Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen that colonizes the lungs of cystic fibrosis (CF) patients. CF lungs often contain a diverse range of P. aeruginosa phenotypes, some of which are likely to contribute to the persistence of infection, yet the causes of diversity are unclear. While the ecological heterogeneity of the lung environment and therapeutic regimes are probable factors, a role for parasitic bacteriophage cannot be ruled out. Parasites have been implicated as a key ecological variable driving the evolution of diversity in host populations. PP7 drove cycles of morphological diversification in host populations of P. aeruginosa due to the de novo evolution of small-rough colony variants that coexisted with large diffuse colony morph bacteria. In the absence of phage, bacteria only displayed the large diffuse colony morphology of the wild-type. Further assays revealed there to be two distinct types of resistant bacteria; these had very different ecological phenotypes, yet each carried a cost of resistance.
Keywords: diversity, host–parasite, cystic fibrosis, Pseudomonas aeruginosa, bacteriophage therapy, experimental evolution
1. Introduction
Pseudomonas aeruginosa is an important opportunistic pathogen with a broad host range (plants, invertebrates, vertebrates; Palleroni 1984) capable also of inhabiting a number of abiotic environments (soil, water; Palleroni 1984). In humans, P. aeruginosa is the most common cause of chronic lung infection in cystic fibrosis (CF) patients (Brennan & Geddes 2002; Lyczak et al. 2002), and a major cause of nosocomial (hospital-borne) infections (Emori & Gaynes 1993). The great diversity of P. aeruginosa phenotypes isolated from the lungs of CF patients has been noted in a number of studies (e.g. Martin et al. 1995; Oliver et al. 2000; Haussler et al. 2003; Kresse et al. 2003). A wide range of colony morphology variants have been observed including small-colony variants (Drenkard & Ausubel 2002; Haussler et al. 2003) and mucoid colonies that overexpress alginate (Govan & Deretic 1996). In addition, isolates with similar colony morphologies have been shown to display variation in other phenotypes including motility, expression of cell-surface components, auxotrophy and resistance to antibiotics (Mahenthiralingam et al. 1994; Oliver et al. 2000; Kresse et al. 2003). Certain colony morphology variants have been shown to be better able to resist antibiotic treatment (Drenkard & Ausubel 2002) or evade detection by the immune system (Mahenthiralingam et al. 1994; Govan & Deretic 1996), raising the possibility that diversity in bacterial populations may prolong infection.
The large amounts of diversity observed in CF lungs are consistent with ecological theory. CF lungs are both spatially and temporally heterogeneous (Oliver et al. 2000), and competition in heterogeneous environments drives diversification (Korona et al. 1994; Rainey & Travisano 1998; Doebeli & Dieckmann 2000; Schluter 2000). However, mortality agents (such as predators, parasites, host immunity and antibiotics) may also promote the origin and maintenance of diversity (Holt 1984; Frank 1993; Thompson 1999). Mortality agents may drive sympatric diversification if there are multiple resistance strategies or fitness costs associated with resistance (Frank 1993; Abrams 2000; Doebeli & Dieckmann 2000; Abrams & Chen 2002). Lysogenic bacteriophages are known to be present in the sputa of CF patients (Tejedor et al. 1982) and have been suggested, among other environmental factors, as a cause of P. aeruginosa mucoid conversion in CF lungs (Martin 1973; Miller & Rubero 1984). While exposure to antibiotics and host immunity is clearly of relevance to CF lung colonizers, the role of bacteriophage in the evolution of P. aeruginosa diversity remains unclear.
Laboratory populations of bacteria and bacteriophage have provided direct evidence that parasites can increase host diversity. Several studies have reported coexistence of bacteriophage and a polymorphic host population containing both resistant and sensitive bacteria, mediated by costs of resistance (Lenski & Levin 1985; Schrag & Mittler 1996; Bohannan & Lenski 1997, 1999, 2000). Other studies have found diversification of bacteria into multiple resistant forms with (Chao et al. 1977; Buckling & Rainey 2002; Brockhurst et al. 2004) and without (Mizoguchi et al. 2003) evidence of costs, or parasite-mediated coexistence of pre-existing spontaneous costless resistance mutants (Lythgoe & Chao 2003).
Here, we investigate the role of a pilus-binding lytic bacteriophage, PP7, in P. aeruginosa diversification. The process by which this and many other phage, including DMS3, a transducing lysogenic phage isolated from a clinical strain of P. aeruginosa (Budzik et al. 2004), infect P. aeruginosa suggests they could drive the evolution of diversity. Moreover, there are probably both costs of resistance to phage and multiple resistance mechanisms, allowing us to explore the role of both these factors in determining parasite-mediated host population dynamics and diversity. PP7 is a small single-stranded RNA-containing lytic bacteriophage belonging to the Leviviridae that binds to the type-IV polar pili of P. aeruginosa (Bradley 1966; Olsthoorn et al. 1995) and infects the cell upon retraction of the pilus (Bradley 1966, 1974). Type-IV polar pili are used for twitching motility (Mattick 2002) and are known to contribute to virulence by allowing adhesion to epithelial cells (Comolli et al. 1999). Mutations conferring resistance to pilus-binding phage change pilus expression thereby preventing phage adsorbtion and infection. Typically, such mutations result in the loss of the pilus (unpiliated bacteria) or loss of pilus retraction (hyperpiliated bacteria) in P. aeruginosa and other bacteria (Bradley 1972a,b, 1974; Lythgoe & Chao 2003). While clearly of benefit in the presence of phage, such resistance mutations will probably carry a cost in the absence of phage because of the loss of pilus function. In apparent contrast, resistance to pilus-dependent phage Φ6 was not found to be costly in Pseudomonas syringae (Lythgoe & Chao 2003). However, these experiments were carried out in homogeneous (shaken) microcosms where motility and adhesion are probably less important than in spatially heterogeneous environments.
To investigate the role of PP7 in the evolution of P. aeruginosa diversity in experimental microcosms, we measured a number of bacterial traits. First, we measured colony morphology, a simple way to measure differences in a variety of ecologically relevant phenotypic traits that has been employed in a number of studies (Korona et al. 1994; Rainey & Travisano 1998; Mizoguchi et al. 2003). Second, we tested for resistance to phage. Third, we measured bacterial swimming motility which, though not directly reliant upon pili, is known to be affected by changes in pilus expression (Deziel et al. 2001; Mattick 2002) and allows for hyperpiliated and unpiliated bacteria to be distinguished. Fourth, we tested for change in niche preference, hyperpiliated P. aeruginosa isolates for example have been shown to grow as biofilms at the air–broth interface in static microcosms (Deziel et al. 2001; Chiang & Burrows 2003). Finally, and most importantly, we measured genotype fitness, or relative growth rates, under a variety of ecological conditions. We hypothesized that bacteriophage PP7 would increase P. aeruginosa diversity if there were multiple mechanisms of phage resistance with associated fitness costs.
2. Materials and methods
(a) Culturing techniques
Fifty-six microcosms (30 ml glass universal bottles containing 6 ml of Luria Broth (LB) media) were inoculated with approximately 105 cells of P. aeruginosa PAO1 (grown for 24 h at 37 °C in an orbital shaker at 200 r.p.m.). Half of the microcosms were also inoculated with approximately 106 clonal particles of PP7, a lytic bacteriophage. All microcosms were then grown in a static incubator, to promote phage binding, at 37 °C. Eight populations (four with phage, four without) were destructively sampled each day for 7 days. On day 7, an aliquot (60 μl) of each of the remaining eight populations was used to inoculate seven fresh microcosms that were then destructively sampled over the next 7 days. Every day, population densities were assayed by counting bacterial colonies on LB plates and counting phage plaques on soft LB agar lawns, containing exponentially growing ancestral PA01, and a sample stored in 20% glycerol at −80 °C.
(b) Resistance assays
The evolution of bacterial resistance to phage was determined daily by streaking 20 independent bacterial colonies across a line of ancestral phage that had previously been streaked on to an agar plate. Agar plates were incubated for 24 h. A colony was defined as resistant if there was no inhibition of growth (i.e. bacterial lysis); otherwise, it was defined as sensitive. As a control, an ancestral bacterial colony was also streaked on every plate.
(c) Measuring diversity
(i) Morphological diversity
In the first instance, host diversity was measured by determining the morphology of at least 100 bacterial colonies per population, per time point. Within-population diversity was then calculated as the complement of Simpson's index of concentration (1−λ; Simpson 1949):
where pi is the proportion of the ith morph and N is the total number of colonies sampled. This measure is the probability that two randomly selected colonies from a single population are morphologically different.
To investigate the link between pilus function, colony morphology and resistance, and to provide broader measures of bacterial diversity, 48 colonies from each population on day 3 of cycle 1 were subjected to the following assays in order.
(ii) Swimming assay
Each colony was inoculated into 1/100 strength semi-solid LB agar plates (0.35% agar) by stabbing directly into the medium. The diameter of bacterial growth was recorded after 6 h.
(iii) Niche preference
Each colony was inoculated into a fresh static microcosm and the region of dominant growth (either biofilm growth at the air–broth interface, or broth growth in the liquid phase) was recorded after 24 h.
(iv) Resistance assay
Each colony was streaked across a line of ancestral phage on an agar plate, as above.
(d) Competition experiments in the presence of phage
On the basis of the diversity assays, two classes of resistant mutant were identified. Six independent colonies representing each resistant class and six sensitive colonies were selected for further investigation. In order to assay the fitness of the resistant mutants, it was necessary to compete them with a marked ancestral strain that could be easily distinguished from wild-type. P. aeruginosa PAO1 ΔpanB (gift from Iain Lamont) requires pantothenate-supplemented media for growth; however, when pantothenate is abundant (i.e. in LB competition media), the deletion incurs only a small cost (mean fitness of P. aeruginosa PAO1 ΔpanB relative to P. aeruginosa PAO1 in LB competition media (W)=0.93±s.e.m. 0.025). Competition experiments between derived types and the marked ancestor were carried out in the presence of phage to determine the selective advantage of resistance.
Competitors were grown separately in microcosms supplemented with 0.0024 mM pantothenic acid for 24 h at 37 °C and shaken at 200 r.p.m., so they were in the same physiological state. A total of 107 cells were then inoculated into static microcosms supplemented with 0.0024 mM pantothenic acid at 1 : 1 ratios of derived : ancestral competing genotypes. Genotypes were competed for 24 h. Relative fitness (W) was calculated from the ratio of the estimated Malthusian parameters (m) of the competitors, m=ln (Nf/N0), where N0 is the starting density and Nf the final density (Lenski et al. 1991). Densities were determined by plate counts of colonies grown on LB agar supplemented with 4.8×10−4 μM pantothenic acid, and colonies grown on M9 agar plates. The M9 count gives the density of the derived type; the M9 count subtracted from the LB count gives the density of the marked ancestor.
(e) Individual growth rate experiments in the absence of phage
Paired competition experiments in the absence of phage were not possible, because some bacteria and phage could not be separated despite numerous attempts using several methods (serial dilution, Virkon treatment; data not shown). Growth rates were therefore measured individually. A single colony of each derived type and six colonies of the marked ancestor were each diluted into 100 μl of LB. Thirty microlitres of each was then inoculated into two separate static microcosms supplemented with 0.0024 mM pantothenic acid. Densities were determined at 4 and 24 h by plate counts of colonies grown on LB agar supplemented with 4.8×10−6 mM pantothenic acid. Growth rates were calculated as Malthusian parameters (m), m=ln (Nf /N0), where N0 is the starting density and Nf the final density (Lenski et al. 1991). Relative fitness (W) was calculated from the ratio of the estimated Malthusian parameters for each derived type relative to the average of the marked ancestor.
3. Results and discussion
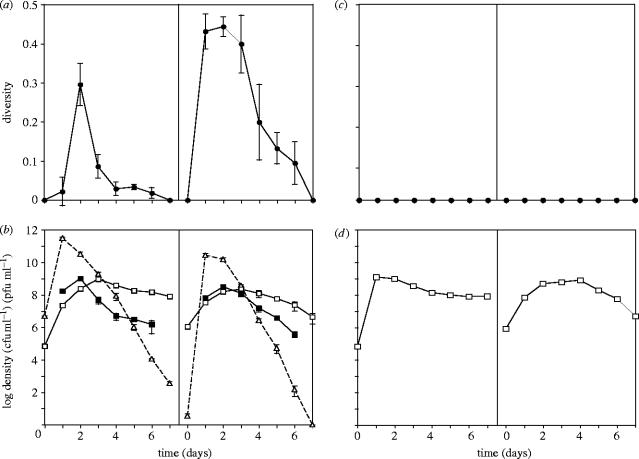
The bacteriophage PP7 caused an increase in morphological diversity in P. aeruginosa populations (figure 1a, panel 1; diversity averaged through time, Mann–Whitney test: w=26, p=0.03), whereas populations evolving in the absence of phage showed no morphological diversification (figure 1c, panel 1). The increase in diversity in populations with phage was due to the rapid evolution and rise to high frequency of small-rough (SR) colony variants. SR variants coexisted at measurable frequency with wild-type-like large-diffuse (LD) colony morphs from days 1 to 6, but then fell below measurable density by day 7 (figure 1b, panel 1). However, this increase in diversity was repeated upon inoculation of fresh microcosms with an aliquot of each day 7 population as the frequency of SR increased again (figure 1b, panel 2, diversity averaged through time, Mann–Whitney test, w=26, p=0.03).
Figure 1.
Mean±s.e.m. sympatric bacterial diversity (closed circles) in the presence (a) and absence (c) of phage. Diversities were calculated as the complement of Simpson's Index, based on the frequencies of colony morphology variants. Mean±s.e.m. densities of bacteria (LD morphology, open squares, SR morphology, closed squares) and phage (open triangles) are shown in the presence (b) and absence (d) of phage. Panel 1 of each figure shows cycle 1, panel 2 shows cycle 2.
The increases in diversity in the presence of phage coincided with viral blooms (figure 1b) and hence are probably the result of evolution of resistant bacteria. Consistent with this hypothesis, all SR variants tested were resistant to PP7, while LD morphs were either resistant (LDR; 39% of total LD bacteria tested for all time points) or sensitive (LDS; 61% of total LD bacteria tested for all time points). Furthermore, all bacteria tested in the early stages of diversification were resistant (100% resistant, days 1 and 2, both cycles), although the resistant proportion decreased through time to leave predominantly sensitive bacteria (LDS) at the end of each diversification cycle (80% LDS cycle 1, 85% LDS cycle 2). The different colony morphologies of SR and LDR bacteria suggest that they may utilise different resistance mechanisms. Conversely, the polymorphism among LD colony morphs indicates that colony morphologies conceal some of the diversity within the bacterial population.
Assays used to provide broader measures of bacterial diversity revealed very different patterns of bacterial motility, resistance and niche preference in the presence and absence of phage. Bacteria propagated in the absence of PP7 displayed little variation in swimming ability (figure 2b; mean variance 0.30). Furthermore, all tested bacteria inhabited the broth phase of microcosms and were sensitive to PP7. By contrast, bacteria propagated in the presence of PP7 showed much greater variation in swimming ability (figure 2a; mean variance 7.86; F-test of swim diameters in presence and absence of phage, F191,191=21.8, p<0.0001). Low range swimmers (0 to 4 mm swim diameter) tended to inhabit the air–broth interface of static microcosms by forming a mat and had SR morphologies. By contrast, mid- to high-range swimmers (8–10 mm swim diameter) tended to inhabit the broth phase, and had LD morphologies. In addition, resistance to PP7 was proportionally greatest in the extreme swimming classes.
Figure 2.
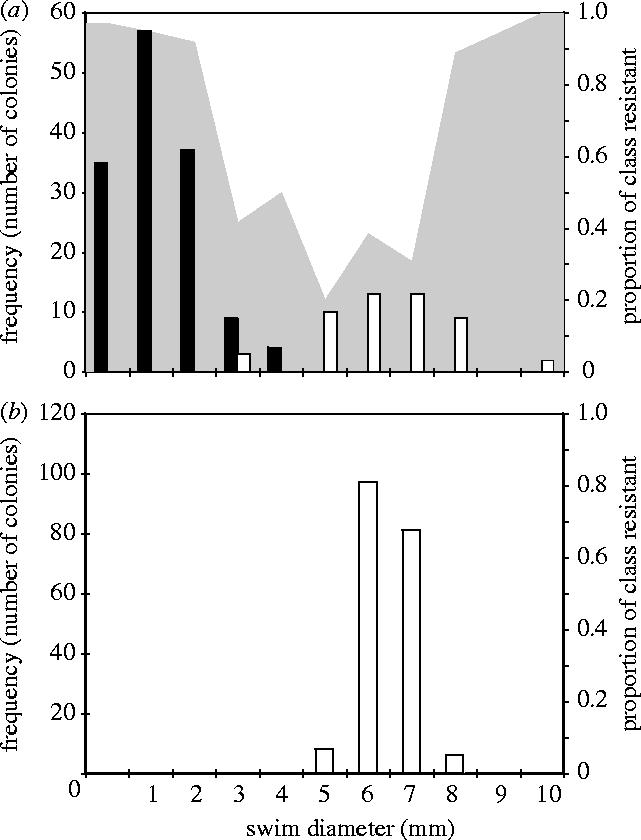
Bacterial diversity on day three of cycle one in the presence (a) and absence (b) of phage. Bars show the frequency of colonies with a given swim diameter that grow predominantly as a biofilm at the air–broth interface (black bars) or in the broth (white bars) of a static microcosm. The grey area shows the proportion of bacteria with a given swim diameter that are resistant to ancestral PP7.
On the basis of these diversity assays, two classes of resistant bacteria were identified. First, mat-forming resistant (MFR) bacteria that inhabit the air–broth interface of the microcosm, have the SR colony morphology on agar plates and small swim diameters relative to wild-type. Secondly, broth dwelling resistant (BDR) bacteria that inhabit the liquid phase of the microcosm, have the LD colony morphology on agar plates and large swim diameters relative to wild-type. Mutations conferring resistance to pilus-dependent phage often change pilus expression, typically resulting in loss of the pilus (unpiliated bacteria) or loss of pilus retraction (hyperpiliated bacteria) in P. aeruginosa and other bacteria (Bradley 1972a,b, 1974; Lythgoe & Chao 2003). The BDR phenotype is consistent with that of an unpiliated mutant (compared with P. aeruginosa ΔPilA; data not shown), while the MFR phenotype is consistent with that of hyperpiliated mutants observed in previous studies (Deziel et al. 2001; Haussler 2004). The phage population dynamics also support the presence of a hyperpiliated mutant; the precipitous decline in phage numbers after day 1 is suggestive of ‘soaking’ whereby phage irreversibly bind to non-retractile pili and are removed from the population. This phenomenon has been repeatedly observed in hyperpiliated Pseudomonas mutants (Bradley 1974; Lythgoe & Chao 2003).
Theory predicts the coexistence of sensitive and resistant types in the presence of parasites because of costs of resistance (Frank 1993; Sasaki 2000; Agrawal & Lively 2003). To investigate this, we measured relative fitness of evolved resistant and sensitive bacteria. Six colonies with MFR characteristics (i.e. SR colony morphology, low-range swimming, biofilm niche preference, resistant), six with BDR characteristics (i.e. LD colony morphology, high-range swimming, broth niche preference, resistant) and six sensitive bacteria (S) were selected for fitness assays. Paired competition experiments between evolved types and a genetically marked ancestor revealed that BDR and MFR-types were significantly fitter, relative to the ancestor, than S in the presence of PP7 (figure 3, BDR-types two-sample t-test, t=3.5, d.f.=5, p=0.009; MFR-types two-sample t-test, t=6.63, d.f.=5, p=0.0006). However, the relative fitness of BDR and MFR in the presence of phage did not differ (figure 3, two-sample t-test, t=0.64, d.f.=8, p=0.5). Individual growth rate experiments over 24 h in the absence of phage revealed BDR-types to be less fit than S types (figure 3, two-sample t-test, t=3.82, d.f.=8, p=0.003), demonstrating a cost of resistance. By contrast, MFR-types showed no fitness cost over 24 h (figure 3, two-sample t-test, t=0.78, d.f.=7, p=0.2).
Figure 3.
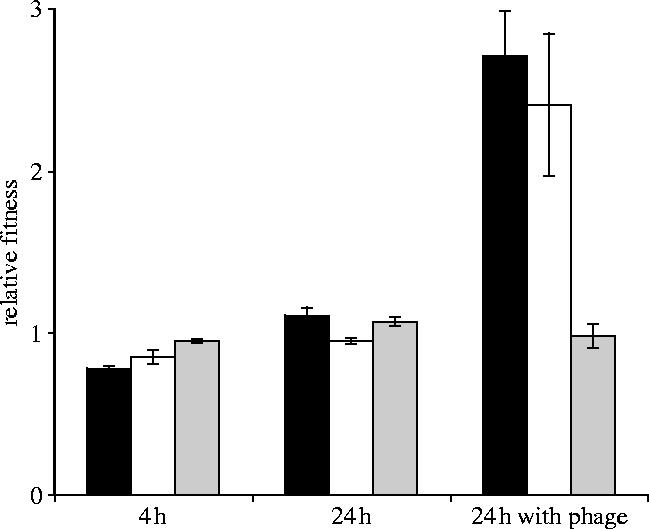
The relative fitness ±s.e.m. of MFR-type resistant mutants (black bars), BDR-type resistant mutants (white bars) and sensitive bacteria (light grey bars). All measurements are relative to a marked ancestor and were carried out over 4 and 24 h in the absence of phage, and 24 h in the presence of phage.
The lack of fitness cost of MFR is surprising and begs the question, why does costly BDR resistance evolve? One possibility is that we were not simply measuring fitness of MFR. LD morphology revertants were observed in all 24 h MFR competition experiments. It is possible that the cost of MFR-resistance was being ameliorated by rapid reversion to sensitivity. To test this, fitness was measured over 4 h to reduce the effect of evolved LD morphs (figure 3). Over the course of 4 h MFR-types were significantly less fit than S (two-sample t-test, t=8.34, d.f.=8, p<0.00001), as were BDR-types (two-sample t-test, t=2.71, d.f.=6, p=0.02).
We carried out reconstruction experiments to explicitly determine the stability of the different resistance mutants. BDR and MFR-types were propagated separately with PP7 in static microcosms for 3 days, after which time 24 colonies from each microcosm were assayed in terms of swimming ability, niche preference and resistance. These experiments revealed that BDR-type bacteria were stable; no reversion to sensitivity was observed among BDR-types and all bacteria tested retained resistance to PP7, high-range swimming and broth niche preference (figure 4b). By contrast, MFR-type bacteria were highly transient and diversified; the 3 day populations contained BDR and MFR type bacteria, as well as sensitive bacteria (figure 4a).
Figure 4.
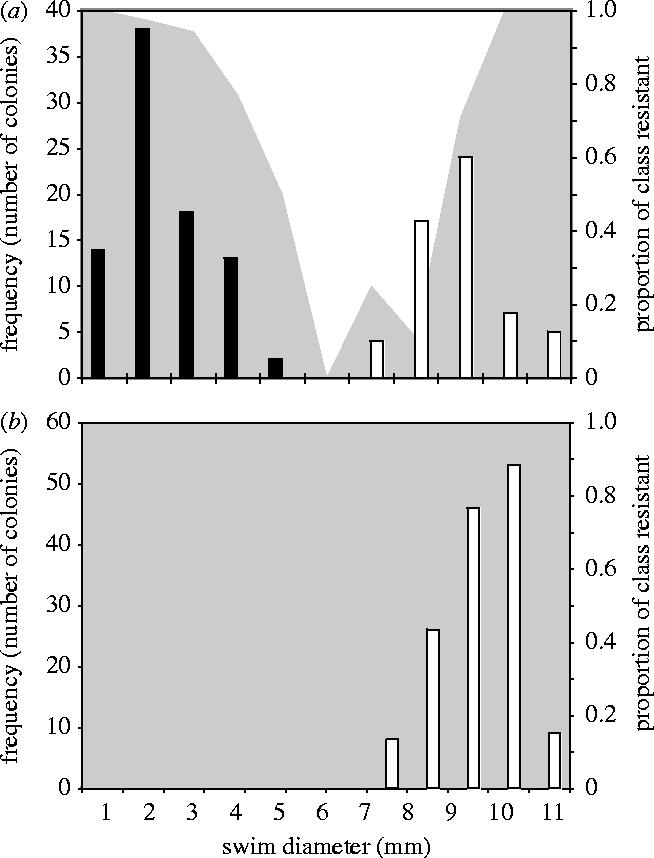
Bacterial diversity after 3 days in static microcosms inoculated with MFR-type resistant mutants (a) and BDR-type resistant mutants (b). Bars show the frequency of colonies with a given swim diameter that grow predominantly as a biofilm at the air–broth interface (black bars) or in the broth (white bars) of a static microcosm. The grey area shows the proportion of bacteria with a given swim diameter that are resistant to ancestral PP7.
The costs of both types of resistance displayed over 4 h suggests that there should be strong selection to revert to sensitivity in the absence of phage. Although the genetic causes of the observed phenotypes are not known, the inability of BDR-type mutants to revert suggests that reverse mutation or compensatory mutations to restore pilus production were not readily available. Conversely, the transience of MFR-type bacteria suggests that reverse mutation or compensatory mutations were readily available. Deziel et al. (2001) also observed reversion to wild-type in non-phage induced hyperpiliated mutants of P. aeruginosa 57RP. They suggested that hyperpiliation might be under the control of a phase variation mechanism. Phase variation generates diverse populations through high frequency, often reversible, genetic events, effectively acting as an on/off molecular switch (Henderson et al. 1999). The assertion is that phase variation regulates the switch from planktonic to biofilm growth, allowing rapid adaptation to low and high nutrient environments respectively (Deziel et al. 2001). The very rapid rise to high frequency of SR colony variants in this study (0 to 97.8%±s.e.m. 1.01 of the phage containing bacterial populations in 24 h), combined with the MFR to S reversion (20.3%±s.e.m. 3.3) observed in the reconstruction experiments is certainly suggestive of high frequency, reversible genetic change. Haussler et al. (2003) also observed reversion among hyperpiliated small-colony P. aeruginosa isolates from CF lungs. However, here strong selection and abundant mutations were thought to account for these dynamics and phase variation was not invoked (Haussler 2004). The rapid evolution and rise to high frequency of Pseudomonas fluorescens ‘wrinkly spreader’ colony mutants in static microcosms provides evidence that random mutation and strong selection alone can cause such evolutionary dynamics (Rainey & Travisano 1998).
If MFR bacteria are indeed hyperpiliated, then it is probable that the ability of MFR-type bacteria to revert to sensitivity carries some degree of risk, especially when parasite prevalence is high and sensitive bacteria will probably become infected. Furthermore, PP7 is known to bind covalently (i.e. irreversibly) to P. aeruginosa type-IV pili (Bradley 1966), and reversion while phage are bound would lead to infection. Phage infection of ancestor and MFR-type mixtures, even after dilution of ambient phage from MFR-type colonies, suggests that such events do occur. In this experiment, PP7 population density decreased over the course of 7 days, reducing the selective pressure to retain resistance. The domination of endpoint populations by sensitive bacteria suggests that in a changeable environment transient resistance allowing reversion to sensitivity may be advantageous. This is consistent with the view that phenotypically plastic characters, which should be made distinct from genetically reversible characters, should be more favoured in variable environments (de Jong & Gavrilets 2000; Agrawal 2001).
Our findings are of potential relevance to the study and treatment of P. aeruginosa as a human pathogen; however, the implications are as yet hypothetical. Although the prevalence of lytic bacteriophage in the lungs of CF patients is unclear, lysogenic bacteriophages have been shown to reach high densities (between 103 and 107 pfu ml−1 of CF patient sputum; Tejedor et al. 1982), and have been shown to induce colony morphology variation (mucoid colonies; Martin 1973; Miller & Rubero 1984). Crucially, a number of studies have described lysogenic bateriophages that, like PP7, infect via the type-IV pilus (Roncero et al. 1990; Budzik et al. 2004), including DMS3, which was isolated from a clinical strain of P. aeruginosa (Budzik et al. 2004). It is possible therefore, in the light of our results, that pilus-dependent phage such as DMS3 may drive the evolution of aberrant type-IV pilus mutants in clinical populations of P. aeruginosa.
Type-IV pili are known to contribute to P. aeruginosa virulence by allowing adhesion to epithelial cells (Hahn 1997; Comolli et al. 1999), yet P. aeruginosa isolates from CF lungs with both unpiliated (Kresse et al. 2003) and hyperpiliated phenotypes (Haussler et al. 2003) have been observed. In addition, type-IV pili are also responsible for bacterial twitching motility (Mattick 2002). Motility appears to decrease as lung infections proceed, one study reporting that 39% of P. aeruginosa isolates from chronic lung infections were non-motile (Mahenthiralingam et al. 1994). Loss of cell surface machinery, such as pili and flagella, may be adaptations to reduce virulence and evade phagocytes, or to conserve energy (Mahenthiralingam et al. 1994). Furthermore, type-IV pili are required for infection of P. aeruginosa by pilus-dependent transducing lysogenic phage (Roncero et al. 1990; Budzik et al. 2004). Transduction of antibiotic resistance by lysogenic phage has been shown to occur in P. aeruginosa (Blahova et al. 1994, 1998, 1999). Hence, type-IV pilus-dependent transducing lysogenic bacteriophages such as DMS3 (Budzik et al. 2004) may contribute to the spread of antibiotic resistance genes in P. aeruginosa populations.
It may be possible therefore to use lytic pilus-dependent phage in early stages of infection as a phage therapy (Levin & Bull 1996, 2004) to both reduce P. aeruginosa virulence in CF lung infections and to reduce the spread of antibiotic resistance genes in nosocomial populations by selecting for aberrant type-IV pili. The tendency for hyperpiliated P. aeruginosa to form biofilms (Deziel et al. 2001; Chiang & Burrows 2003; Haussler et al. 2003) may impair treatment as biofilms are known to display increased resistance to antibiotics (Drenkard & Ausubel 2002; Stewart 2002). However, the risk of reversion inherent to hyperpiliation (Deziel et al. 2001; Haussler et al. 2003) suggests that a continuous high dose of lytic pilus-dependent phage may favour the evolution of unpiliated bacteria that are unable to form a biofilm and are likely to display reduced virulence.
Acknowledgments
We thank Andrew Spiers and John Pannell for useful discussions; Iain Lamont for the kind gift of P. aeruginosa; Julie Stansfield for technical assistance; three anonymous reviewers for comments on the manuscript. We gratefully acknowledge funding from the Wellcome Trust.
References
- Abrams P.A. Character shifts of prey species that share predators. Am. Nat. 2000;156(Suppl. X):S45–S61. doi: 10.1086/303415. [DOI] [PubMed] [Google Scholar]
- Abrams P.A, Chen X. The effect of competition between prey species on the evolution of their vulnerabilities to a shared predator. Evol. Ecol. Res. 2002;4:897–909. [Google Scholar]
- Agrawal A.F. Phenotypic plasticity in the interactions and evolution of species. Science. 2001;294:321–326. doi: 10.1126/science.1060701. [DOI] [PubMed] [Google Scholar]
- Agrawal A.F, Lively C.M. Modelling infection as a two step process combining gene-for-gene and matching-allele genetics. Proc. R. Soc. B. 2003;270:323–334. doi: 10.1098/rspb.2002.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahova J, Hupkova M, Krcmery V., Sr. Phage F-116 transduction of antibiotic resistance from a clinical isolate of Pseudomonas aeruginosa. J. Chemother. 1994;6:184–188. doi: 10.1080/1120009x.1994.11741150. [DOI] [PubMed] [Google Scholar]
- Blahova J, Kralikova K, Krcmery V, Mikovicova A, Sr., Bartonikova N. Two high-frequency-transduction phage isolates from lysogenic strains of Pseudomonas aeruginosa transducing antibiotic resistance. Acta Virol. 1998;42:175–179. [PubMed] [Google Scholar]
- Blahova J, Kralikova K, Krcmery V, Sr., Jezek P. Transduction of antibiotic resistance in Pseudomomas aeruginosa: relationship between lytic and transducing activity of phage isolate AP-423. Acta Virol. 1999;43:395–398. [PubMed] [Google Scholar]
- Bohannan B.J.M, Lenski R.E. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology. 1997;78:2303–2315. [Google Scholar]
- Bohannan B.J.M, Lenski R.E. Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am. Nat. 1999;153:73–82. doi: 10.1086/303151. [DOI] [PubMed] [Google Scholar]
- Bohannan B.J.M, Lenski R.E. The relative importance of competition and predation varies with productivity in a model community. Am. Nat. 2000;156:329–340. doi: 10.1086/303393. [DOI] [PubMed] [Google Scholar]
- Bradley D.E. The structure and infective process of a Pseudomonas aeruginosa bacteriophage containing ribonucleic acid. J. Gen. Microbiol. 1966;45:83–96. [Google Scholar]
- Bradley D.E. Evidence for the retraction of Pseudomonas aeruginosa RNA phage pili. Biochem. Biophys. Res. Commun. 1972a;47:142–149. doi: 10.1016/s0006-291x(72)80021-4. [DOI] [PubMed] [Google Scholar]
- Bradley D.E. A study of pili on Pseudomonas aeruginosa. Genet. Res. 1972b;19:39–51. [Google Scholar]
- Bradley D.E. The adsorbtion of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with non-contractile pili. Virology. 1974;58:149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- Brennan A.L, Geddes D.M. Cystic fibrosis. Curr. Opin. Infect. Dis. 2002;15:175–182. doi: 10.1097/00001432-200204000-00013. [DOI] [PubMed] [Google Scholar]
- Brockhurst M.A, Rainey P.B, Buckling A. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. B. 2004;271:107–111. doi: 10.1098/rspb.2003.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Rainey P.B. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- Budzik J.M, Rosche W.A, Rietsch A, O'Toole G.A. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 2004;186:3270–3273. doi: 10.1128/JB.186.10.3270-3273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Levin B.R, Stewart F.M. A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology. 1977;58:369–378. [Google Scholar]
- Chiang P, Burrows L.L. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 2003;185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli J.C, Hauser A.R, Waite L, Whitchurch C.B, Mattick J.S, Engel J.N. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 1999;67:3625–3630. doi: 10.1128/iai.67.7.3625-3630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G, Gavrilets S. Maintenance of genetic variation in phenotypic plasticity: the role of environmental variation. Genet. Res. 2000;76:295–304. doi: 10.1017/s0016672300004729. [DOI] [PubMed] [Google Scholar]
- Deziel E, Comeau Y, Villemur R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 2001;183:1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebeli M, Dieckmann U. Evolutionary branching and sympatric speciation caused by different types of ecological interactions. Am. Nat. 2000;156(Suppl. X):S77–S101. doi: 10.1086/303417. [DOI] [PubMed] [Google Scholar]
- Drenkard E, Ausubel F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- Emori G.T, Gaynes R.P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.A. Evolution of host parasite diversity. Evolution. 1993;47:1721–1732. doi: 10.1111/j.1558-5646.1993.tb01264.x. [DOI] [PubMed] [Google Scholar]
- Govan J.R.W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H.P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- Haussler S. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 2004;6:546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- Haussler S, Ziegler I, Lottel A, von Gotz F, Rohde M, Wehmhohner D, Saravanamuthu S, Tummler B, Steinmetz I. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 2003;52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- Henderson I.R, Owen P, Nataro J.P. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- Holt R.D. Spatial heterogeneity, indirect interactions, and the coexistence of prey species. Am. Nat. 1984;124:377–406. doi: 10.1086/284280. [DOI] [PubMed] [Google Scholar]
- Korona R, Nakatsu C.H, Forney L.J, Lenski R.E. Evidence for multiple adaptive peaks from populations of bacteria evolving in a structured habitat. Proc. Natl Acad. Sci. USA. 1994;91:9037–9041. doi: 10.1073/pnas.91.19.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse A.U, Dinesh S.D, Larbig K, Romling U. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 2003;47:145–158. doi: 10.1046/j.1365-2958.2003.03261.x. [DOI] [PubMed] [Google Scholar]
- Lenski R.E, Levin B.R. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 1985;125:585–602. [Google Scholar]
- Lenski R.E, Rose M.R, Simpson S.C, Tadler S.C. Long term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2000 generations. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- Levin B.R, Bull J.J. Phage therapy revisited: the population biology of a bacterial infection and its treatment with bacteriophage and antibiotics. Am. Nat. 1996;147:881–898. [Google Scholar]
- Levin B.R, Bull J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004;2:166–173. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- Lyczak J.B, Cannon C.L, Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe K.A, Chao L. Mechanisms of coexistence of a bacteria and bacteriophage in a spatially homogenous environment. Ecol. Lett. 2003;6:326–334. [Google Scholar]
- Mahenthiralingam E, Campbell M.E, Speert D.P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.R. Mucoid variation in Pseudomonas aeruginosa induced by the action of phage. J. Med. Microbiol. 1973;6:111–118. doi: 10.1099/00222615-6-1-111. [DOI] [PubMed] [Google Scholar]
- Martin C, Ichou M.A, Massicot P, Goudeau A, Quentin R. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J. Clin. Microbiol. 1995;33:1461–1466. doi: 10.1128/jcm.33.6.1461-1466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Miller R.V, Rubero V.J. Mucoid conversion by phages of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J. Clin. Microbiol. 1984;19:717–719. doi: 10.1128/jcm.19.5.717-719.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi K, Morita M, Fischer C.R, Yoichi M, Tanji Y, Unno H. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 2003;69:170–176. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A, Canton R, Campo P, Banquero F, Bazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R.C, Garde G, Dayhuff T, Atkins J.F, Van Duin J. Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa: kinship to coliphages and conservation of regulatory RNA structures. Virology. 1995;206:611–625. doi: 10.1016/s0042-6822(95)80078-6. [DOI] [PubMed] [Google Scholar]
- Palleroni N.J. Pseudomonadaceae. In: Kreig N.R, Holt J.G, editors. Bergey's manual of systematic bacteriology. Williams & Wilkins Co; Baltimore, MD: 1984. pp. 141–199. [Google Scholar]
- Rainey P.B, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Roncero C, Darzins A, Casadaban M.J. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J. Bacteriol. 1990;172:1899–1904. doi: 10.1128/jb.172.4.1899-1904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. Host–parasite coevolution in a multilocus gene-for-gene system. Proc. R. Soc. B. 2000;267:2183–2188. doi: 10.1098/rspb.2000.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Ecological character displacement in adaptive radiation. Am. Nat. 2000;156(Suppl.):S4–S16. [Google Scholar]
- Schrag S.J, Mittler J.E. Host–parasite coexistence: the role of spatial refuges in stabilizing bacteria–phage interactions. Am. Nat. 1996;148:348–377. [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Stewart P.S. Mechnisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- Tejedor C, Foulds J, Zasloff M. Bacteriophages in sputum of patients with bronchopulmonary Pseudomonas infections. Infect. Immun. 1982;36:440–441. doi: 10.1128/iai.36.1.440-441.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.N. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 1999;153(Suppl.):S1–S14. [Google Scholar]