Abstract
The Cape Verde kite (Milvus milvus fasciicauda) is considered to be one of the rarest birds of prey in the world and at significant risk of extinction. For this reason there is great interest in both the taxonomic and the population status of this group. To help resolve its taxonomic status, we provide phylogenetic analyses based on three mitochondrial genes for a sampling of kites in the genus Milvus, including a broad geographical sampling of black kites (Milvus migrans), red kites (Milvus milvus), Cape Verde kite museum specimens collected between 1897 and 1924, and five kites trapped on the Cape Verde Islands during August 2002. We found that the historical Cape Verde kites, including the type specimen, were non-monophyletic and scattered within a larger red kite clade. The recently trapped kites from the Cape Verde Islands were all phylogenetically diagnosed as black kites. Our findings suggest that the traditional Cape Verde kite is not a distinctive evolutionary unit, and the case for species status, as recently suggested by others, is not supported. We do find support for recognition of at least one clade of yellow-billed kites, traditionally considered as a black kite subspecies, as a distinctive phylogenetic species.
Keywords: mitochondrial DNA, species, subspecies, reciprocal monophyly, phylogeography, conservation
1. Introduction
Delimiting species has important conservation implications. However, the criteria for doing so are actively debated (Claridge et al. 1997; Wheeler & Meier 2000; Hey 2001), and the availability of data and the application of those criteria, even among vertebrates, vary broadly (de Queiroz 1998; Soltis & Gitzendanner 1998; Wiens & Servedio 2000; Moritz 2002; Sites & Marshall 2003; Agapow et al. 2004; Zink 2004). We depend on defined taxa as indicators of evolutionarily significant entities that are roughly comparable within primary groups of organisms for use in allocating finite resources for conservation. Despite difficulties, recognition of species is essential and should be based on repeatable scientific analyses. As such, the use of molecular methods in assessing monophyletic groups has become a useful tool in helping to establish priorities for species conservation (Frankham et al. 2002; Avise 2004; Purvis et al. 2004).
The Cape Verde kite (Milvus milvus fasciicauda) is considered by some to be the rarest raptor in the world (Sangster 2000) and is restricted to the Cape Verde archipelago, approximately 500 km off the coast of Senegal. The Cape Verde kite was initially described by Hartert (1914) as a subspecies of red kite (Milvus milvus), possessing intermediate characteristics of plumage and size between red and black (Milvus migrans) kites. However, de Naurois (1987) hypothesized that the intermediate characters noted by Hartert (1914) were plesiomorphic and that the Cape Verde kite may have existed prior to the speciation of the present day red and black kites. The current distribution of the red kite is restricted to the western Palearctic and northern Africa, whereas black kites, consisting of numerous subspecies, are quite common throughout Europe, Australasia, and Africa, including the Cape Verde Islands. It has recently been proposed that the Cape Verde kite should be considered a distinct species (Milvus fasciicauda), endemic to the Cape Verde Islands, based on its geographical isolation and plumage characteristics (Hazevoet 1995; Sangster et al. 2003); however, any plumage distinctiveness remains to be demonstrated. Despite the lack of quantified morphological character differences, some recent ornithological sources have treated the Cape Verde kite as a distinct species (e.g. Furgeson-Lees & Christie 2001). To date, no thorough phylogenetic analysis has been conducted for taxa within the genus Milvus.
The traditional geographical range of the Cape Verde kite is distinct from that of red kites, though it is shared with black kites on the Cape Verde Islands (Hazevoet 1995). The historical distribution of the two kite taxa on the Cape Verde Islands as described in the literature are uncertain due to difficulties in field identification (Hazevoet 1995; Hille & Thiollay 2000), especially for juveniles. The potential existence of hybrids also complicates field identification (Hazevoet 1995). Unfortunately, over the past 50 years both Cape Verde kites and black kites have declined dramatically in abundance on the archipelago, and concern exists about the extinction of the Cape Verde kite (Bannerman & Bannerman 1968; Ortlieb 1988; Hille 1998; Hille & Thiollay 2000; Sangster 2000). After an extensive survey of most islands, Hille & Thiollay (2000) reported only two individual Cape Verde kites and a single black kite. Given their precipitous decline and Hazevoet's (1995) suggestion that Cape Verde kites be designated a distinct species, an effort to initiate a captive breeding programme developed and five birds were trapped and brought into captivity in August 2002. As a result of increasing concern about endangerment and possible extinction of the Cape Verde kite, we have undertaken the present study, which uses molecular methods to assess the genetic distinctiveness of Cape Verde kite specimens collected between 1897 and 1924, and compare them phylogenetically with five kites recently trapped on the Cape Verde Islands, as well as a geographically extensive sampling of red and black kites.
2. Material and methods
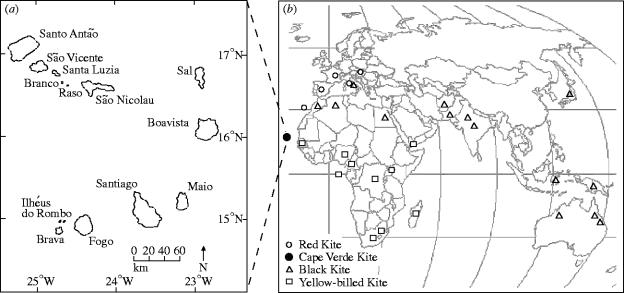
To infer phylogenetic relationships among Milvus taxa, a total of 43 individuals were sampled from 27 geographical locations (figure 1; Electronic Appendix part A). At least one specimen was sampled from each of the recognized subspecies of black and red kites (Mayr & Cottrell 1979), with the exception of Milvus migrans formosanus from Taiwan. Seven museum specimens of Cape Verde kite collected between 1897 and 1924, including the type specimen (AMNH cat# 531873), were included in the analyses to help ascertain the identity of five birds recently trapped (August 2002) on the islands of Maio and Boavista from the Cape Verde Islands (figure 1). Buteo buteo (mitochondrial DNA (mtDNA) genome, GenBank accession AF380305), Buteo jamaicensis (cytB, AY274044; ND2, AY987156), Haliastur indus (cytB, AY987309; ND2, AY987131) and Haliastur sphenurus (cytB, AY987310; ND2, AY987132; Lerner & Mindell, in press) were used as out-group taxa for the mitochondrial cytB and ND2 phylogenetic analyses.
Figure 1.
(a) Map of the Cape Verde Islands and (b) Milvus sample localities. Locality and species information are given in figure 2 and Electronic Appendix part A.
Total genomic DNA was extracted from blood or from toe pad tissue for museum specimens using a DNeasy tissue extraction kit (QIAGEN, Inc.). All work with museum samples was conducted in a facility used only for ancient DNA work at the University of Michigan Museum of Zoology, with protocols developed for ancient DNAs (Cooper & Poinar 2000). PCR amplifications were performed with Platinum Taq (Invitrogen) using primers designed for mitochondrial cytochrome B (cytB), NADH dehydrogenase subunit 2 (ND2), and the 5′ end of the control region (primer sequences are given in Electronic Appendix part B). We selected these relatively rapidly evolving mitochondrial genes based on their demonstrated suitability in other studies (Mindell & Thacker 1996; Yuri & Mindell 2002). We obtained cytB and ND2 sequences from 43 representative individuals, and control region sequences from a subset of 26 individuals (see Electronic Appendix part A). Potential contamination was monitored through the use of multiple extraction and PCR controls. PCR products were directly sequenced in both directions with ABI Big Dye Terminator chemistry, resolved on an ABI 3730 automated sequencer (Applied Biosystems), and deposited in GenBank (accession numbers AY994391–AY994502).
Sequences were aligned by eye. No indels were observed in cytB or ND2, and the few indels observed in the control region were readily resolved. We used both maximum parsimony (MP) and Bayesian inference using Markov Chain Monte Carlo (MCMC) sampling approaches to reconstruct phylogenies. MP trees were inferred using PAUP* 4.0b10 (Swofford 2003), and all character-state changes were equally weighted. All MP analyses were heuristic, with starting trees obtained by random taxa addition with 100 replicates, tree bisection and reconnection branch swapping, and support values for clades were calculated from 1000 bootstrap replicates. The relationship between mtDNA control region haplotypes was visualized with a minimum spanning cladogram estimated using the program TCS v. 1.17 (Clement et al. 2000) that provides the most parsimonious branch connections between haplotypes. Gaps in the mitochondrial control region were treated as a fifth character state.
Bayesian analyses were implemented using MrBayes v.3.0B4 (Huelsenbeck & Ronquist 2001). The best-fit model of evolution was determined by the hierarchical log-likelihood ratio test in ModelTest v.3.5 (Posada & Crandall 1998), which identified the TrN+G model for complete cytB and ND2 sequences combined. Because MrBayes cannot implement the TrN+G model, a similar model, (GTR+G), was used for the cytB and ND2 data. The general time reversible (GTR) model allows rates to vary across all six bidirectional DNA substitution types, and incorporates heterogeneity in base composition (Rodriguez et al. 1990). All Bayesian analyses were run for about five million generations following a burn-in period of about 50 000 generations, and phylogenetic hypotheses were sampled every 300 generations. Four chains in the Bayesian MCMC analyses were used in each of four independent runs. Each of the independent runs converged on similar optimal log likelihood scores and identical tree topologies. We use the criterion of monophyly for diagnosing distinctive evolutionary units, as have others, in assessing the status of threatened or endangered taxa (see Avise 2004).
3. Results
(a) Phylogeny for the genus Milvus
Based on 2146 bp of mitochondrial cytB and ND2 combined, 31 unique haplotypes were distinguished for the 43 Milvus samples, including 106 variable sites (98 transitions and 13 transversions). Based on 547 bp of mtDNA control region data, 22 unique haplotypes were found for 26 individual kites, including 48 variable sites (42 transitions, 3 transversions and 3 indels). MP and Bayesian analyses produced identical tree topologies in both separate and combined analyses of the cytB and ND2 datasets. The only differences between MP and Bayesian analyses involved levels of statistical support (figure 2). This similarity probably reflects the low levels of homoplasy in data for these relatively recently diverged taxa.
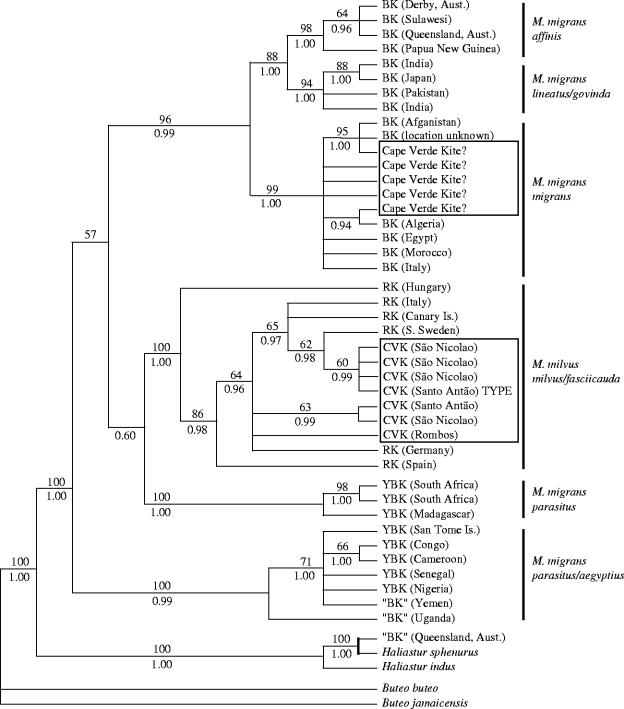
Figure 2.
Phylogeny for Milvus taxa based on mitochondrial cytB and ND2 including seven Cape Verde kite museum specimens (CVK) and five birds recently trapped on Cape Verde (‘Cape Verde kite?’), indicated by two boxes within the phylogeny. The topology shown is the Bayesian inference majority rule tree. MP bootstrap nodal support values (grater than 50%) are above branches and Bayesian posterior probability values are shown below. BK, black kite (M. migrans); YBK, yellow-billed kite (M. migrans parasitus/aegyptius); RK, red kite (M. milvus). The ‘BK’ from Queensland, Australia was determined to have been misidentified as BK and properly identified as Haliastur sphenurus.
The Milvine kite cytB and ND2 haplotypes form a series of clades that correspond only partially to the species and subspecies categories traditionally recognized for this genus (figure 2). The red kite (M. milvus) formed a single clade, while black kites (M. migrans) did not. Black kites exclusive of the yellow-billed subspecies (Milvus migrans parasitus/aegyptius) were monophyletic, with monophyly supporting three to four generally recognized, geographical subspecies. Yellow-billed kites are found in two well supported but non-sister clades (South Africa/Madagascar and African clades). Two museum specimens labelled as black kites (Milvus migrans migrans) from Yemen and Uganda were apparently misidentified as to subspecies since they cluster with high statistical support within a yellow-billed kite clade (M. m. parasitus/aegyptius; see §4).
GTR divergence estimates (cytB and ND2 dataset) between black kites were 0.6% between the Australian/New Guinea clade and a southeast Asian clade and 0.8% between a northern African/southern Europe/Afghanistan clade and the eastern Asia/Australian clade. However, the two yellow-billed kite clades (Africa and South Africa/Madagascar) were as divergent from the remaining black kite clades (1.7 and 1.9% GTR divergence, respectively) as the black kites were divergent from red kites (1.8% GTR divergence), and the two yellow-billed kite clades were as divergent from each other (1.8% GTR divergence) as they were from the other Milvus taxa. Within clade GTR divergence ranged from 0.14% for Milvus migrans affinis, 0.18% for Milvus migrans lineatus/govinda, 0.15% for M. migrans migrans, 0.98% for M. m. parasitus/aegyptius (or 0.26% for Africa and 0.12% for South Africa/Madagascar), and 0.24% for Milvus milvus milvus. Species level relationships were not strongly supported.
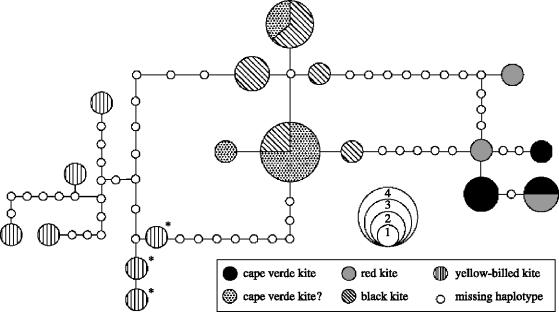
The mitochondrial control region data shown as a minimum spanning cladogram was consistent with the tree found in analyses of cytB and ND2 (figure 2), with a large number of intermediate haplotypes required to connect red, black and yellow-billed kite haplotype clusters (figure 3). Control region analysis showed two separate clusters of yellow-billed kite samples similar to what we observed with cytB and ND2; however, their association with either red or black kites differs, with yellow-billed kites being closer to black kites than to red kites (figure 3).
Figure 3.
A minimum spanning cladogram inferred from maximum parsimony based on mitochondrial control region. Each circle represents a single haplotype where the size of the circle corresponds to the number of individuals observed with that particular haplotype. Solid lines represent most parsimonious connections between haplotypes with a probability higher than 95%. Each connection between circles corresponds to a single point-mutation and open circles represent intermediate haplotypes missing in the sample. The three yellow-billed kite haplotypes represented with asterisks are from South Africa and Madagascar. Individuals sampled from the Cape Verde Islands in 2002 are designated ‘Cape Verde kite?’.
(b) Phylogenetic placement of the five birds recently caught on Cape Verde Islands
When we included the five birds recently trapped on Cape Verde Islands in the phylogenetic analyses for cytB and ND2, the topology and the support values for the other Milvus taxa remained largely unchanged in both MP and Bayesian analyses. All five birds clustered within the black kite clade of the subspecies M. migrans migrans; however, they did not form a monophyletic group themselves (figure 2). In fact, of the three haplotypes observed, none of the haplotypes were unique to the Cape Verde Islands. The ND2 and cytB GTR divergence estimate within the black kite clade (M. migrans migrans) including the five captive kites from Cape Verde was 0.11%. Analyses of control region sequences were consistent with the findings in figure 2, all five birds clustered within the black kite clade with three birds having identical haplotypes with two black kites and a fourth sharing an identical haplotype with two other black kites (figure 3).
(c) Phylogenetic placement of the Cape Verde kite
All seven museum specimens representing the Cape Verde kite, including the type specimen used in first describing the taxon, clustered within the red kite cytB and ND2 clade, although they did not form a monophyletic group (figure 2). Average pairwise GTR divergence within the ND2 and cytB red kite clade, including the seven museum Cape Verde kite specimens, was 0.17%. The mtDNA control region data analyses produced similar results with historical Cape Verde specimens being non-monophyletic and placed within the red kite clade (figure 3). One of the museum specimens had an identical control region haplotype observed with a red kite collected from the Canary Island population, which went extinct in the 1960s (Furgeson-Lees & Christie 2001).
4. Discussion
(a) Genetic distinctiveness of the Cape Verde kite?
The five birds recently trapped on the islands of Maio and Boavista of the Cape Verde Island archipelago are not members of the Cape Verde kite group originally described by Hartert (1914), but are more closely related to black kites from southern Europe and northern Africa (M. migrans migrans). This is well supported by an extensive phylogeographic survey of the genus Milvus including multiple representatives of all species and nearly all subspecies currently recognized. Furthermore, we have included seven museum specimens of Cape Verde kite, including the type specimen (AMNH cat# 531873), all of which were collected between 1897 and 1924. All of the Cape Verde kite museum specimens cluster with high statistical support with red kites based on cytB and ND2 (figure 2). The average pairwise GTR divergence between the two sets of birds collected on the Cape Verde Islands (museum versus contemporary samples; cytB and ND2 dataset) is 1.8%, which is similar to the divergence observed between red and black kites overall (1.8%).
Based on our molecular phylogenetic analyses, the historical Cape Verde kite samples do not denote a monophyletic group. Thus, our analyses do not support species status for the Cape Verde kite (M. fasciicauda) as recently proposed by Hazevoet (1995). The historical samples, including the type specimen, are nested within the red kite clade, and it is possible that there are no longer any descendents of this lineage breeding on the Cape Verde Islands. In this case, kites currently breeding on the Cape Verde Islands and regional black kites represent similar priorities for management and conservation. Both the historical Cape Verde samples from 1897 to 1924 and the recently collected samples (2002) denote non-monophyletic groups and neither warrants species recognition based on these analyses. Consequently, the initiation of a captive breeding programme for Cape Verde kites should be reevaluated.
We used a large number of museum specimens (i.e. dried skins) in our analyses and we were unable to amplify nuclear markers due to lower yields and greater degradation of nuclear DNAs. It would be ideal to have sequence data from both nuclear and mitochondrial genomes; however, mtDNA markers have been shown to recover phylogenetic relationships among taxa routinely in congruence with nuclear markers (e.g. Gains et al. 2005). Further, the coalescence time needed to produce monophyletic relationships using nuclear markers is likely to be longer than observed with mitochondrial markers in a random mating population (Moore 1995; Avise 2004; for exceptions see Hoelzer 1997). In fact, a recent study conducted with nuclear encoded allozymes found a very small genetic distance (D=0.009) between black and red kites in Germany (Schreiber et al. 2000). Nevertheless, it would be useful to confirm the findings reported here with nuclear markers, if additional samples become available. Further indicating the utility of mitochondrial data, our cytB and ND2 sequences were able to show that a museum specimen identified as a black kite (AMNH cat# 824394) was actually a whistling kite (H. sphenurus), subsequently confirmed by morphological traits, and two black kite specimens (AMNH cat# 789260 and 532038) were misidentified as to subspecies and were actually juvenile yellow-billed kites (figure 2), which are commonly misidentified because juveniles have black coloured bills (Furgeson-Lees & Christie 2001).
(b) Milvine phylogeography
Our initial objective was to assess the genetic distinctiveness of the Cape Verde kite based on historical and contemporary samples. However, as our study progressed, we found support for a number of phylogenetic subdivisions, variably in agreement or disagreement with geographical clustering and traditional taxonomic designations within the genus Milvus (figure 2). Most of the phylogenetic subdivisions were observed within the black kite species complex, which is not surprising given that this species is quite common and has a broad geographical distribution throughout the Old World (Furgeson-Lees & Christie 2001), and most of these corresponded with traditional taxonomic designations and geography.
Yellow-billed kites, however, yielded unexpected results. Traditionally, yellow-billed kites comprise two black kite subspecies (M. m. aegyptius and M. m. parasitus), which breed in northeastern and southern Africa, respectively. We found two well supported yellow-billed kite clades, with individuals from South Africa and Madagascar being readily distinguished from individuals collected further north in Africa, although these clades do not correspond to traditional taxonomy (figure 2). The divergence estimate between the two yellow-billed kite clades (1.8%) was similar to that between yellow-billed and other named black kite subspecies (1.7–1.9%), and similar to divergence estimates between red and black kites (1.8%) as well. The significant subdivision between black kites and yellow-billed kites is not surprising given that these two groups of birds possess observable morphological differences, most obviously the colour of the bill in adults. However, we are unaware of any studies suggesting a possible phylogenetic distinction between the two yellow-billed kite groups observed in this study. Further investigations using additional markers and more geographical sampling is warranted to verify this subdivision and help clarify sister relationships among the major groups in this genus. Our analysis indicates black kites as traditionally configured to be non-monophyletic (figure 2).
(c) Molecular phylogenetics and conservation priority
The Cape Verde kite type specimen and others like it collected during the late nineteenth and early twentieth century from the Cape Verde Islands denote a non-monophyletic group within a more inclusive red kite clade. It is not known whether or not the lineage represented by the type specimen is extinct, although kites of any kind are now uncommon to rare on the Cape Verde Islands, and the five kites trapped and sampled in 2002 are readily diagnosed as members of the black kite clade. Our recommendation is to continue monitoring of kites on the Cape Verde Islands, and to periodically assess their phylogenetic distinctiveness and the possibility of unique morphological or behavioural traits. Captive breeding efforts for black kites from the Cape Verde Islands, however, is unwarranted.
Application of the phylogenetic species concept, in which phylogenetically distinct groups are recognized as both evolutionary units and conservation units, has the potential to significantly increase the numbers of species vying for attention. Indeed, many phylogenetic analyses have identified evolutionarily distinctive taxa within groups previously thought to be undifferentiated (Ryan & Bloomer 1999; Omland et al. 2000; Burbidge et al. 2003; Zink & Weckstein 2003; Ravaoarimanana et al. 2004). However, if biologists are consistent with this approach, instances in which named species and subspecies are found to lack empirical, phylogenetic support can also be expected. This is the case for the Cape Verde kite based on our analyses presented here, and has been found in analyses of other taxa as well (e.g. Karl & Bowen 1999; Zink et al. 2000; Palkovacs et al. 2003; Rubinoff & Sperling 2004; Zink 2004). We use mtDNA as an empirical measure of temporal isolation between taxa (see Rosenburg 2003). We do not claim mtDNA to be a perfect arbiter of evolutionary distinctiveness or species status. A variety of phenomena can complicate interpretation of mtDNA results, including hybridization, male-biased gene flow and insensitivity to rapid adaptive change at other loci. It is, however, a useful marker readily compared among diverse species. We can say conclusively that mtDNA characters for the historical Cape Verde kite samples failed to demonstrate the monophyly and distinctiveness expected of historically and reproductively isolated taxa.
By including a geographically extensive sampling of black and red kites, we observed higher divergence between black kite subspecies (i.e. M. m. affinis and M. m. govinda/lineatus) than between red kites and historical Cape Verde kite samples. Further, we have identified two phylogenetically distinctive groups of black kites (yellow-billed kites from South Africa and Madagascar and from northern and central Africa) that were not recognized prior to this study and deserve further study and consideration as species.
Acknowledgments
We thank Jim Willmarth, Simon Thomsett, Mia Jessen, Andrew Heath and Sabine Hille for their help in obtaining samples, and gratefully acknowledge the American Museum of Natural History, New York; the University of Michigan Museum of Zoology; World of Birds Wildlife Sanctuary, South Africa; Center for Rehabilitation of Wildlife, South Africa and Jemima Parry-Jones from the National Birds of Prey Centre for allowing us to obtain samples for DNA work from study specimens in their care. Heather Lerner and Matt Klaver assisted in laboratory work. We thank the Peregrine Fund and the U.S. National Science Foundation for funding.
Supplementary Material
References
- Agapow P.-M, Bininda-Edwards O.R.P, Crandall K.A, Gittleman J.L, Mace G.M, Marshall J.C, Purvis A. The impact of species concept on biodiversity studies. Q. Rev. Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- Avise J.C. Sinauer Associates, Inc; Sunderland, MA: 2004. Molecular markers, natural history, and evolution. [Google Scholar]
- Bannerman D.A, Bannerman W.M. Oliver & Boyd; Edinburgh: 1968. History of the birds of the Cape Verde Islands. [Google Scholar]
- Burbidge M.L, Colbourne R.M, Robertson H.A, Baker A.J. Molecular and other biological evidence supports the recognition of a least three species of brown kiwi. Conserv. Genet. 2003;4:167–177. [Google Scholar]
- Claridge M.F, Dawah H.A, Wilson M.R, editors. Species: the units of biodiversity. Chapman & Hall; London: 1997. [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cooper A, Poinar H. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- de Naurois R. Les oiseaux de l'archipel du Cap Vert, peuplements, adaptations, endémisme. Bulletin de la Société Zoologique de France. 1987;112:307–326. [Google Scholar]
- de Queiroz K. The general lineage concept of species, species criteria, and the process of speciation: a conceptual unification and terminological recommendation. In: Howard D.J, Berlocher S.H, editors. Endless forms, species and speciation. Oxford University Press; Oxford: 1998. pp. 57–75. [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge: 2002. An introduction to conservation genetics. [Google Scholar]
- Furgeson-Lees J, Christie D.A. Christopher Helm; London: 2001. Raptors of the World. [Google Scholar]
- Gains C.A, Hare M.P, Beck S.E, Rosenbaum H.C. Nuclear markers confirm taxonomic status and relationships among highly endangered and closely related right whale species. Proc. R. Soc. B. 2005;272:533–542. doi: 10.1098/rspb.2004.2895. 10.1098/rspb.2004.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartert E. Milvus milvus fasciicauda subsp. n. Bull. Br. Ornithol. Club. 1914;33:89–91. [Google Scholar]
- Hazevoet C.J. BOU check-list No. 13. British Ornithologists’ Union; Tring, UK: 1995. The birds of the Cape Verde Islands. [Google Scholar]
- Hey J. Oxford University Press; New York: 2001. Genes, categories and species. [Google Scholar]
- Hille S. Zur Situation der Milane Milvus milvus fasciicauda (Hartert, 1915) und Milvus m. migrans (Boddaert, 1783) auf den Kapverdischen Inseln. J. Ornithol. 1998;139:73–75. [Google Scholar]
- Hille S, Thiollay J.-M. The imminent extinction of the kites Milvus milvus fasciicauda and Milvus m. migrans on the Cape Verde Islands. Bird Conserv. Int. 2000;10:361–369. [Google Scholar]
- Hoelzer G.A. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees revisited. Evolution. 1997;51:622–626. doi: 10.1111/j.1558-5646.1997.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F.R. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Karl S.A, Bowen B.W. Evolutionary significant units versus geopolitical taxonomy: molecular systematics of an endangered sea turtle (genus Chelonia) Conserv. Biol. 1999;13:990–999. [Google Scholar]
- Lerner, H. R. L. & Mindell, D. P. In Press. Phylogeny of eagles, Old World vultures and other Accipitridae based on nuclear and mitochondrial DNA. Mol. Phylogenet. Evol. [DOI] [PubMed]
- Mayr E, Cottrell G.W. Check-list of the birds of the World. 2nd edn. vol. 1. Museum of Comparative Zoology, Harvard University; Cambridge, MA: 1979. [Google Scholar]
- Mindell D.P, Thacker C.E. Rates of molecular evolution: phylogenetic issues and applications. Ann. Rev. Ecol. Syst. 1996;27:279–303. [Google Scholar]
- Moore W.S. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution. 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Omland K.E, Tarr C.L, Marzluff J, Boarman W, Fleischer R.C. Cryptic genetic variation and paraphyly in ravens. Proc. R. Soc. B. 2000;267:2475–2482. doi: 10.1098/rspb.2000.1308. 10.1098/rspb.2000.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlieb H. Milvus milvus fasciicauda in danger of extinction? Newsl. World Working Group Birds Prey Owls. 1988;8:12. [Google Scholar]
- Palkovacs E.P, Marschner M, Ciofi C, Gerlach J, Caccone A. Are the native giant tortoises from the Seychelles really extinct? A genetic perspective based on mtDNA and microsatellite data. Mol. Ecol. 2003;12:1403–1413. doi: 10.1046/j.1365-294x.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Posada D.A, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Purvis A, Brooks T, Gittleman J. Cambridge University Press; Cambridge: 2004. Phylogeny and conservation. [Google Scholar]
- Ravaoarimanana I.B, Tiedmann B, Montagnon D, Rumpler Y. Molecular and cytogenetic evidence for cryptic speciation within a rare endemic Malagasy lemur, the northern sportive lemur (Lipilemur septentrionalis) Mol. Phylogenet. Evol. 2004;31:440–448. doi: 10.1016/j.ympev.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Oliver J.F, Marin A, Medina J.R. The general stochastic model of nucleotide substitution. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Rosenburg N.A. The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution. 2003;57:1465–1477. doi: 10.1111/j.0014-3820.2003.tb00355.x. [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Sperling F.A.H. Mitochondrial DNA sequence, morphology and ecology yield contrasting conservation implications for two threatened buckmoths (Hemileuca: Saturniidae) Biol. Conserv. 2004;118:341–351. [Google Scholar]
- Ryan P.G, Bloomer P. The long-billed lark complex: a species mosaic in southwestern Africa. Auk. 1999;116:194–208. [Google Scholar]
- Sangster G. Taxonomic stability and avian extinctions. Conserv. Biol. 2000;14:579–581. [Google Scholar]
- Sangster G, van den Berg A.B, van Loon A.J, Roselaar C.S. Dutch avifaunal list: taxonomic changes in 1999–2003. Ardea. 2003;91:279–286. [Google Scholar]
- Schreiber A, Stubbe M, Stubbe A. Red kite (Milvus milvus) and black kite (M. migrans): minute genetic interspecies distance of two raptors breeding in a mixed community (Falconiformes: Accipitridae) Biol. J. Linn. Soc. 2000;69:351–365. [Google Scholar]
- Sites J.W, Marshall J.C. Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 2003;18:462–470. [Google Scholar]
- Soltis P.S, Gitzendanner M.A. Molecular systematics and the conservation of rare species. Conserv. Biol. 1998;13:471–483. [Google Scholar]
- Swofford D.L. 4th edn. Sinauer; Sunderland, MA: 2003. PAUP*, phylogenetic analysis using parsimony (* and other methods) [Google Scholar]
- Wiens J.J, Servedio M.R. Species delimitation in systematics: inferring diagnostic differences between species. Proc. R. Soc. B. 2000;267:631–636. doi: 10.1098/rspb.2000.1049. 10.1098/rspb.2000.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler Q.D, Meier R, editors. Species concepts and phylogenetic theory. Columbia University Press; New York: 2000. [Google Scholar]
- Yuri T, Mindell D.P. Molecular phylogenetic analysis of nine-primaried oscines (Aves: Passeriformes: Fringillidae) Mol. Phylogenet. Evol. 2002;23:229–243. doi: 10.1016/S1055-7903(02)00012-X. [DOI] [PubMed] [Google Scholar]
- Zink R.M. The role of subspecies in obscuring avian biological diversity and misleading conservation policy. Proc. R. Soc. B. 2004;271:561–564. doi: 10.1098/rspb.2003.2617. 10.1098/rspb.2003.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink R.M, Weckstein J.D. Recent evolutionary history of the fox sparrows (genus: Passerella) Auk. 2003;120:522–527. [Google Scholar]
- Zink R.M, Barrowclough G.F, Atwood J.L, Blackwell-Rago R.C. Genetics, taxonomy, and conservation of the threatened California gnatcatcher. Conserv. Biol. 2000;14:1394–1405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.