumably reflects the key role of histones and their modifications in chromatin maintenance and remodelling (Vermaak et al. 2003).
Many functional systems show a distinct pattern, with a dense core of central components of a relatively high status and low adaptability, and a sparse periphery of more adaptable, lower-status genes. This pattern is illustrated in figure 3d for the RNA processing and modification systems. As a class, these have a relatively high average status and low adaptability as it is characteristic of information processing systems in general (table 3 and table 3S of the electronic supplementary material). However, a closer examination reveals a tight, high-status–low-adaptability cluster that is enriched for core subunits of the spliceosome and the mRNA cleavage–polyadenylation complex and a scattered cloud with a significantly lower average status and a wide range of adaptability values consisting of diverse proteins involved in various forms of RNA processing and modification (figure 3d).
Different functional groups of genes also display distinct adaptability–reactivity patterns, e.g. low–low for RNA processing and modification; low–high for translation, ribosomal structure and biogenesis; high–low for signal transduction systems; and high–high for carbohydrate transport and metabolism; figure 4 and table 3S of the electronic supplementary material). These patterns might reflect different functional–evolutionary modalities of these categories of genes. For example, both the translation systems components and those of signal transduction systems are involved in various forms of environmental response but the latter are characterized by a high level of functional back-up as opposed to the former.
Figure 4.
Adaptability and reactivity of four functional classes of genes: 1, carbohydrate transport and metabolism; 2, signal transduction mechanisms; 3, replication, RNA processing and modification; and 4, translation, ribosomal structure and biogenesis. Ellipses encompass the area of 3 s.d. of the mean for the corresponding KOG sets in the two-dimensional adaptability (PC2)–reactivity (PC3) space. Blue dots show the rest of the 3912 KOGs.
4. Conclusions
The analysis described here suggests that the relationships between phenotypic and evolutionary characteristics of genes can be meaningfully described with composite variables (PCs), which seem to reflect the biological role and ‘importance’ of a gene, and its functional and evolutionary modes. This is one of the rare cases where the top PCs appear to be amenable to appealing biological interpretations. Clustering of genes in the PC space has the potential to reveal previously unnoticed functional links.
The notion of a gene's status could have an additional meaning. Since phenotypic variables contribute positively to the status and evolutionary variables contribute negatively, this notion provides a useful generator of null hypotheses on the sign of the correlations between variables associated with functioning and evolution of genes. Any deviation from the expected pattern of correlation calls for attention—to the quality of the data, the nature of the analysed relationship, or both.
Acknowledgements
We thank Sergei Maslov, Dmitry Chklovsky, Mikhail Gelfand, Alex Kondrashov and members of the Koonin group for useful discussions.
Supplementary Material
Details of the methods used and the results of this study, particularly the analysis demonstrating the robustness of the conclusions
References
- Altschul S.F, Madden T.L, Schaffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris-Brosou S. Determinants of adaptive evolution at the molecular level: the extended complexity hypothesis. Mol. Biol. Evol. 2005;22:200–209. doi: 10.1093/molbev/msi006. doi:10.1093/molbev/msi006 [DOI] [PubMed] [Google Scholar]
- Bailey T.L, Noble W.S. Searching for statistically significant regulatory modules. Bioinformatics. 2003;19(Suppl. 2):II16–II25. doi: 10.1093/bioinformatics/btg1054. [DOI] [PubMed] [Google Scholar]
- Bloom J.D, Adami C. Apparent dependence of protein evolutionary rate on number of interactions is linked to biases in protein–protein interactions data sets. BMC Evol. Biol. 2003;3:21. doi: 10.1186/1471-2148-3-21. doi:10.1186/1471-2148-3-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulomb S, Bauer M, Bernard D, Marsolier-Kergoat M.C. Gene essentiality and the topology of protein interaction networks. Proc. R. Soc. B. 2005;272:1721–1725. doi: 10.1098/rspb.2005.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. doi:10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld S, Littauer S.B. McGraw-Hill; New York, NY: 1964. Introduction to statistical methods. [Google Scholar]
- Fraser H.B. Modularity and evolutionary constraint on proteins. Nat. Genet. 2005;37:351–352. doi: 10.1038/ng1530. doi:10.1038/ng1530 [DOI] [PubMed] [Google Scholar]
- Fraser H.B, Hirsh A.E, Steinmetz L.M, Scharfe C, Feldman M.W. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. doi:10.1126/science.1068696 [DOI] [PubMed] [Google Scholar]
- Fraser H.B, Wall D.P, Hirsh A.E. A simple dependence between protein evolution rate and the number of protein–protein interactions. BMC Evol. Biol. 2003;3:11. doi: 10.1186/1471-2148-3-11. doi:10.1186/1471-2148-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Walhout A.J, Vidal M. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. doi: 10.1016/j.tig.2003.08.009. doi:10.1016/j.tig.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Herbeck J.T, Wall D.P. Converging on a general model of protein evolution. Trends Biotechnol. 2005;23:485–487. doi: 10.1016/j.tibtech.2005.07.009. doi:10.1016/j.tibtech.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Hirsh A.E, Fraser H.B. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. doi:10.1038/35082561 [DOI] [PubMed] [Google Scholar]
- Hurst L.D, Smith N.G. Do essential genes evolve slowly? Curr. Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. doi:10.1016/S0960-9822(99)80334-0 [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason S.P, Barabasi A.L, Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. doi:10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- Jordan I.K, Rogozin I.B, Wolf Y.I, Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. doi:10.1101/gr.87702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K, Wolf Y.I, Koonin E.V. No simple dependence between protein evolution rate and the number of protein–protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol. Biol. 2003;3:1. doi: 10.1186/1471-2148-3-1. doi:10.1186/1471-2148-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K, Marino-Ramirez L, Wolf Y.I, Koonin E.V. Conservation and coevolution in the scale-free human gene coexpression network. Mol. Biol. Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. doi:10.1093/molbev/msh222 [DOI] [PubMed] [Google Scholar]
- Kamath R.S, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. doi:10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Koonin E.V. Systemic determinants of gene evolution and function. Mol. Syst. Biol. 2005 doi: 10.1038/msb4100029. doi:101038/msb4100029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. doi:10.1186/gb-2004-5-2-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren Y, Carmel L. Robust linear dimensionality reduction. IEEE Trans. Visual. Comput. Graph. 2004;10:459–470. doi: 10.1109/TVCG.2004.17. doi:10.1109/TVCG.2004.17 [DOI] [PubMed] [Google Scholar]
- Krylov D.M, Wolf Y.I, Rogozin I.B, Koonin E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. doi:10.1101/gr.1589103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears J.A, Cannone J.J, Stagg S.M, Gutell R.R, Agrawal R.K, Harvey S.C. Modeling a minimal ribosome based on comparative sequence analysis. J. Mol. Biol. 2002;321:215–234. doi: 10.1016/s0022-2836(02)00568-5. doi:10.1016/S0022-2836(02)00568-5 [DOI] [PubMed] [Google Scholar]
- Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc. Natl Acad. Sci. USA. 2005;102(Suppl. 1):6630–6635. doi: 10.1073/pnas.0501984102. doi:10.1073/pnas.0501984102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A. Protein content of minimal and ancestral ribosome. RNA. 2005;11:1400–1406. doi: 10.1261/rna.2180205. doi:10.1261/rna.2180205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst L.D. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst L.D. Genomic function: rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. discussion 497-8. [DOI] [PubMed] [Google Scholar]
- Pennisi E. How will big pictures emerge from a sea of biological data? Science. 2005;309:94. doi: 10.1126/science.309.5731.94. doi:10.1126/science.309.5731.94 [DOI] [PubMed] [Google Scholar]
- Provart N.J, McCourt P. Systems approaches to understanding cell signaling and gene regulation. Curr. Opin. Plant Biol. 2004;7:605–609. doi: 10.1016/j.pbi.2004.07.001. doi:10.1016/j.pbi.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Rocha E.P, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol. Biol. Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. doi:10.1093/molbev/msh004 [DOI] [PubMed] [Google Scholar]
- Tatusov R.L, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. doi:10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Ahmad K, Henikoff S. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 2003;15:266–274. doi: 10.1016/s0955-0674(03)00043-7. doi:10.1016/S0955-0674(03)00043-7 [DOI] [PubMed] [Google Scholar]
- Wall D.P, Hirsh A.E, Fraser H.B, Kumm J, Giaever G, Eisen M.B, Feldman M.W. Functional genomic analysis of the rates of protein evolution. Proc. Natl Acad. Sci. USA. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. doi:10.1073/pnas.0501761102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.C, Carlson S.S, White T.J. Biochemical evolution. Annu. Rev. Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. doi:10.1146/annurev.bi.46.070177.003041 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang J, He X. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol. Biol. Evol. 2005;22:1147–1155. doi: 10.1093/molbev/msi101. doi:10.1093/molbev/msi101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the methods used and the results of this study, particularly the analysis demonstrating the robustness of the conclusions