Abstract
Sister chromatid exchange (SCE) can occur by several recombination mechanisms, including those directly initiated by double-strand breaks (DSBs), such as gap repair and break-induced replication (BIR), and those initiated when DNA polymerases stall, such as template switching. To elucidate SCE recombination mechanisms, we determined whether spontaneous and DNA damage-associated SCE requires specific genes within the RAD52 and RAD3 epistasis groups in Saccharomyces cerevisiae strains containing two his3 fragments, his3-Δ5′ and his3-Δ3′::HOcs. SCE frequencies were measured after cells were exposed to UV, X-rays, 4-nitroquinoline 1-oxide (4-NQO) and methyl methanesulfonate (MMS), or when an HO endonuclease-induced DSB was introduced at his3-Δ3′::HOcs. Our data indicate that genes involved in gap repair, such as RAD55, RAD57 and RAD54, are required for DNA damage-associated SCE but not for spontaneous SCE. RAD50 and RAD59, genes required for BIR, are required for X-ray-associated SCE but not for SCE stimulated by HO-induced DSBs. In comparison with wild type, rates of spontaneous SCE are 10-fold lower in rad51 rad1 but not in either rad51 rad50 or rad51 rad59 double mutants. We propose that gap repair mechanisms are important in DNA damage-associated recombination, whereas alternative pathways, including a template switch pathway, play a role in spontaneous SCE.
INTRODUCTION
Sister chromatid exchange (SCE) results from recombination between two essentially identical DNA duplexes that are generated when a chromosome undergoes DNA replication. Sister chromatids are thus ideal templates for accurate gap repair of double-strand breaks (DSBs). The frequency of SCE is increased in both yeast (1) and mammalian cells (2) after exposure to DNA-damaging agents that generate DSBs.
Specific types of DNA damage impede replication fork progression and initiate SCE through complex pathways (3). DSBs resulting from replication fork collapse can initiate SCE and re-establish a replication fork; such a mechanism is referred to as break-induced replication (BIR) (4). Other proposed SCE mechanisms include replication fork reversal, which leads to template switching and repair that is initiated by single-strand gaps (5). Identifying eukaryotic DNA repair (RAD) genes required for SCE is one approach by which to elucidate these complex recombinational pathways.
We studied SCE in Saccharomyces cerevisiae, an organism in which double and triple mutants can be constructed easily to define epistastic relationships. RAD genes involved in recombinational repair of DSBs belong to the RAD52 epistasis group (6,7). RAD52 is required for most homologous recombination pathways involved in gene conversion or reciprocal exchange (8), as well as those involved in BIR (4,9). Genes required for gap repair mechanisms but which are not necessary for BIR include the recA homologs RAD51, RAD55, RAD57 (10), and RAD54, which encodes a Swi4/Snf1-related kinase (11); these genes are referred to as the RAD51 subgroup. Genes involved in BIR include RAD50 and RAD59 (4,9). Mammalian RAD50, RAD51, RAD52 and RAD54 orthologs, and RAD51 paralogs also function in DSB repair (12,13); however, there is no known mammalian RAD59 ortholog (13). Thus, understanding the genetic interactions of the yeast RAD genes in SCE may elucidate the genetics of SCE in mammals.
Genes in the RAD3 epistasis group (excision repair pathway), such as RAD1 and RAD10, also participate in recombinational repair of direct (14–16) and inverted repeats (10). Rad1/Rad10 nuclease cleaves 3′ single-stranded ends adjacent to double-stranded DNA (17); this activity functions in either the resolution or the stabilization of recombination intermediates that result from single-strand annealing (SSA) (18) or occur in the initiation of recombination (19).
Previous studies found that SCE requires genes belonging to both the RAD52 (1) and the RAD3 epistasis groups (5). In rad52 mutants, spontaneous SCE was reduced at least 10-fold (1,20), and DNA damage-associated SCE was abolished (5). However, rad51 mutants did not exhibit lower rates of spontaneous unequal SCE but were defective in DNA damage-associated SCE (1). Although rates of spontaneous SCE are similar in rad1 and wild-type strains, rad1 rad52 mutants exhibited a synergistic decrease in the rate of spontaneous SCE (5).
In this study, we determined the RAD dependence of unequal SCE in yeast. We found that the genetic requirements for DNA damage-associated SCE depend on the type of DNA lesion and that there are distinct RAD1-dependent and RAD51-dependent pathways for spontaneous SCE.
MATERIALS AND METHODS
Standard media for yeast culture
SC (synthetic complete, dextrose), SC-TRP (synthetic complete lacking tryptophan), SC-HIS (synthetic complete lacking histidine), SD (synthetic dextrose), YP (yeast extract, peptone), YPD (yeast extract, peptone, dextrose) and sporulation media are described by Sherman et al. (21). YPL medium contains YP medium with 2% lactate (pH 5.5); YPGlu medium contains YP medium with 2% ultra-pure glucose; YPGal medium contains YP medium with 2% ultra-pure galactose (Sigma, St Louis, MO). YPD (Kan) is YPD supplemented with 200 µg/ml of kanamycin sulfate. YPD (HU) is supplemented with 50 mM hydroxyurea (HU).
Yeast strains
A list of yeast strains and their relevant sources is shown in Table 1. All strains are derivatives of S288c. Strain YB203 is a Leu– derivative of the YB163 strain and contains tandem his3 fragments, his3-Δ5′ and his3-Δ3′::HOcs, at the trp1 locus. Plasmids used for the gene disruptions included pSTL11 (Δrad55::LEU2), pSM51 (Δrad57::LEU2) (22) and pNKY83 (Δrad50::hisG-URA3-hisG) (23). To make rad55, rad57 and rad50 null mutants, a HindIII fragment of pSTL11, a SacI fragment of pSM51 and an EcoRI–BglII fragment of pNKY83, respectively, were introduced into YB203 by one-step gene disruption (24), and the appropriate prototrophs were selected. We confirmed that the rad55, rad57 and rad50 mutants were sensitive to both X-rays and HU.
Table 1. Yeast strains.
| Lab name | Genotype | Source (synonym) |
|---|---|---|
| YA165 | MATα ura3-52 his3-Δ200 trp1-Δ1 leu2-Δ1 | F. Winston (FY250) |
| YA166 | MATa ura3-52 his3-Δ200 ade2-101 trp1-Δ1 leu2-Δ1 | F. Winston (FY251) |
| YA187 | MATa ura3-Δ0 his3-Δ1 lys2-Δ0 leu2-Δ0 met15-Δ0 rad59::KanMX | Resgen company (3756) |
| YA188 | MATα ura3-Δ0 his3-Δ1 lys2-Δ0 leu2-Δ0 rad59::KanMX | Resgen company (13756) |
| YA189 | MATα ura3-Δ0 his3-Δ1 lys2-Δ0 leu2-Δ0 rad4::KanMX | Resgen company (16158) |
| | ||
| Strains to measure SCEa | ||
| YB163 | MATa-inc ura3-52 his3-Δ200 ade2-101 lys2-801 trp1-Δ1 gal3– trp1::[his3-Δ3′::HOcs, his3-Δ5′] | This lab |
| YB213 | MATa-inc rad50::URA3 | rad50::URA3 disruption in YB163 |
| YB177 | MATa-inc rad51::URA3 | This lab |
| YB203 | MATa-inc ADE2 leu2-Δ1 | From cross of YB163 with YA165 |
| YB204 | MATα leu2-Δ1 | From cross of YB177 with YA165 |
| YB205 | MATα leu2-Δ1 rad51::URA3 | From cross of YB177 with YB204 |
| YB206 | MATa-inc ADE2 rad55::LEU2 | rad55::LEU2 disruption in YB203 |
| YB207 | MATα rad55::LEU2 | From cross of YB206 with YB204 |
| YB208 | MATa-inc ADE2 rad57::LEU2 | rad57::LEU2 disruption in YB203 |
| YB209 | MATα ADE2 rad57::LEU2 | From cross of YB208 with YB204 |
| YB211 | MATa-inc ADE2 rad55::LEU2 rad57::LEU2 | From cross of YB206 with YB209 |
| YB212 | MATa-inc ADE2 rad51::URA3 rad55::LEU2 rad57::LEU2 | From cross of YB211 with YB205 |
| YB214 | MATa-inc ADE2 rad51::URA3 rad50::URA3 | From cross of YB213 with YB205 |
| YB215 | MATα rad50::URA3 | From cross of YB213 with YB205 |
| YB216 | MATa-inc ADE2 rad54::KanMX | rad54::KanMX disruption in YB203 |
| YB217 | MATa-inc rad50::URA3 rad54::KanMX | From cross of YB216 with YB215 |
| YB218 | MATa-inc ADE2 ura3 rad59::KanMX | From cross of YB163 with YA188 |
| YB219 | MATa-inc ADE2 ura3 rad51::URA3 rad59::KanMX | From cross of YB205 with YA187 |
| YB220 | MATa-inc ura3 ADE2 rad50::URA3 rad59::KanMX | From cross of YB218 with YB215 |
| YB221 | MATa-inc ADE2 rad1::KanMX | rad1::KanMX disruption inYB203 |
| YB222 | MATa-inc rad50::URA3 rad1::KanMX | From cross of YB221 with YB215 |
| YB223 | MATα ADE2 rad51::URA3 rad1::KanMX | From cross of YB221 with YB205 |
| YB224 | MATa-inc rad50::URA3 rad51 rad1::KanMX | From cross of YB222 with YB223 |
| YB225 | MATa-inc ura3 rad4::KanMX | From cross of YB163 with YA189 |
| YB226 | MATa-inc ura3 rad4::KanMX rad51::URA3 | From cross of YB225 with YB205 |
aIf not indicated, the genotype is the same as YB163; mating-type added for clarity.
To delete RAD1 and RAD54 in YB203, genomic DNA from the corresponding ResGen yeast deletion strains was used as a template for PCR amplification; the appropriate DNA fragments were introduced into YB203 by one-step gene disruption (24); and kanamycin-resistant transformants were selected. The primers used to amplify the rad1::KanMX fragment were 5′-CTTTATTTTGCGACTTTTCTTCATC-3′ and 5′-TAATGAATATGATTGTGCGCTTCTA-3′. The primers used to amplify the rad54::KanMX fragment were 5′-CTTTATTTTGCGACTTTTCTTCATC-3′ and 5′-TAATGA ATATGATTGTGCGCTTCTA-3′. We confirmed that the rad1 and rad54 mutants were sensitive to UV and X-rays, respectively.
Strains containing a rad59::KanMX gene disruption were derived by a diploid cross of the ResGene strain 13756 (YA188) with the MATa-inc strain YB203, and the appropriate kanamycin-resistant meiotic segregant was obtained by tetrad dissection. We confirmed that the rad59 mutant was sensitive to X-rays and methyl methanesulfonate (MMS). The presence of the his3-Δ200 allele was confirmed by PCR analysis using the primers 5′-CACGGCAGAGACCAAT CAGTA-3′ and 5′-GCACTCCTGATTCCGCTAATA-3′.
Other single, double and triple mutants were constructed by crossing the appropriate haploid strains and screening the meiotic segregants for prototrophic phenotypes, antibiotic resistance and radiation sensitivities.
Determining rates of spontaneous recombination and frequencies of DNA damage-associated recombinants
The rates (events per cell division) of spontaneous SCE were determined by the method of median (25), using 11 independent colonies for each rate calculation as described previously (26). At least three independent rate calculations were done for each strain, and the significance of the differences was determined by the Mann–Whitney U-test (27).
Protocols used to measure the recombinogenicity of MMS, 4-nitroquinoline 1-oxide (4-NQO), UV and X-rays have been described previously (26). At least three independent experiments were performed at 30°C for each DNA-damaging agent. Because radiation resistance in rad55 and rad57 mutants is enhanced at 30°C, we also measured the MMS and 4-NQO stimulation of SCE in rad55 and rad57 at room temperature. After exposure to the DNA-damaging agent at room temperature, cells were plated on the appropriate medium and incubated at 30°C.
Induction of HO endonuclease
pGHOT-GAL3 (28), containing the HO gene under GAL control, was introduced into rad51, rad50, rad54, rad55, rad57 and rad59 strains by selecting for Trp+ transformants. After growth in SC-TRP medium, cells were diluted 1:10 in YPL and incubated for a minimum of 12 h. At a density of 107 cells/ml, glucose or galactose was added to a final concentration of 2%, and cells were incubated for 2 h, as previously described (1).
Characterizing repair events after HO induction
Three different mechanisms may participate in the repair of the DSB generated by HO endonuclease digestion at the trp1::his3-Δ3′::HOcs locus. These include non-homologous end joining (NHEJ), SSA and homologous recombination. SSA events generate a chromosomal deletion and a single his3 fragment lacking both 3′ and 5′ ends, rendering cells unable to generate His+ recombinants (Fig. 1). To determine frequencies of chromosomal deletions (SSA), cells were plated on YPD plates after HO induction. The surviving colonies were replica plated onto SC-HIS to measure the number of His– colonies that cannot generate His+ recombinants, and onto SC-TRP to measure the number of colonies that maintain the pGHOT-GAL3 plasmid. The presence of the single his3 fragment was confirmed by the presence of a 1.4 kb PCR product using primers 5′-CACGGCAGAGACCAATCAGTA-3′ and 5′-GCACTCCTGATTCCGCTAATA-3′ (Fig. 1).
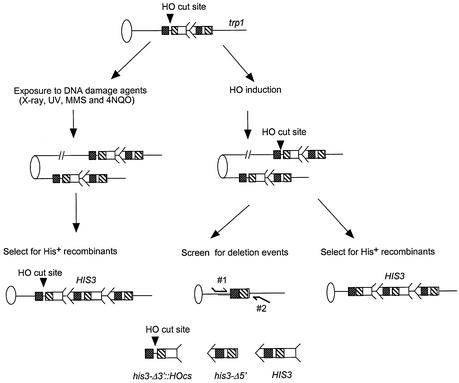
Figure 1.
Recombination assays used in this study. Ovals represent centromeres and lines represent chromosomes. For simplicity, the left arms of chromosomes are not included. An arrow and feathers together denote HIS3. As indicated on the bottom of the figure, the 5′ deletion lacks the feather and the 3′ deletion lacks the arrow. The two regions of the sequence identity shared by the his3 fragments are indicated by decorated boxes; dot-filled boxes indicate a region of 167 bp, and the hatched boxes indicate a region of ∼300 bp. The 117 bp HO-cut site (HOcs), as indicated by an arrowhead, is located between these sequences within the his3-Δ3′::HOcs fragment. The his3-truncated fragments are integrated into the trp1 locus on chromosome IV. His+ recombinants resulting from SCE were selected after exposure to DNA-damaging agents (left panel) or after the induction of HO endonuclease (right panel). The left panel indicates that the HO-cut site remains when there is no induction of HO endonuclease. The right panel indicates that HO endonuclease-induced DSB can be repaired by either the gap repair pathway, which generates SCE, or by the SSA pathway, which generates intrachromosomal deletions. The arrows indicate the sequences that are participating in gap repair. SCE events are measured by selecting for His+ recombinants while intrachromosomal deletions are screened, since the deletion renders cells unable to generate His+ recombinants. The primers are designed from regions flanking the trp1 locus, as indicated by the thin curved lines labeled 1 and 2, and their sequences are identified in Materials and Methods.
RESULTS
Recombination assays
Our aim was to elucidate the genetic pathways for SCE by measuring spontaneous and DNA damage-associated SCE in particular rad mutants that are defective in recombinational repair. To measure unequal SCE, we selected His+ recombinants that resulted from mitotic recombination between two tandem truncated his3 fragments (29) (Fig. 1). To target DSBs at the site of recombinational substrate, the HO-cut site was inserted in his3-Δ3′ at the trp1 locus as previously described (28). For each rad mutant, we first measured rates of spontaneous SCE and then measured frequencies of DNA damage-associated SCE after exposure to UV, X-rays, MMS and 4-NQO, or after HO induction. The rate of spontaneous SCE in the wild-type strain (YB163) was 1.2 × 10–6, which is consistent with previously reported data (1,29,30).
Spontaneous SCE does not require RAD55, RAD57, RAD54 or RAD59 and is modestly dependent on RAD50
Previous studies found that spontaneous SCE is not reduced in the rad51 null mutant (1). We measured the rate of spontaneous recombination in other rad mutants that are defective in gap repair of DSBs (Table 2). The rates of spontaneous SCE in rad51 (YB177), rad55 (YB206), rad57 (YB208), rad54 (YB216) single mutants, the rad55 rad57 double mutant (YB211), and the rad51 rad55 rad57 (YB212) triple mutant were the same as in wild type (YB163) (P > 0.05). These results are consistent with the idea that RAD51, RAD55, RAD57 and RAD54 participate in a single pathway for gene conversion (13), which, however, is not necessary for spontaneous SCE.
Table 2. Spontaneous rates of SCEs in S.cerevisiae rad mutants.
| Genotype (strains)a | Rate of recombination (×106)b | Ratioc |
|---|---|---|
| RAD (YB163) | 1.2 ± 0.2 | 1 |
| |
|
|
| RAD51 group | ||
| rad51 (YB177) | 1.5 ± 0.2 | 1.3 |
| rad54 (YB222) | 0.9 ± 0.1 | 0.8 |
| rad55 (YB206) | 1.2 ± 0.2 | 1.0 |
| rad57 (YB208) | 1.3 ± 0.2 | 1.1 |
| rad55 rad57 (YB211) | 1.3 ± 0.2 | 1.1 |
| rad51 rad55 rad57 (YB212) | 1.1 ± 0.1 | 0.9 |
| |
|
|
| RAD3 group | ||
| rad1 (YB219) | 2.6 ± 0.2 | 2.2 |
| rad4 (YB225) | 1.4 ± 0.1 | 1.1 |
| rad1 rad50 (YB220) | 1.8 ± 0.1 | 1.5 |
| rad4 rad51 (YB226) | 2.1 ± 0.1 | 1.6 |
| rad1 rad51 (YB221) | 0.1 ± 0.02 | 0.1 |
| |
|
|
| RAD50 and RAD59 | ||
| rad50 (YB215) | 0.7 ± 0.1 | 0.6 |
| rad59 (YB216) | 1.3 ± 0.2 | 1.1 |
| rad50 rad59 (YB218) | 0.6 ± 0.2 | 0.5 |
| rad50 rad54 (YB223) | 0.6 ± 0.1 | 0.5 |
| rad50 rad51 (YB214) | 0.6 ± 0.1 | 0.5 |
| rad51 rad59 (YB217) | 0.5 ± 0.06 | 0.4 |
| rad50 rad51 rad1 (YB224) | 0.05 ± 0.01 | 0.04 |
aAll strains are S288c(s) background. RAD disruptions are indicated. For full genotype, see Table 1.
bRate, number of events per cell division; n = 3.
cRatio, rate of SCE in mutant/rate of recombination in wild type.
One explanation for the RAD51 independence of SCE is that the major pathway for spontaneous SCE is either RAD59 or RAD50 dependent, because both genes have been shown to function in several RAD51-independent pathways for DSB repair, including BIR (31) and SSA (32). We measured spontaneous rates of SCE in rad50 (YB213) and rad59 (YB218) single mutants and in rad50 rad59 (YB220), rad50 rad51 (YB214) and rad59 rad51 (YB219) double mutants. The rate of spontaneous SCE in rad59 was the same as in wild type (P > 0.05) (Table 2), whereas in the rad51 rad59 double mutant, the rate of spontaneous SCE was reduced between 2- and 3-fold (P < 0.05) (Table 2). This indicates that RAD51 and RAD59 participate in independent SCE pathways. We found that the rates of spontaneous SCE in the rad50 single mutant, as well as in rad51 rad50, rad54 rad50 (YB217) and rad59 rad50 double mutants were reduced 2-fold compared with wild type (P < 0.05) (Table 2). Thus, mutations in rad50 conferred a modest reduction in rates of spontaneous SCE in combination with either rad59 mutations or null mutations in the RAD51 subgroup, implying that RAD50 functions in all pathways for spontaneous SCE.
The rate of spontaneous SCE is reduced 10-fold in rad1 rad51 mutants and 25-fold in rad1 rad51 rad50 mutants
An alternative RAD51-independent pathway involves RAD1, a gene that participates in both intrachromosomal (15) and ectopic recombination events (33). There is a 2-fold higher rate of spontaneous SCE (P < 0.05) in the rad1 mutant compared with wild type, possibly resulting from the mutant’s inability to excise potentially recombinogenic DNA adducts. The rate of spontaneous SCE in the rad1 rad50 mutant was the same as in wild type, whereas the rate of spontaneous SCE in the rad1 rad51 double mutant was reduced 10-fold compared with wild type. To determine whether RAD51-independent SCE requires other genes that participate in UV excision repair, spontaneous rates of SCE were measured in both rad4 (YB225) and the rad51 rad4 (YB226) double mutant; these rates were the same as those of wild type (Table 2). Because RAD4 participates in the initial steps of UV excision repair (34,35), these data suggest that the RAD1-dependent recombination pathway does not require most genes in the UV excision repair pathway and might result from Rad1 nuclease activity. Thus, there is a strong requirement for RAD1 in RAD51-independent SCE.
Although the rad50 mutation conferred a modest decrease in SCE when combined with rad1, rad51, rad50 and rad59 mutations, the rad1 rad51 rad50 triple mutant exhibited a 25-fold decrease in spontaneous SCE compared with wild type. These data indicate that in the absence of both RAD51 and RAD1, a RAD50-dependent pathway, such as BIR, might play a major role in spontaneous SCE.
X-ray-associated SCE requires RAD50, RAD54, RAD55 and RAD57 and is reduced in the rad59 mutant
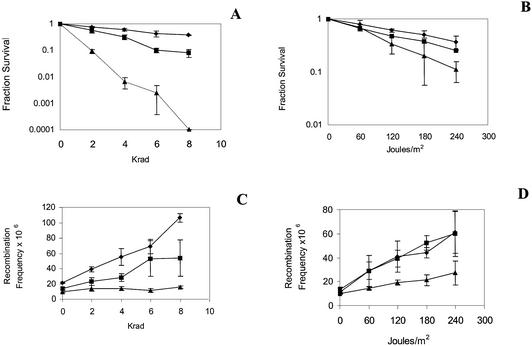
Previous studies have demonstrated that chemical agents and radiation, including 4-NQO, MMS, UV and X-rays, stimulate SCE by RAD51-dependent (1) and RAD52-dependent recombination mechanisms (1,29,30). We investigated whether other genes within the RAD52 epistasis group are also required for the stimulation of SCE after exposure to DNA-damaging agents. Because all the genes within the RAD52 epistasis group initially were defined as conferring haploid resistance to ionizing radiation (7), it would be expected that all of the corresponding rad mutants would also be defective in stimulating SCE after X-ray exposure. As in previous studies (1), we observed a significant increase in SCE frequencies after wild-type cells were exposed to 4, 6 or 8 krads of X-rays (Fig. 2); a maximum 5-fold increase in SCE frequencies was observed after 8 krad exposure in the wild-type strain. However, no significant increase in SCE frequencies was observed after the rad55, rad57, rad54 (data not shown) and rad50 mutants were exposed to X-rays (Fig. 2). All these mutants were very X-ray sensitive. The modest increase in SCE frequencies in the rad59 mutant after exposure to X-rays also correlates with the moderate X-ray sensitivity of the rad59 mutant (Fig. 2). These results indicate that there is a correlation between X-ray resistance and X-ray-associated SCE in rad mutants defective in recombinational repair.
Figure 2.
DNA damage-associated SCE after wild type, rad59 and rad50 strains were exposed to UV and X-rays. Data on the left were obtained when strains were exposed to X-rays. Data on the right were obtained when strains were exposed to UV. (A) Fraction survival after cells were exposed to X-rays. (B) Fraction survival after cells were exposed to UV. (C) Frequency of His+ recombinants (number of His+ recombinants/c.f.u.) plotted against the X-ray dose. (D) Frequency of His+ recombinants (number of His+ recombinants/c.f.u.) plotted against the UV dose. Filled diamonds = wild type (YB163); filled squares = rad59 mutant (YB218); filled triangles = rad50 mutant (YB213).
UV-associated SCE requires RAD54, RAD55, RAD57 and RAD50, but not RAD59
Whereas recombinational repair mutants are only modestly sensitive to UV, the rad51 mutant exhibits reduced levels of UV-associated SCE (1). We therefore measured UV-associated SCE in rad50, rad59, rad54, rad55 and rad57 mutants. After exposure to UV, we observed a maximum 5-fold increase in SCE frequencies in wild type. No significant increase in SCE frequencies was observed in the rad54, rad55, rad57 (data not shown) and rad50 mutants after exposure to UV (Fig. 2). However, frequencies of SCE were increased in the rad59 mutant at all levels of UV exposure (Fig. 2). Thus, there are different genetic requirements for UV-associated SCE and X-ray-associated SCE.
MMS- and 4-NQO-associated SCE requires RAD54, RAD55, RAD57 and RAD50, but is modestly defective in the rad59 mutant
4-NQO and MMS are considered to be DNA-damaging agents that are UV mimetic and X-ray mimetic, respectively (35,36). We therefore investigated whether the chosen rad mutants exhibited higher SCE frequencies after exposure to 4-NQO and MMS. In wild type, the frequencies of His+ recombinants increased a maximum of 11- and 8-fold above the spontaneous frequency after exposure to 4-NQO and MMS, respectively. rad54, rad55 and rad57 mutants are all defective in 4-NQO- and MMS-associated SCE compared with wild type (Tables 3 and 4). Compared with frequencies of spontaneous SCE (Tables 3 and 4), frequencies of DNA damage-associated SCE were 3-fold higher (P < 0.05) in the rad55 mutant and in the rad55 rad57 double mutant after exposure to 2 mM MMS, and 3-fold (P < 0.05) higher in the rad55 rad57 double mutant after exposure to 10 µM 4-NQO. This slight increase is probably related to temperature because, when the rad55 single mutant and the rad55 rad57 double mutant were exposed to either MMS or 4-NQO at room temperature, DNA damage-associated SCE frequencies were increased <2-fold (Tables 3 and 4). No increase in DNA damage-associated SCE was observed when the rad51 rad55 rad57 triple mutant was exposed to either MMS or 4-NQO (Tables 3 and 4). These results are consistent with the idea that DNA damage-associated SCE occurs by a gap repair mechanism that requires the RAD51 subgroup (13) but, in the absence of RAD55, a modest stimulation of recombination is still possible.
Table 3. Stimulation of SCE by 4-NQO in rad50, rad54, rad51, rad55, rad57 and rad59 mutant strains.
| Genotype (strain)a | His+/c.f.u. × 106 (average)b | ||||
|---|---|---|---|---|---|
| Control | 2 µM 4-NQO | Fold | 10 µM 4-NQO | Fold | |
| |
|
(survival%) |
increasec |
(survival%) |
increasec |
| RAD (YB163) | 15 ± 4 | 74 ± 20 (71) | 5 | 164 ± 48 (30) | 11 |
| |
|
|
|
|
|
| RAD51 group | |||||
| rad51 (YB177) | 20 ± 4 | 19 ± 4 (54) | <1 | 22 ± 8 (13) | 1.1 |
| rad54 (YB216) | 10 ± 2 | 13 ± 4 (53) | 1.1 | 17 ± 2 (14) | 1.7 |
| rad55 (YB206) | 15 ± 4 | 16 ± 1 (62) | 1.1 | 23 ± 6 (10) | 1.5 |
| rad57 (YB208) | 17 ± 10 | 14 ± 1 (70) | <1 | 24 ± 11 (10) | 1.4 |
| rad55 rad57 (YB211) | 14 ± 3 | 23 ± 1 (70) | 1.6 | 39 ± 5 (9) | 2.8 |
| rad55 rad57 (23°C) | 9 ± 2 | 10 ± 3 (68) | 1.1 | 11 ± 3 (10) | 1.2 |
| rad51 rad55 rad57 (YB212) | 15 ± 2 | 22 ± 3 (39) | 1.5 | 14 ± 4 (4) | <1 |
| |
|
|
|
|
|
| RAD50 and RAD59 | |||||
| rad50 (YB213) | 9 ± 4 | 17 ± 8 (52) | 1.9 | 46 ± 19 (6) | 5.1 |
| rad59 (YB218) | 16 ± 1 | 40 ± 10 (80) | 4.0 | 64 ± 23 (21) | 6.2 |
aFor complete genotype, see Table 1.
bFrequency is events/viable cells; experiments were performed at 30°C unless otherwise stated; n = 3.
cHis+ frequency with agent/spontaneous His+ frequency.
Table 4. Stimulation of SCE by MMS in rad50, rad54, rad51, rad55, rad57 and rad59 mutant strains.
| Genotype (strain)a | His+/c.f.u. × 106 (average)b | ||||
|---|---|---|---|---|---|
| Control (survival%) | 2 mM MMS increasec | Fold (survival%) | 10 mM MMS increasec | Fold | |
| RAD (YB163) | 15 ± 5 | 59 ±12 (92) | 4 | 118 ± 27 (60) | 8 |
| | |||||
| RAD51 group | |||||
| rad51 (YB177) | 21 ± 9 | 35 ± 10 (48) | 1.7 | 37 ± 12 (4) | 1.8 |
| rad54 (YB216) | 13 ± 2 | 19 ± 4 (50) | 1.5 | 17 ± 1 (4) | 1.3 |
| rad55 (YB206) | 10 ± 2 | 24 ± 6 (65) | 2.6 | 31 ± 8 (11) | 3.1 |
| rad55 (23°C) | 9 ± 2 | 15 ± 2 (64) | 1.7 | 21 ± 4 (4) | 2.3 |
| rad57(YB208) | 17 ± 8 | 33 ± 9 (50) | 1.9 | 22 ± 7 (11) | 1.3 |
| rad55 rad57 (YB211) | 13 ± 3 | 39 ± 5 (64) | 3 | 24 ± 5 (12) | 1.8 |
| rad55 rad5(23°C) | 9 ± 1 | 13 ± 4 (64) | 1.4 | 12 ± 5 (6) | 1.3 |
| rad51 rad55 rad57 (YB212) | 17 ± 4 | 25 ± 3 (39) | 1.5 | 36 ± 16 (2) | 2.1 |
| | |||||
| RAD50 and RAD59 | |||||
| rad50 (YB213) | 10 ± 3 | 22 ± 13 (22) | 2.2 | 38 ± 15 (1.3) | 3.8 |
| rad59 (YB218) | 16 ± 1 | 76 ± 4 (66) | 4.8 | 64 ± 23 (35) | 4.0 |
aFor complete genotype, see Table 1.
bFrequency is events/viable cells; experiments were performed at 30°C unless otherwise stated; n = 3.
cHis+ frequency with agent/spontaneous His+ frequency.
We next measured SCE frequencies in rad50 and rad59 after exposure to 4-NQO and MMS and compared them with those obtained after UV and X-ray exposure. DNA damage-associated SCE was defective in rad50 after mutants were exposed to low concentrations of MMS or 4-NQO, which is consistent with the observations that rad50 mutants are defective in UV- and X-ray-stimulated recombination (Tables 3 and 4). After rad59 cells were exposed to low concentrations of MMS (2 mM) and 4-NQO (2 µM), we observed SCE frequencies that were 5- and 4-fold higher, respectively, than spontaneous SCE frequencies, similar to increases that are observed in wild type (Tables 3 and 4). However, after rad59 cells were exposed to high concentrations of 4-NQO (10 µM) and MMS (10 mM), a more modest increase was observed compared with wild type. Thus, RAD50 is required for all DNA damage-associated recombination; however, DNA damage-associated SCE can be detected in rad59 mutants.
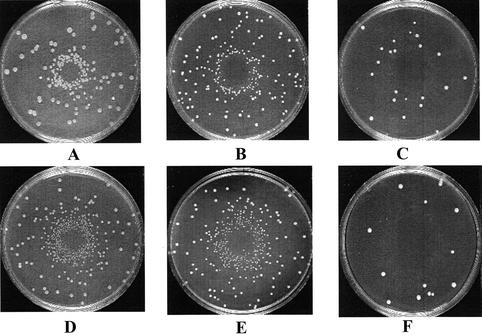
To visualize the 4-NQO- and MMS-associated SCE events in rad59 mutants, we performed a simple plate assay. Wild type, rad50 and rad59 cells were plated on SC-HIS plates, and the chemical compound was diffused from the center of the plate. We observed a halo of His+ recombinants on plates inoculated with wild type or rad59 cells, but not on plates containing rad50 cells (Fig. 3), even when lower amounts of 4-NQO (<3 nmol) and MMS (<22 nmol) were diffused from the center of the plates (data not shown). The halo of recombinants is observed when there are more viable recombinants per total number of inoculated cells, not when there are simply more recombinants per viable cell. These results support the idea that exposure to MMS and 4-NQO can stimulate SCE events in rad59 but not in rad50 mutants.
Figure 3.
Plate assay demonstrating the DNA damage inducibility of SCE in wild-type and rad59 strains but not in rad50 strains. Each SC-HIS plate contains a lawn of 107 cells, and the DNA-damaging agent was placed in the center. (A) Wild-type strain (YB163) + 0.2 µl of 4-NQO (15 mM). (B) rad59 mutant + 0.2 µl of 4-NQO (15 mM). (C) rad50 mutant + 0.2 µl of 4-NQO (15 mM). (D) Wild-type strain + 0.2 µl of MMS (10.5 M). (E) rad59 mutant + 0.2 µl of MMS (10.5 M). (F) rad50 mutant + 0.2 µl of MMS (10.5 M).
Stimulation of SCE by HO endonuclease-induced DSBs requires RAD51, RAD55 and RAD57, but not RAD50 and RAD59
Because exposure to UV, X-ray, 4-NQO and MMS generates a variety of DNA lesions and confers significant lethality in all rad mutants, we examined whether stimulation of SCE by a site-specific DSB generated by an inducible HO endonuclease would be defective in rad mutants. The DSB could be repaired by SSA, SCE or NHEJ (Fig. 1). All the rad strains contain the MATa-inc allele so that there is no HO endonuclease digestion at the MAT locus.
We observed an ∼13-fold increase in the SCE frequency in wild type after HO induction, but no significant increases in rad54, rad55 and rad57 mutants (Table 5). For all these mutants, the percentage viability (mean, 77 ± 4%) after HO induction is similar. Thus, similar to rad51 mutants, stimulation of SCE by HO-induced DSBs is defective in rad55, rad54 and rad57 mutant strains.
Table 5. Stimulation of SCE by HO-induced DSBs in rad50, rad54, rad55, rad57 and rad59 mutants.
| Genotype (strain)a | % viability after | His+ recombinants/Trp+ c.f.u. × 105 | Trp+ c.f.u./total c.f.u. (%) | |||
|---|---|---|---|---|---|---|
| HO inductionb | Before HO inductionc | After HO inductiond | Fold increasee | Before HO induction | After HO induction | |
| RAD (YB163) | 84 ± 18 | 5.8 ± 1 | 76 ± 13 | 13 | 94 ± 3 | 91 ± 6 |
| | ||||||
| RAD59 and RAD50 | ||||||
| rad59 (YB218) | 70 ± 13 | 7.4 ± 1 | 137 ± 21 | 19 | 95 ± 1 | 92 ± 3 |
| rad50 (YB213) | 41 ± 3 | 14 ± 4 | 217 ± 25 | 16 | 96 ± 1 | 83 ± 2 |
| | ||||||
| RAD51 group | ||||||
| rad51 (YB177) | 77 ± 14 | 8.2 ± 2 | 7 ± 3 | <1 | 83 ± 8 | 75 ± 6 |
| rad54 (YB216) | 77 ± 17 | 8.2 ± 2 | 7 ± 3 | <1 | 93 ± 2 | 91 ± 1 |
| rad55 (YB206) | 84 ± 14 | 1.1 ± 0.2 | 1.3 ± 0.5 | 1.2 | 93 ± 5 | 90 ± 8 |
| rad57 (YB208) | 75 ± 25 | 1.5 ± 0.4 | 1.3 ± 0.3 | <1 | 96 ± 3 | 92 ± 5 |
aFor complete genotype, see Table 1.
bTrp+ c.f.u. after HO induction/Trp+ c.f.u. before HO induction × 100%.
cHis+ recombinants before HO induction/Trp+ c.f.u. before HO induction.
dHis+ recombinants after HO induction/Trp+ c.f.u. after HO induction.
eHis+ frequency after HO induction/His+ frequency before HO induction.
Equivalent lethality of HO-induced DSBs in wild type and in rad54, rad55 and rad57 mutants suggests that alternative pathways for repair of the DSB at the HO-cut site, such as NHEJ or SSA, are the preferred repair pathways. SSA would generate a deletion and a single his3 fragment (Fig. 1) that could be confirmed by PCR; thus, His– colonies would not be able to generate His+ recombinants. We therefore determined whether unselected Trp+ His– c.f.u.s arising after HO induction could generate His+ recombinants, and we observed that ∼77% (average) could not generate His+ recombinants (Table 6). Thus, SSA was stimulated in both wild type and in rad54, rad55 and rad57 mutants. These data suggest that the HO-induced DSB at trp1::his3-Δ3′ is repaired by similar mechanisms in rad54, rad55, rad57 and rad51 mutants.
Table 6. Stimulation of intrachromosomal deletions by HO-induced DSBs in rad50, rad51, rad54, rad55, rad57 and rad59 mutant strains.
| Genotype (strain)a | His– c.f.u. containing deletions (%)b (total His– c.f.u./ total Trp+ c.f.u.) | Fold differencec |
|---|---|---|
| RAD (YB163) | 75 (124/165) | 1.0 |
| |
|
|
| RAD59 and RAD50 | ||
| rad59 (YB218) | 77 (381/499) | 1.0 |
| rad50 (YB213) | 89 (371/416) | 1.2 |
| |
|
|
| RAD51 group | ||
| rad51 (YB177) | 76 (93/123) | 1.0 |
| rad54 (YB216) | 79 (244/309) | 1.1 |
| rad55 (YB206) | 68 (137/202) | 0.9 |
| rad57 (YB208) | 73 (87/119) | 1.0 |
aFor complete genotype, see Table 1.
bDeletion events after HO induction/total Trp+ c.f.u. before HO induction × 100%.
cDeletion % in mutant strain/deletion % in wild-type strain.
Although both rad59 and rad50 mutants are defective in the X-ray stimulation of SCE, we found that HO-induced DSBs stimulated SCE in both rad50 and rad59 mutants. In rad59 and rad50 mutants, there was a 19- and 16-fold increase, respectively, in SCE frequencies after HO induction (Table 5). Of the unselected Trp+ His– colonies that appeared after HO induction in rad59 and rad50 mutants, 77% and 89% of them, respectively, did not generate His+ recombinants (Table 6). This finding suggests that HO-induced DSBs can be repaired by SSA in both mutants. However, the level of viability after HO induction in the rad50 mutant decreased significantly, while the rad59 mutant exhibited a level of viability similar to rad51, rad55, rad57 and rad54 mutants. We suggest that this higher lethality of HO-induced DSBs in rad50 mutants may result from the rad50 defect in NHEJ, as previously documented (37).
DISCUSSION
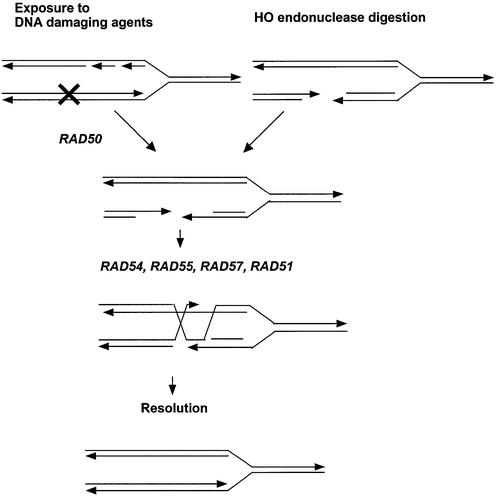
SCE has been suggested to occur by several mechanisms, including template switching, gap repair and BIR. In this study, we defined genetic pathways for spontaneous and DNA damage-associated SCE by measuring rates of spontaneous SCE and frequencies of DNA damage-associated SCE in S.cerevisiae rad mutants. We suggest that DNA damage-associated SCE occurs mainly by gap repair and requires genes within the RAD51 group, but the requirement for RAD50 or RAD59 depends on the DNA-damaging agent. Although genes in the RAD51 group are not required for spontaneous SCE, rad1 rad51 double mutants showed a synergistic decrease in recombination, whereas a rad59 deletion conferred only a modest reduction in RAD51-independent SCE. Together, these results suggest that BIR is not a major mechanism for spontaneous SCE, and we speculate that spontaneous SCE may involve as yet undefined mechanisms involving template switching (Fig. 4).
Figure 4.
A model for the RAD51, RAD54, RAD55 and RAD57 pathway for DNA damage-associated SCE. Each line represents a single strand of DNA, and the arrow designates the 3′ end. A DNA lesion occurs after progression of the DNA replication fork. Left: in radiation-associated SCE, RAD50 is required to process the site of DNA damage to reveal 3′ single-strand tails. Right: HO-induced DSBs already have 3′ overhangs and are not RAD50 dependent. RAD51, RAD54, RAD55 and RAD57 are then required for gap repair of both types of DSBs.
The RAD50 and RAD59 requirements for DNA damage-associated SCE depend on the DNA-damaging agent and might reflect the requirement for different nucleases to process HO-induced DSBs (38), X-ray-induced DNA damage (39) and UV-induced DNA damage for recombinogenic repair. Because UV does not induce DSBs directly, we speculate that DSBs are not obligate intermediates for UV-associated SCE. A DNA replication-dependent SCE pathway involving template switching has been proposed previously (5) and could be consistent with the RAD51-dependent recombination pathway that we propose for spontaneous SCE (Fig. 5). Genetic differences between UV-associated SCE and DSB-initiated recombination are observed in Escherichia coli, in which the RecO, RecR and RecF proteins promote UV-associated SCE; whereas RecBC functions in DSB repair (40).
Figure 5.
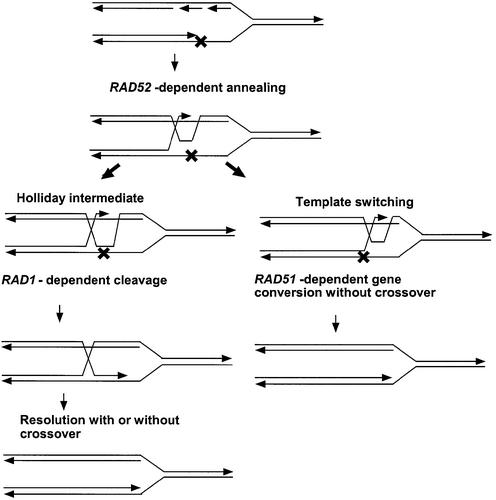
A model of the RAD1 and RAD51 pathways for spontaneous SCE. Each line represents a single strand of DNA, and the arrow designates the 3′ end. A DNA lesion blocks DNA polymerase progression that results in a recombinogenic single-strand gap. Rad52 then catalyzes the initial strand invasion of the sister chromatid, forming a transient replication intermediate that can bypass the replication block. Left: Rad1 and associated proteins (Rad10) cleave the transient replication intermediate resulting in the stable formation of the Holliday structure, which can be resolved either with or without crossing over. Right: in the absence of Rad1, the Holliday structure is unstable, but replication can continue and branch migration of the replication intermediate is Rad51 dependent. This process may be similar to synthesis-dependent strand annealing and will generate gene conversion events.
Genetic requirements for spontaneous SCE are different from those for either DSB-initiated gap repair or BIR. DSB-initiated gap repair requires all the genes in the RAD51 group (13), whereas BIR initiated at the MAT locus requires RAD59 and RAD50 but not RAD1 (4,9). However, we observed a 10-fold decrease in rates of spontaneous SCE in the rad1 rad51 but not in the rad59 rad51 or rad50 rad51 double mutants. The minor role of BIR in spontaneous SCE agrees with observations that cell cycle checkpoints prevent replication fork collapse and thus minimize BIR (41).
The genetics of spontaneous SCE are consistent with a template switch mechanism, first proposed for mammalian cells (42). In our model (Fig. 5), recombination is initiated by a single-strand gap after a replication fork stalls. Recombination forms a transient replication intermediate that allows polymerase progression after the replication block. A stable Holliday intermediate is then generated by the Rad1/Rad10 nuclease and is resolved to generate either crossovers or non-crossovers. The RAD1-independent pathway is RAD51 dependent and cannot result in crossovers. Although we could not measure crossovers directly, Kadyk and Hartwell (5) did demonstrate that most UV-associated SCE in rad1 mutants occurs by gene conversion not associated with crossing over. The synergistic decrease in SCE observed in rad1 rad51 mutants is consistent with a proposed RAD1-dependent pathway for plasmid integration (43). The initiation of SCE by single-strand gaps has also been used to explain the phenotypes of sgs1 and srs2 yeast mutants (44).
Considering that yeast and mammalian RAD genes have significant similarities, our studies may reveal new insights into the genetics of SCE in mammalian cells. For example, when Rad51 protein is inhibited by a dominant negative, the effect on the frequency of spontaneous SCE is variable and depends on the cell type (45,46). We speculate that the variable effect of inhibiting RAD51 gene expression may reflect the requirement for RAD51-independent recombination on ERCC1 (RAD10), which is required for crossing over in gene targeting experiments (13,47).
In summary, the genetic requirements of both spontaneous and DNA damage-associated SCE indicate that there are multiple pathways for SCE. Additional studies in mammalian cells are needed to find similar genetic relationships.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Lorraine Symington for the gift of plasmids and strains, and Cinzia Cera and Mingzeng Sun for carefully reading this manuscript. This work was supported by grant CA70105 from the National Cancer Institute of the National Institutes of Health.
REFERENCES
- 1.Fasullo M., Giallanza,P., Dong,Z., Cera,C. and Bennett,T. (2001) Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics, 158, 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latt S.A., Schreck,R.R., Loveday,K.S., Dougherty,C.P. and Shuler,C.F. (1980) Sister chromatid exchanges. Adv. Hum. Genet., 10, 267–331. [DOI] [PubMed] [Google Scholar]
- 3.Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- 4.Malkova A., Ivanov,E.L. and Haber,J.E. (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA, 93, 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadyk L.C. and Hartwell,L.H. (1993) Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics, 133, 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Game J.C. and Mortimer,R.K. (1974) A genetic study of X-ray sensitive mutants in yeast. Mutat. Res., 24, 281–292. [DOI] [PubMed] [Google Scholar]
- 7.Resnick M.A. (1969) Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics, 62, 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson J.A. and Fink,G.R. (1981) Gene conversion between duplicated genetic elements in yeast. Nature, 292, 306–311. [DOI] [PubMed] [Google Scholar]
- 9.Signon L., Malkova,A., Naylor,M.L., Klein,H. and Haber,J.E. (2001) Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol., 21, 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rattray A.J. and Symington,L.S. (1995) Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics, 139, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery H.S., Schild,D., Kellogg,D.E. and Mortimer,R.K. (1991) Sequence of RAD54, a Saccharomyces cerevisiae gene involved in recombination and repair. Gene, 104, 103–106. [DOI] [PubMed] [Google Scholar]
- 12.Jackson S.P. (2001) Detecting, signalling and repairing DNA double-strand breaks. Biochem. Soc. Trans., 29, 655–661. [DOI] [PubMed] [Google Scholar]
- 13.Symington L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev., 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein H.L. (1988) Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics, 120, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiestl R.H. and Prakash,S. (1988) RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol., 8, 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiestl R.H. and Prakash,S. (1990) RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol., 10, 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailly V., Sommers,C.H., Sung,P., Prakash,L. and Prakash,S. (1992) Specific complex formation between proteins encoded by the yeast DNA repair and recombination genes RAD1 and RAD10. Proc. Natl Acad. Sci. USA, 89, 8273–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishman-Lobell J. and Haber,J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- 19.Prado F. and Aguilera,A. (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10 and RAD52 genes. Genetics, 139, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadyk L.C. and Hartwell,L.H. (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics, 132, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Appendix A. In Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 163–168.
- 22.Lovett S.T. and Mortimer,R.K. (1987) Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics, 116, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alani E., Subbiah,S. and Kleckner,N. (1989) The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics, 122, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- 25.Lea D.E. and Coulson,C.A. (1949) The distribution of the numbers of mutants in bacterial populations. J. Genet., 49, 264–284. [DOI] [PubMed] [Google Scholar]
- 26.Fasullo M., Dave,P. and Rothstein,R. (1994) DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae. Mutat. Res., 314, 121–133. [DOI] [PubMed] [Google Scholar]
- 27.Zar J.H. (1996) Biostatistical Analysis. Prentice Hall, Englewood Cliffs, NJ.
- 28.Fasullo M., Bennett,T., AhChing,P. and Koudelik,J. (1998) The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol. Cell. Biol., 18, 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fasullo M.T. and Davis,R.W. (1987) Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl Acad. Sci. USA, 84, 6215–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus E., Leung,W.Y. and Haber,J.E. (2001) Break-induced replication: a review and an example in budding yeast. Proc. Natl Acad. Sci. USA, 98, 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov E.L., Sugawara,N., Fishman-Lobell,J. and Haber,J.E. (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics, 142, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailis A.M. and Rothstein,R. (1990) A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics, 126, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batty D.P. and Wood,R.D. (2000) Damage recognition in nucleotide excision repair of DNA. Gene, 241, 193–204. [DOI] [PubMed] [Google Scholar]
- 34.Jansen L.E., Verhage,R.A. and Brouwer,J. (1998) Preferential binding of yeast Rad4·Rad23 complex to damaged DNA. J. Biol. Chem., 273, 33111–33114. [DOI] [PubMed] [Google Scholar]
- 35.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 36.Ramotar R., Belanger,E., Brodeur,I., Masson,J.Y. and Drobetsky,E.A. (1988) A yeast homologue of the human phosphotyrosyl phosphatase activator PTPA is implicated in protection against oxidative DNA damage induced by the model carcinogen 4-nitroquinoline 1-oxide. J. Biol. Chem., 273, 21489–21496. [DOI] [PubMed] [Google Scholar]
- 37.Moore J.K. and Haber,J.E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov E.L., Sugawara,N., White,C.I., Fabre,F. and Haber,J.E. (1994) Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau S., Morgan,E.A. and Symington,L.S. (2001) Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics, 159, 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keller K.L., Overbeck-Carrick,T.L. and Beck,D.J. (2001) Survival and induction of SOS in Escherichia coli treated with cisplatin, UV-irradiation, or mitomycin C are dependent on the function of the RecBC and RecFOR pathways of homologous recombination. Mutat. Res., 486, 21–29. [DOI] [PubMed] [Google Scholar]
- 41.Sogo J.M., Lopes,M. and Foiani,M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science, 297, 599–602. [DOI] [PubMed] [Google Scholar]
- 42.Higgins N.P., Kato,K. and Strauss,B. (1976) A model for replication repair in mammalian cells. J. Mol. Biol., 101, 417–425. [DOI] [PubMed] [Google Scholar]
- 43.Saffran W.A., Greenberg,R.B., Thaler-Scheer,M.S. and Jones,M.M. (1994) Single strand and double strand DNA damage-induced reciprocal recombination in yeast. Dependence on nucleotide excision repair and RAD1 recombination. Nucleic Acids Res., 22, 2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabre F., Chan,A., Heyer,W.D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/ Mms4 and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stark J.M., Hu,P., Pierce,A.J., Moynahan,M.E., Ellis,N. and Jasin,M. (2002) ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem., 277, 20185–20194. [DOI] [PubMed] [Google Scholar]
- 46.Lambert S. and Lopez,B.S. (2000) Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J., 19, 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niedernhofer L.J., Essers,J., Weeda,G., Beverloo,B., de Wit,J., Muijtjens,M., Odijk,H., Hoeijmakers,J.H. and Kanaar,R. (2001) The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J., 20, 6540–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]