Abstract
Adaptive speciation has gained popularity as a fundamental process underlying the generation of diversity. We tested whether populations respond to similar forms of disruptive selection by diversifying in similar or parallel ways by investigating diversified populations of Escherichia coli B evolved in glucose and glucose–acetate environments. In both environments, the populations have differentiated into two phenotypes, named for their characteristic colony morphologies: large (L) and small (S). Each type is heritable and this polymorphism (or ‘diversified pair’) appears to be maintained by negative frequency dependence. The L and S phenotypes from different environments are convergent in their colony morphology and growth characteristics. We tested whether diversification was parallel by conducting competition experiments between L and S types from different environments. Our results indicate that replicate diversified pairs from different environments have not diversified in parallel ways and suggest that subtle differences in evolutionary environment can crucially affect the outcome of adaptive diversification.
Keywords: Escherichia coli, frequency-dependent selection, parallel evolution, adaptive diversification, convergent evolution, disruptive selection
1. Introduction
In theory, disruptive selection arising as a consequence of frequency-dependent processes (e.g. competition for limited resources) can result in adaptive diversification, that is, the splitting of an ancestral lineage into distinct descendent lineages as a consequence of natural selection acting on individuals within the population (Dieckmann & Doebeli 1999; Friesen et al. 2004). This idea highlights the central role of ecology in the speciation process. Empirical support for adaptive speciation is accumulating (Via 2001; Dieckmann et al. 2004) although experimental support has, in general, been lacking (but see Rainey & Travisano 1998; Friesen et al. 2004). Evolution experiments using microbial systems are useful for testing ideas of adaptive diversification because microbes have large population sizes, short generation times and are easy to culture in the laboratory (Lenski 1991; Rosenzweig et al. 1994; Travisano et al. 1995; Travisano & Lenski 1996; Rainey & Travisano 1998; Treves et al. 1998; Rainey et al. 2000; Rainey & Rainey 2003). In an attempt to study diversification experimentally, 36 lines of Escherichia coli B were evolved under seasonal conditions (i.e. daily serial batch transfer into fresh media) for 1000 generations in different nutritional environments (Saxer et al. in preparation; Friesen et al. 2004). One of the main findings of these experiments was that the ancestor had repeatedly diversified into (at least) two types that can be identified by the size of the colony formed on agar plates. One type formed large colonies (L), which were visible after 24 h, and the second type formed small colonies (S), which were visible after 48 h. This polymorphism was observed in lines that evolved in glucose as well as in lines that evolved in a mixture of glucose and acetate, two environmental conditions that are actually very similar (see §3). In both environments, the L–S polymorphism, which we refer to as a diversified pair, appeared to be stable over both evolutionary and ecological time-scales. Friesen et al. (2004) suggested that the L and S colonies corresponded to two ecological types, a glucose specialist and a fast-switcher (or acetate) specialist (named after their differential growth performance in glucose–acetate environments as detailed below), which were maintained by negative frequency-dependent selection (Levin 1988); that is, each type had the advantage when rare.
Being a consequence of natural selection, adaptive diversification is a deterministic process and is therefore often associated with ‘repeatability’. For example, parallel diversification is often recognized as evidence for adaptive diversification (or ecological speciation; Schluter 2000) because parallelism implies the action of natural selection. However, does adaptive diversification imply parallelism? In other words, if one sees adaptive diversification repeatedly under similar ecological conditions, does this imply parallel diversification? Or can subtle differences in the environment lead to qualitatively different pairs of diversified types? We address this question by using a biological system—the L–S polymorphism—for which we are confident that adaptive processes have generated diversification (Friesen et al. 2004). Saxer et al. (in preparation) argue that there is good evidence for parallel diversification among all replicate microcosms that evolved in the glucose–acetate mixture. Here we ask whether diversification in glucose alone, a very similar environment, occurred in parallel with diversification in the glucose–acetate mixture. To do this, we conducted competition experiments between L and S strains in order to determine if the outcome of competition between L and S strains from diversified pairs that evolved in different environments was analogous to the outcome of competition between L and S strains that evolved in the same environment.
2. Background: evolved strains
All bacterial lines were initiated by Saxer et al. (in preparation) from, alternately, Ara− or Ara+ variants of a common ancestral strain of E. coli B that differed from one another only in their ability to metabolize l-arabinose. This difference served as a useful cross-line contamination check (Lenski 1991). We briefly review their experimental protocol and results (see also Friesen et al. 2004). The bacterial strains used reproduce asexually and contain no plasmids (Lenski 1991). Replicate cultures were grown in 10 mL of Davis Minimal (DM) media and supplemented with either 410 μg mL−1 glucose (DMGLU), 410 μg mL−1 acetate (DMACE), or 205 μg mL−1 glucose and 205 μg mL−1 acetate (DMMIX) as the additional source(s) of carbon in 50 mL flasks at 37 °C and 120 r.p.m. Each culture underwent a 100-fold dilution during transfer to fresh media once every 24 h, resulting in 6.6 binary divisions per day.
By 1000 generations (ca. 150 days), a stable polymorphism had appeared in 6 of 12 DMGLU replicates and 6 of 12 DMMIX replicates. In both environments, the ancestral type had diversified into two types with respect to colony morphology. One type formed large colonies (L) within 24 h after plating on arabinose-free agar and a second type formed small colonies (S) between 24 and 48 h after plating. L and S types also differed in their growth characteristics in liquid media (Friesen et al. 2004). Similar diversity has previously been described (Helling et al. 1987; Rosenzweig et al. 1994; Turner et al. 1996; Rozen & Lenski 2000).
When bacteria are grown in liquid media containing two resources, they exhibit diauxie, whereby an initial growth phase is correlated with use of the preferred metabolite (e.g. glucose) and a subsequent growth phase is correlated with use of the less preferred metabolite (e.g. acetate). This diauxic growth pattern is therefore expected in the DMMIX environment, as two resources are present at the onset of the transfer period. However, diauxie also occurred in DMGLU replicates, for reasons that can be found in the metabolic details: the aerobic breakdown of glucose in glycolysis generates acetate, which is available for uptake and catabolism by E. coli once the glucose in the media has been exhausted. Because anaerobic metabolism of glucose yields acetate, the DMGLU environment progressively becomes more like the DMMIX environment during a single season (batch) and differs only in the temporal availability of acetate and the absolute and relative amounts of glucose and acetate throughout the course of the season. We exploited this similarity of nutritional environments to test the repeatability of parallel diversification.
Friesen et al. (2004) proposed that diauxic growth is the basis for the L and S polymorphism and that the polymorphism is maintained by frequency-dependent competition for limited resources and a trade-off between initial growth on glucose and subsequent growth on acetate (see also Turner et al. 1996). Within the course of one transfer period, L appears to grow faster on glucose (i.e. L is a glucose specialist) but takes longer to switch to acetate metabolism than S (i.e. S is a fast switching (or acetate) specialist). Frequency dependence restores the balance between the two types whenever one type becomes too common; that is, whenever competition for the resource on which the common type specializes becomes intense. In the glucose-only environment, the observed polymorphism appears to be related to the crossfeeding polymorphisms that have been found in chemostat cultures of E. coli (Rosenzweig et al. 1994; Turner et al. 1996; Treves et al. 1998).
(a) Why competition experiments?
Preliminary investigations revealed that a diversified pair mixed in different starting proportions of L and S and observed over ecological time (up to 14 days or about 100 generations) consistently reached a stable, intermediate frequency (Friesen et al. 2004). When rare, each type had an advantage and increased in frequency until a stable frequency was obtained. Thus, it appeared that frequency-dependent selection played an important role in generating and maintaining the stable polymorphism within replicates. We use this result as a basis of comparison to determine if diversified types were functionally equivalent (in their ‘competitive ability’) across replicates within and between nutrient environments. We use the term competitive ability as a composite term encompassing all traits required to extract and metabolize resources (glucose and acetate) from the environment in the face of competition with the complementary partner of a diversified pair. We employ competition experiments as an ecological assay for parallelism because of the importance of competition for generating frequency-dependent selection and, ultimately, for the maintenance of diversity.
3. Methods
(a) Competition experiments
All colonies in this experiment were selected from cultures evolved by Saxer et al. (in preparation). We assayed for diversity on arabinose-free plates, and isolated single L and S genotypes from DMGLU-derived population 10 and DMMIX-derived populations 31 and 33 by plating the diversified strains on arabinose-free agar plates and picking single colonies as representatives of each diversified pair (identified by characteristic colony morphology). We suspended and grew these in 10 mL of DMGLU or DMMIX for 24 h (37 °C at 250 r.p.m.) and then mixed L and S in three different proportions to initiate our competition experiments. Starting proportions of L were 90, 50 and 10% (with S making up the complementary proportion) and were mixed by correcting proportionate volumes for cell density (determined by optical density using a spectrophotometer). We added 100 μL of this mixture to 10 mL of liquid media (DMMIX or DMGLU) and subsequently transferred 100 μL of stationary phase culture (24 h) to new media for 8, 10 or 12 days. At each transfer, we assayed cultures by plating on four to six arabinose-free plates (providing us with 500–1000 colonies per assay) to determine the density of L and S. S and L ecotypes were identified as detailed above.
We replicated each treatment three or four times and recorded the means of proportion L (±s.e.m.). In addition to the mixed cultures, we initiated pure cultures of L or S to examine the possibility that the complementary type in a diversified pair (S or L, respectively) might arise by mutation and become established in cultures within the timeframe of our study. We periodically checked for cross-contamination between cultures.
In our first assay, we repeated earlier work (Friesen et al. 2004) to establish a baseline of comparison for our subsequent assays. Thus, we mixed L and S from a single diversified pair in different starting proportions to confirm that mixed cultures would equilibrate to an intermediate frequency of L due to negative frequency dependence (with each type having an advantage when rare). The evolutionary environment for each player and the ecological environment of the assay were the same: DMMIX. Our second assay involved mixing an L from one diversified pair in DMMIX with an S from a different diversified pair in DMMIX (and vice versa). Therefore, both players evolved in and competed against one another in the DMMIX environment; however, the players came from different replicate microcosms. Finally, we mixed one partner from a diversified pair that evolved in DMMIX with its complementary partner from another diversified pair that evolved in DMGLU, and vice versa. Comparing results from this last set of assays with results from the other assays would therefore shed light on whether diversified pairs that evolved in different environments underwent parallel diversification. Specifically, we would conclude that diversification of the phenotypes underlying competitive ability had occurred in parallel if the dynamics of competition between L and S, when L and S come from different diversified pairs that evolved in different environments (assay 3 above), are similar to the dynamics when L and S come from either the same diversified pair (assay 1) or from different diversified pairs that evolved in the same environment (assay 2). However, if those dynamics were different we would conclude that diversification had not occurred in a parallel manner.
4. Results
Our first assay involved competing L and S from diversified pairs of Populations 31 and 33 which were initiated at different starting proportions and evolved and competed in the DMMIX environment. Regardless of starting proportions, all trials between strains from population 31 diversified pairs levelled off at a stable, intermediate frequency by day 8. If we define the frequency at the end of each trial as the equilibrium frequency, then the equilibrium frequencies from different trials were indistinguishable from one another (proportion large=0.49, F2,6=0.7647, p=0.506; figure 1a). Likewise, diversified pairs from population 33 reached intermediate equilibrium frequencies by day 8 that were also indistinguishable from one another (proportion large=0.80, F2,6=0.1552, p=0.860; figure 1b). Combining all trials involving population 31 and 33 diversified pairs, we tested whether the equilibrium frequency was similar between populations 31 and 33. We determined that the equilibrium frequencies were in fact different from one another (t=8.76, p<0.0001). Treatments initiated with only L or S types in both population 31 and 33 remained pure throughout the entire study (results not shown).
Figure 1.
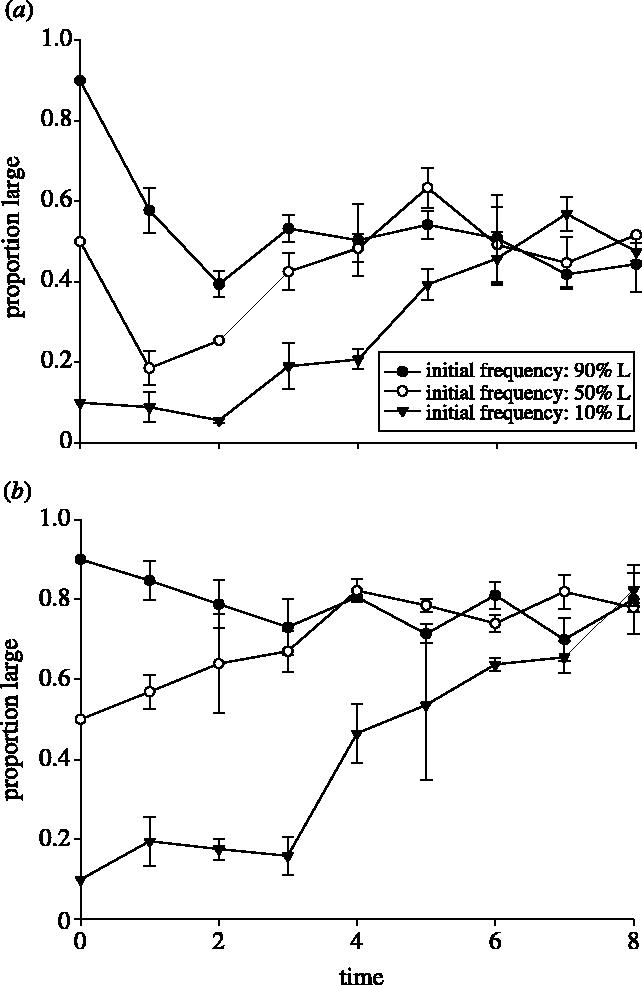
The means of proportion Large (±s.e.m.) for three starting mixtures of (a) LMIX-31 and SMIX-31 versus time (days) in DMMIX and (b) LMIX-33 and SMIX-33 versus time (days) in DMMIX. In both cases, the proportions stabilized at an intermediate frequency by day eight in all treatments.
Our second assay involved competing complementary partners (L versus S) from two different diversified pairs, albeit that both evolved in the DMMIX environment. Figure 2a illustrates the dynamics of competition between LMIX-31 versus SMIX-33. Again, regardless of starting proportion, all trials converged to an intermediate equilibrium frequency by day 10 (proportion large =0.79, F2,6=2.074, p=0.2067). Similarly, when we competed LMIX-33 versus SMIX-31, we observed convergence to an intermediate equilibrium frequency within 8 days (proportion large =0.37, F2,6=0.6047, p=0.5764; figure 2b). Because our first assay revealed that sets of diversified pairs were fine-tuned to different intermediate equilibrium frequencies (see above), we had little reason to expect that the equilibrium frequencies attained in our mixed-partner trials would be equal and, indeed, they were not (t=7.845, p<0.0001).
Figure 2.
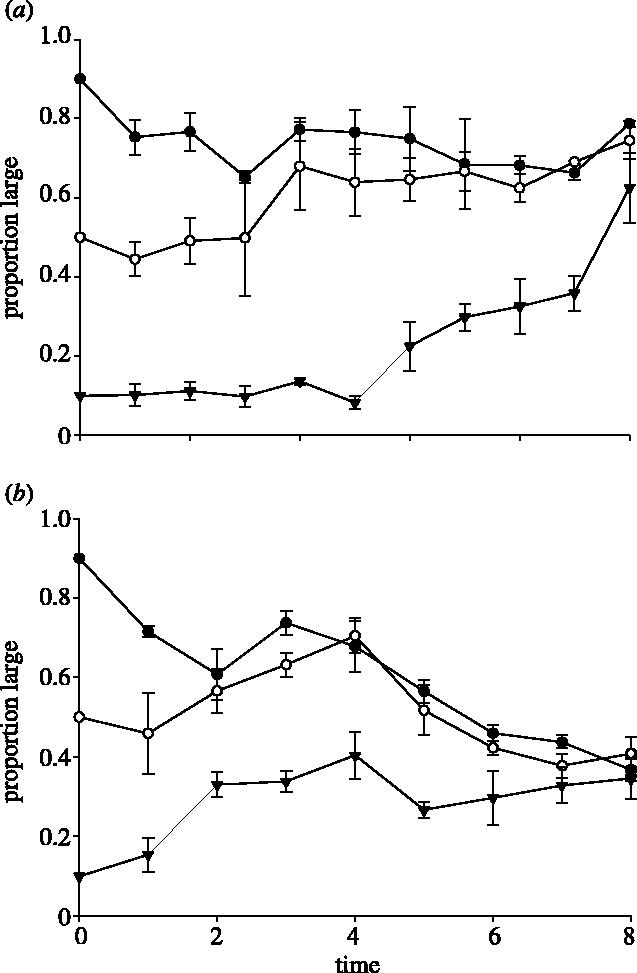
The means of proportion large (±s.e.m.) for three starting mixtures of (a) LMIX-31 and SMIX-33 versus time (days) in DMMIX and (b) LMIX-33 and SMIX-31 versus time (days) in DMMIX. The proportions stabilized at an intermediate frequency within eight–ten days of competition. Symbols are the same as for figure 1.
Third, complementary partners that had evolved in different environments were placed in a competitive environment. Figures 3a,b and 4a,b cogently summarize the results of these mixed partner/mixed evolutionary environment assays: the competitive dynamics are qualitatively different from those attained above and from one another, with outcomes depending on both the partners and the environments.
Figure 3.
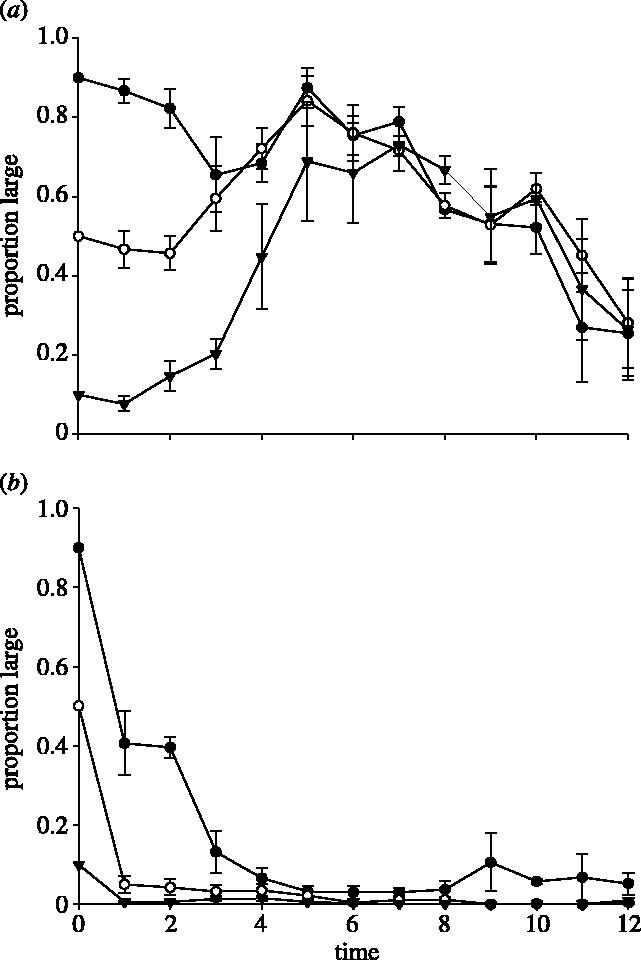
The means of proportion large (±s.e.m.) for three starting mixtures of (a) LMIX-33 and SGLU-10 versus time (days) and (b) LGLU-10 and SMIX-33 versus time (days) in DMMIX. Symbols are the same as for figure 1.
Figure 4.
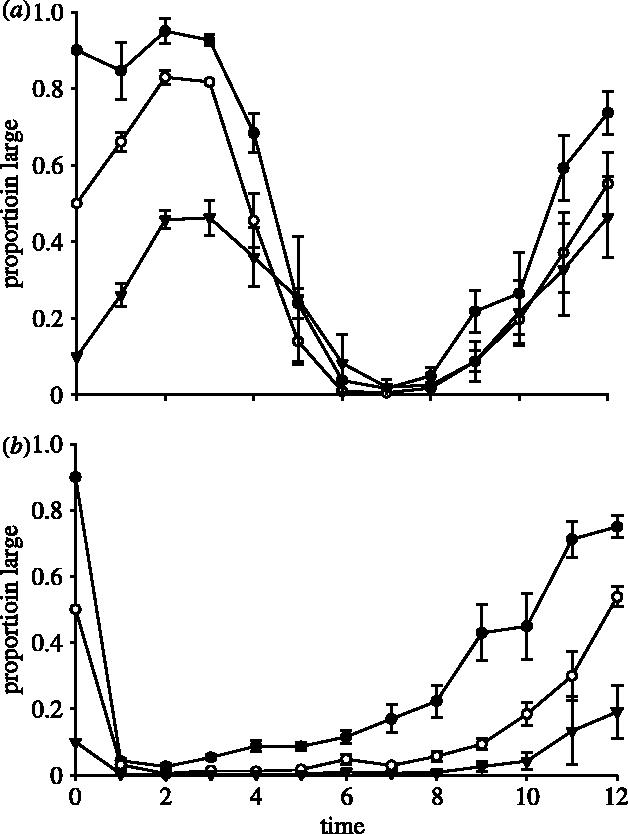
The means of proportion large (±s.e.m.) for three starting mixtures of (a) LMIX-33 and SGLU-10 versus time (days) and (b) LGLU-10 and SMIX-33 versus time (days) in DMGLU. The proportions did not stabilize within the duration of the experiment. Symbols are the same as for figure 1.
Figure 3a illustrates the competitive dynamics between LMIX-33 and SGLU-10 in the DMMIX environment. Though the dynamics appeared to converge to similar frequency dynamics for different initial conditions, the frequencies tend to oscillate over the 12-day duration of the assay rather than level off at some intermediate frequency. In contrast, when the partners came from reciprocal environments (LGLU-10 versus SMIX-33), L almost vanished in all trials, regardless of initial conditions (figure 3b). Because one of the partners had evolved in DMGLU, we set up analogous competition assays in the DMGLU environment. When we placed LMIX-33 and SGLU-10 in a competitive environment, we observed a convergence in frequency dynamics for different initial conditions but oscillatory dynamics over time (figure 4a). Finally, when we placed LGLU-10 and SMIX-33 in DMGLU, we observed an initial decline in the proportion L and then a slow recovery over the duration of the assay (figure 4b). Overall, the dynamics observed when L and S from diversified pairs that evolved in different environments were placed in competition are qualitatively different from the equilibrium dynamics that result when L and S from diversified pairs that evolved in the same environment were placed in competition.
5. Discussion
We competed diversified strains of E. coli with each other to examine the degree of parallel diversification that had occurred in replicate populations, within and between evolutionary environments. Our results (first assay) agree with earlier studies (Helling et al. 1987; Rosenzweig et al. 1994; Turner et al. 1996; Friesen et al. 2004; Saxer et al. in preparation) that have demonstrated that diversified types can be maintained by negative frequency-dependent selection. This empirical result formed the basis of comparison for our next two experiments. Evidence for parallel diversification would consist of a similar convergence to intermediate frequency when complementary partners selected from different replicates were pitted against one another.
In our second set of assays, we competed complementary partners from different replicates, each derived in identical evolutionary environments (LMIX-31 versus SMIX-33 and LMIX-33 versus SMIX-31, both derived in DMMIX). Because L and S converged to intermediate frequencies, we conclude that parallel diversification has occurred. However, when we selected complementary partners from different evolutionary environments for competition, our results were significantly different. In one outcome, we observed the near elimination of one partner; in three others, we observed oscillatory dynamics with no evidence of convergence to a stable intermediate frequency.
No simple pattern emerged to help explain the range of competitive outcomes we observed when we competed complementary partners from disparate evolutionary environments. Although SMIX-33 was clearly superior to LGLU-10 in the DMMIX environment, the advantage was not so clear in the DMGLU environment, suggesting perhaps that ‘home turf’ (i.e. competition occurring in the environment that one partner had evolved in) confers some competitive advantage—or at least restored some semblance of balance to the system. There were no other indications that one type was absolutely superior in competition, or that home turf afforded any advantages.
It is important to note that the two environments in which the microcosms evolved and diversified are actually quite similar. In the glucose–acetate mixture (DMMIX), the acetate is present from the start and bacteria use the two resources sequentially. In the glucose-only environment (DMGLU), only glucose is present at the start of each batch culture, but acetate is produced during glucose metabolism and is available for uptake and catabolism once the glucose has been exhausted. Thus, the DMGLU environment differs from the DMMIX environment only in the absolute and relative amounts of glucose and acetate throughout the course of the season.
The similarity of the environments is reflected in the fact that our diversified populations have apparently evolved in parallel with respect to colony morphology (diameter and form). Because we observed similar phenotypic patterns of diversification in the two environments, and because in both environments, diversification appears to be a result of similar mechanisms (owing to the existence of a tradeoff between metabolic efficiency on glucose and acetate), one might expect that diversification has occurred in parallel. However, our results suggest that only replicates that shared identical environments diversified in parallel with respect to competitive ability. Thus, in general, whether we find evidence of parallel diversification may be contingent on the traits selected for experimentation. It would be interesting to quantify why some traits are indifferent to the details of the disruptive selection (e.g. colony morphology), while other traits appear to be more sensitive (e.g. competitive ability).
Functional variation (in competitive ability) within each type suggests that there is more than one way to diversify as an L or S partner within a diversified pair. We find this result somewhat sobering, as researchers using this system (and others, e.g. Pseudomonas fluorescens; Rainey & Travisano 1998) often rely on colony morphology to identify diversified types within their cultures. We suspect that employing colony phenotype to estimate diversification underestimates the variation present in diversified cultures.
It is interesting to consider the unstable dynamics observed in figures 3a and 4a, and speculate about the cause of such patterns. Ultimately, we need to account for the observation of a change in the trajectory of population composition (increasing versus decreasing proportion L) given the same initial frequencies of L within the population, differing only in the time period in which they were observed. Hypotheses to explain this variation fall into two broad categories, genetic variation and phenotypic variation:
(a) Genetic hypotheses
This category of hypotheses depends on genetic variation (for competitive ability) existing within the competitive microcosms that is masked by the characteristic morphological traits. This requires that genetic variation has arisen within the timeframe of the experiment, as L and S types were isolated from single (presumably genetically uniform) colonies. Imagine there are two types of L, L1 and L2. Then one could envisage that competition within the L category (L1 versus L2), as well as competition among the L and S categories, drives the observed dynamics. Thus, when L is rare (relative to S), L1 out competes L2; however, when L is common, L2 out competes L1. We might expect, then, for the population to consist of different types of L at different times throughout the competitive experiment. If we isolated L colonies at different times, and competed them against the S from their reciprocal times, then we would expect to see a reversal in the trajectory of population composition. One difficulty with testing hypotheses of this nature is that they rely on no mutations arising within the course of the test, yet the hypothesis of genetic variation requires variation to have arisen within the timeframe of the original experiment in the first place (since L and S cultures were isolated from a single colony).
(b) Phenotypic hypotheses
This category of hypotheses depends on phenotypic variation driving the observed cyclical dynamics. One possibility is that gene expression is sensitive to extreme frequencies of L. Crossing such a threshold alters gene expression, shifting the dynamics between L and S. No doubt variations on this hypothesis exist. They await future formulation and testing.
This study adds to the body of work that highlights the importance of frequency dependence for generating and maintaining diversity (reviewed in Levin 1988; Rainey et al. 2000). In our system, negative frequency dependence maintains the balance between different types specializing on different resources. However, one general implication of our study is that polymorphisms maintained by negative frequency dependence may be highly sensitive to the environmental conditions and evolutionary histories of the players involved. Thus, even slight changes in conditions may shift the system from a state of stability to one of periodicity or extinction. For example, other forms of frequency dependence involving spatial structure may also be sensitive to subtle shifts in the environment (Rainey & Travisano 1998; Rozen & Lenski 2000). Our results also have implications for conservation management plans. Reintroductions of ‘similar’ species (ecotypes) in restoration projects after the loss of a species (e.g. swapping one type for another) could result in similar patterns of instability or even system collapse.
Further, our results suggest that while diversification in one trait (e.g. colony morphology) may be robust across environments, diversification in other traits may not be (e.g. competitive ability), because we observed parallelism with respect to colony morphology, but not with respect to competitive ability. Parallelism has often been used as a test of the role of ecology in speciation (or adaptation in diversification; Schluter 2000). We are confident that our microcosms have diversified due to similar adaptive processes (Friesen et al. 2004), yet we failed to see parallel diversification when we used competitive ability as our assay of parallelism. Thus, studies that observe non-parallel patterns should not necessarily conclude that ecology has played little role in the diversification process. Unparallel diversification may simply reflect subtle differences between otherwise similar environments.
Acknowledgments
We thank G. Takhar, J. Briscoe and S. Merali for laboratory assistance, R. Redfield, C. Spencer, M. Bertrand and S. Kahlon for technical advice and members of the Doebeli and SOWD laboratory groups for fruitful discussions and feedback on early versions of this manuscript. The McDonnell Foundation Grant and NSERC provided financial assistance to M.D.
References
- Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Doebeli M, Metz J.A.J, Tautz D, editors. Adaptive speciation. Cambridge University Press; Cambridge: 2004. [Google Scholar]
- Friesen M.L, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- Helling R, Vargas C.H, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R, Rose M.R, Simpson S.C, Tadler S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- Levin B. Frequency-dependent selection in bacterial populations. Phil. Trans. R. Soc. B. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- Rainey P.B, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. Nature. 2003;425:72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- Rainey P, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rainey P, Buckling A, Kassen R, Travisano M. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol. Evol. 2000;15:243–247. doi: 10.1016/s0169-5347(00)01871-1. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Sharp R, Treves D, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D, Lenski R.E. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Saxer, G., Friesen, M., Doebeli, M., Travisano, M. In preparation The repeatability of adaptive radiation during long-term experimental evolution of Escherichia coli in a mixed nutrient environment. [DOI] [PMC free article] [PubMed]
- Schluter D. Oxford series in ecology and evolution. Oxford University Press; Oxford: 2000. The ecology of adaptive radiation. [Google Scholar]
- Travisano M, Lenski R.E. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Vasi F, Lenski R.E. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution. 1995;49:189–200. doi: 10.1111/j.1558-5646.1995.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Treves D, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 1998;15:787–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Turner P, Souza V, Lenski R.E. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology. 1996;77:2119–2129. [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]


