Abstract
In tobacco, 30 of 34 sites in chloroplast transcripts that undergo C-to-U RNA editing can be grouped into clusters of 2–5 sites based on sequence similarities immediately 5′ to the edited C. According to a previous transgenic analysis, overexpression of transcripts representing one cluster member results in reduction in editing of all cluster members, suggesting that members of an individual cluster share a trans-factor that is present in limiting amounts. To compare leaves and roots, we quantified the editing extent at 34 sites in wild-type tobacco and at three sites in spinach and Arabidopsis. We observed that transcripts of most NADH dehydrogenase subunits are edited inefficiently in roots. With few exceptions, members of the same editing site cluster co-varied in editing extent in chloroplasts versus non-green root plastids, with members of most clusters uniformly exhibiting either a high or low editing extent in roots. The start codon of the ndhD transcript must be created by editing, but the C target is edited inefficiently in roots, and no NDH-D protein could be detected upon immunoblotting. Our data are consistent with the hypothesis that cluster-specific trans-factors exist and that some are less abundant in roots, limiting the editing extent of certain sites in root plastids.
INTRODUCTION
Transcripts of higher plant organelles are modified by C-to-U editing (1–4). The chloroplast genomes of investigated vascular plants typically contain about 30 editing sites (5,6), while 441 and 491 sites were discovered, respectively, in the Arabidopsis and rice mitochondrial genomes (7,8). Start and stop codons may be created by C-to-U editing, and editing often results in amino acid substitutions, which usually restore the conserved amino acid encoded by orthologous genes (3,6,9). These conserved amino acids have been shown to be essential for proper gene product function in several cases (10,11). Furthermore, editing appears sometimes to be necessary to restore recognition sequences that allow intron removal (12–14). Thus RNA editing primarily appears to be a correction mechanism for T to C mutations that would prevent proper gene function. In plastids, only one silent editing site, which does not affect the encoded amino acid, has been found, in the gene atpA (15). A number of silent editing events can be documented in plant mitochondria, and these sites are more likely to be partially, rather than fully, edited in the transcript populations that have been examined (16). A few sites are present in intergenic regions in both organelles (4,17).
Because editing occurs in an albino mutant lacking plastid ribosomes (18), any protein trans-factors needed for chloroplast editing must be imported from the nucleus. Despite the availability of in vivo (19) and in vitro systems (20,21) for studying plastid editing, none of the components of the editing machinery have yet been identified. These editing systems have, however, been used to define the minimal surrounding sequences required to support in vivo or in vitro editing. For those sites analyzed, typically fewer than 150 nt of surrounding RNA sequences are necessary to support editing, with more sequence required 5′ than 3′ of the C target of editing. The tobacco psbL-1 site requires only 16 nt 5′ and 5 nt downstream of the C target to support >50% editing in vivo.
Though no consensus sequence can be detected by simultaneously comparing the sequences surrounding 34 editing sites in tobacco, conserved nucleotides can be detected in clusters of 2–5 chloroplast editing sites, and can also be seen in subgroups of mitochondrial RNA editing sites (22). When we overexpressed two editing sites in tobacco transgenic chloroplasts, we observed that two clusters of editing sites, each exhibiting conserved cis-elements, were impaired in editing efficiency. These in vivo competition experiments are consistent with the hypothesis that the same, or closely related, trans-factors recognize members of the same editing cluster. Because protein trans-factors must be nuclear encoded, such factors may be subject to developmental regulation, as are known nuclear-encoded factors that affect plastid gene function (23). We therefore considered the impact of developmental regulation of editing trans-factors on the editing status of plastid transcripts in different tissues. If trans-factor abundance is a limiting factor in a tissue, and one such factor recognizes multiple editing sites, then we would expect that members of the same editing site cluster should co-vary in editing efficiency in different tissues. However, little is known about the developmental regulation of RNA editing in different plant organs and tissues. Only in maize has a thorough survey of editing efficiencies been carried out (9). Previously, most editing sites were thought to be fully edited in chloroplasts, with the exceptions of atpA-2, ndhD-1 and rpoA-1 found to be partially edited in green leaves of tobacco (15,24,25). Tobacco rpoA-1 was found to be 70% edited in leaf but only 50% edited in cultured cells (24). Editing of tobacco atpA-2 and ndhD-1 was found to be impaired after antibiotic treatment of seedlings and in cultured cells (15,25). Heat stress, antibiotics and growth in complete darkness were reported to modulate the editing extent of several sites in ndh gene transcripts (26–28).
Here we report a study of the editing efficiency of 34 tobacco plastid sites in leaves versus roots, using the quantitative poisoned primer extension (PPE) method. We also analyzed the leaf and root editing extent of members of a cluster conserved in spinach and Arabidopsis. We selected these two tissues as plastid developmental extremes that were likely to vary in nuclear gene expression. Of the 34 editing sites analyzed, we found that transcripts encoding NADH dehydrogenase were most likely to be reduced in editing efficiency in roots. Partial editing of NADH dehydrogenase transcripts in roots probably has no functional consequence; immunoblotting with anti-NDH-D antibody indicates that the enzyme complex is not present in detectable quantity in roots. Consistent with our hypothesis of common trans-factors for multiple sites, we find that most editing site clusters detected by sequence inspection also co-vary developmentally in editing efficiency in roots versus leaves.
MATERIALS AND METHODS
Young leaves were harvested from mature tobacco plants (Nicotiana tabacum cv. Petit Havana) grown in a greenhouse. Roots were collected from 1-month-old plants grown in liquid MS medium (29) without agar. Etiolated seedlings were obtained after growing tobacco seeds in complete darkness for 15 days while control plants were growing in a 8/16 h dark–light cycle. Arabidopsis thaliana (cv. WS) plants were grown on Metromix soil in a growth chamber. Spinach leaves and roots were obtained from a local grocery store.
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) and treated with a DNA-free kit (Ambion). First-strand cDNA was synthesized from 1.5 µg of DNA-free RNA for 1 h at 37°C with an Omniscript kit (Qiagen) using random hexamers following the manufacturer’s protocol. Reactions without reverse transcriptase were performed to check genomic DNA contaminations. cDNA samples were amplified by a standard protocol (5 min at 94°C followed by 40 cycles of 94°C for 30 s, 50–55°C for 30 s, 72°C for 1 min) in a PTC-200 thermal cycler (MJ Research).
PPE of RT–PCR products and determination of editing efficiency were conducted as previously described (9). To confirm the results, all experiments were performed from at least two different RNA extractions per stage or tissue, and PPE was done at least twice from the same RNA sample. Primers used for PCR and PPE have been described by us previously (22) or are listed in Table 1.
Table 1. Oligonucleotides used.
| Name | Sequence 5′–3′ | Purpose |
|---|---|---|
| F1ndhD1 | AATATTTTGAGCACGGGTTTTTA | PCR ndhD-1,2 5′a |
| R1ndhD1 | TGTGCTTCTCCATGGGTATCTG | PCR ndhD-1,2 3′ |
| F2ndhD1 | CAAGTGTATCTTGTCTTTAC | PCR ndhD-1,2 5′ |
| R2ndhD1 | AAAATTTAATGTTGGTTC | PCR ndhD-1,2 3′ |
| FndhD2 | CCATAAAGGAAATAGGGTAAT | PCR ndhD-3,4 5′ |
| RndhD2 | ATAGAATGGGCATG GGTAATA | PCR ndhD-3,4 3′ |
| NdhD-3(G) | GAATATTATTTCTAAAACCACAGGATATGACTG | PPE ndhD-3b |
| NdhD-4(T) | TGGATTTTTTATTGCTTTTGCTGTCAAAT | PPE ndhD-4 |
| Rps2-1(C) | CGCCTTATATTTCTGCAAAGCGTAAGGGTATTC | PPE rps2-1 |
| CH36 | CTTCCAGTACCTATTTTACTAGGAGTTGG | PCR ndhF At 5′ |
| CH37 | CTCAGGTATCCTTGATCATGCG | PCR ndhF At 3′ |
| CH38 | CAGAACCAAAATCCCAACAGTTGT | PPE ndhF-2 At |
| CH52 | GAGTACGCGTTCTTTGGACCTGGTG | PCR ndhD At 5′ |
| CH53 | GTAGCCGAATACAGACGTTTCTTTC | PCR ndhD At 3′ |
| CH54 | GAAAAACAATTATTGTTAACCAAGG | PPE ndhD-1 At |
| CH26 | CTTTCGTTTACTTGGGTCACTGG | PCR atpF At 5′ |
| CH27 | CACGCAGTTCTTCTGAATTTCGAATAG | PCR atpF At 3′ |
| CH28 | GATTTAATACCGATATTTTAGCAACAAATC | PPE atpF-1 At |
| FndhDso | TTTCCTTTTGGGTACGGGTTTTT | PCR ndhD So 5′ |
| RndhDso | CCATGTGAGATACGGAGGAATAGG | PCR ndhD So 3′ |
| SondhD-1(A) | ACTACAATTGTTGTTAACCAGGGAAAAGAA | PPE ndhD-1 So |
| FndhFso | CCCAAGTATATCTTGTCTTTATC | PCR ndhF So 5′ |
| RndhFso | GCACTATACATCGCTAACATC | PCR ndhF So 3′ |
| SondhF-2(G) | TATAAATAAGAACCAGAATTGCAACAGTAG | PPE ndhF-2 So |
aPCR: oligonucleotides used to amplify fragments containing editing sites (not indicated and Nt, Nicotiana tabacum; So, Spinacia oleacera; At, Arabidopsis thaliana).
bPPE: oligonucleotides used in poisoned primer extension.
Total proteins from leaf and root tissues were prepared by homogenization in 100 mM Tris–HCl pH 7.5, 5 mM EDTA, 40 mM 2-mercaptoethanol and complete protease inhibitor used according to the manufacturer’s instructions (Roche). Membrane-associated proteins were solubilized by adding Triton X-100 to a final concentration of 2% and incubating for 30 min at room temperature. Soluble proteins were recovered after a 10 min centrifugation to remove insoluble material. Protein concentration of the samples was determined with a Protein Assay kit (BioRad) using bovine serum albumin as a standard. SDS–PAGE, transfer and immunoblotting were performed as previously described (30). The anti-NDH-D antibody was kindly provided by Mercedes Martin. NDH-D polypeptide was visualized using a 1:1000 dilution of this antibody (31).
RESULTS
Plastid DNA editing sites in tobacco
We analyzed the 34 C-to-U editing sites that have been reported to date on the tobacco chloroplast genome. Table 2 lists all the editing sites following the nomenclature proposed by Tsudzuki et al. (5), updated since the recent report of a third site in the tobacco ndhD transcript (6) and additional sites in rps2 and ndhD (R.Maier, personal communication). As an example of the nomenclature, the ndhD transcript encodes the NDH-D subunit, and the sixth edited C from the 5′ end of the ndhD trancript that has been detected in any angiosperm to date will be referred to as ‘ndhD-6’. Editing sites are distributed on transcripts of 15 different genes; 16 sites out of 34 are located in transcripts encoding four subunits of the NADH dehydrogenase complex (subunits A, B, D and F). Of these, the ndhB gene contains nine sites out of the 34 edited Cs. The editing site clusters that have been detected as a result of the finding of the three additional sites since our earlier publication (22) are shown in Figure 1.
Table 2. RNA editing sites in tobacco chloroplasts.
| Site | Position | Codon | Amino acid change |
|---|---|---|---|
| atpA-1 | 791 | cCc | P to L |
| atpA-2 | 795 | ucC | No (S to S) |
| atpF-1 | 92 | cCa | P to L |
| ndhA-2 | 341 | uCa | S to L |
| ndhA-5 | 1073 | uCc | S to F |
| ndhB-1 | 149 | uCa | S to L |
| ndhB-2 | 467 | cCa | P to L |
| ndhB-3 | 586 | Cau | H to Y |
| ndhB-4 | 611 | uCa | S to L |
| ndhB-6 | 737 | cCa | P to L |
| ndhB-7 | 746 | uCu | S to F |
| ndhB-8 | 830 | uCa | S to L |
| ndhB-9 | 836 | uCa | S to L |
| ndhB-10 | 1481 | cCa | P to L |
| ndhD-1 | 2 | aCg | T to M |
| ndhD-2 | 383 | uCa | S to L |
| ndhD-3 | 599 | uCa | S to L |
| ndhD-4 | 674 | uCg | S to L |
| ndhF-2 | 290 | uCa | S to L |
| petB-1 | 611 | cCa | P to L |
| psbE-1 | 214 | Ccu | P to S |
| psbL-1 | 2 | aCg | T to M |
| rpl20–1 | 308 | uCa | S to L |
| rpoA-1 | 830 | uCa | S to L |
| rpoB-1 | 338 | uCu | S to F |
| rpoB-2 | 473 | uCa | S to L |
| rpoB-3 | 551 | uCa | S to L |
| rpoB-6 | 2000 | uCu | S to F |
| rpoC1-1 | 62 | uCa | S to L |
| rpoC2-2 | 3743 | uCa | S to L |
| rps2-1 | 134 | aCa | T to I |
| rps2-2 | 248 | uCa | S to L |
| rps14-1 | 80 | uCa | S to L |
| rps14-2 | 149 | cCa | P to L |
Figure 1.
The three recently discovered editing sites of tobacco, rps2-1, ndhD-3 and ndhD-4, can be grouped into clusters. Bold letters represent conserved nucleotides between members of the cluster. Gaps (–) were introduced to show similarities. C, C target of editing.
Editing efficiency in wild-type tobacco leaf chloroplasts and root plastids
We selected leaves of mature plants and roots as the two tissues to compare in order to determine whether any of the clusters exhibit coordinate developmental variation. No comprehensive study of the editing extent of all sites in tobacco plastids in these two tissues has been undertaken previously. We chose to compare leaves and roots because they represent distinct tissue types where nuclear gene expression is likely to vary; also, in our previous study of editing in maize (9), we found that the editing extent of a number of C targets of editing was lower in roots than in leaves.
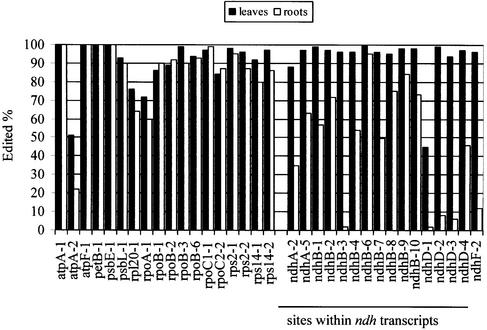
The editing efficiency of all editing sites was determined on transcripts isolated from young leaves of mature tobacco plants (cv. Petit Havana). We used PPE to quantify the editing extent of each site. No error bars are shown in Figure 2 because the variations between samples and assays were very small (never greater than 5%). An example of an actual PPE experiment is shown in Figure 3.
Figure 2.
Editing extent of the 34 sites in young leaf chloroplasts of mature tobacco plants and in root plastids of 1-month-old tobacco plants. The percentage of edited transcripts was determined on PPE products by quantifying the radioactivity associated with edited and unedited sites using ImageQuant software (Molecular Dynamics). The x-axis represents the 34 editing sites listed in Table 2.
Figure 3.
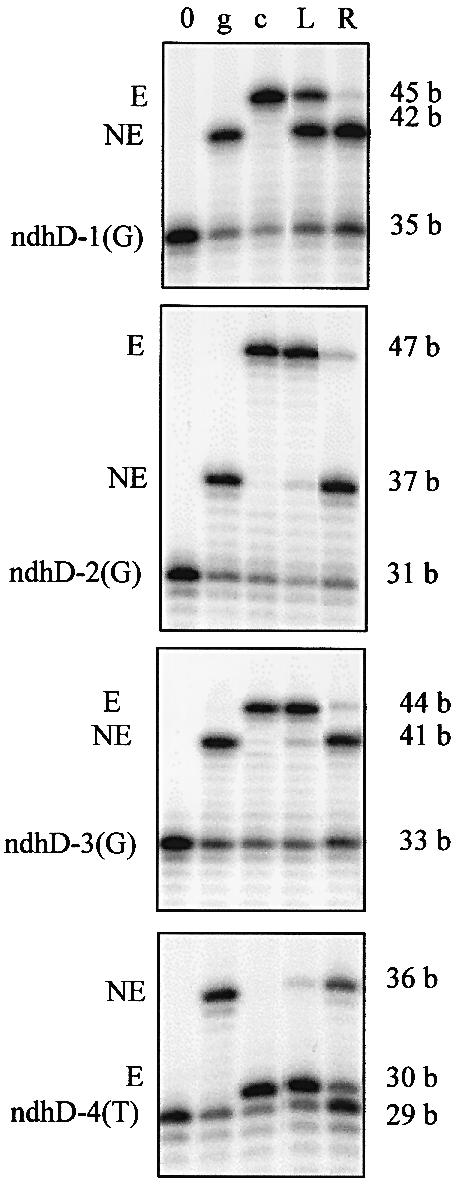
PPE assays showing the different editing extents of the four sites located within ndhD transcripts in leaves (L) and roots (R) of tobacco. PPE was performed on site-specific RT–PCR products, from radiolabeled ndhD-1(G), ndhD-2(G), ndhD-3(G) and ndhD-4(T). The primer extension was poisoned by ddNTP incorporation, ddGTP for ndhD-1, -2 and -3 and ddTTP for ndhD-4. PPE products were resolved on 12% sequencing gels, which were then exposed on a phosphorimager screen. 0, PPE without template indicating the size of the radiolabeled oligonucleotide; g, PPE with a cloned (genomic) unedited PCR product; c, PPE with a cloned edited RT–PCR product; L, PPE made from leaf extracts of mature tobacco plants; R, PPE made from root extracts of young tobacco; E, PPE product corresponding to the edited transcript; NE, PPE product corresponding to the unedited transcript.
Most sites are nearly fully edited in leaves of mature tobacco plant (Fig. 2). Some are edited by 80–90% (rpoB-1, rpoB-2, rpoC2-2 and ndhA-2), and four sites are clearly partially edited (atpA-2, rpl20-1, rpoA-1 and ndhD-1). Unedited transcripts of rpl20-1 and rpoA-1 would encode a protein containing a serine rather than the leucine that would be present in proteins translated from edited transcripts. The lowest editing efficiency in leaves was found in ndhD-1, editing of which creates the AUG codon initiating the translation of NDH-D in all dicots investigated and in Liliaceae and Aloaceae (25,32,33). Because editing of atpA-2 does not affect the predicted amino acid, partial editing of this site has no consequence on the amino acid composition of the protein. Two sites differ in editing extent between the mature plant leaves reported here and the immature leaves analyzed in our earlier report (22). AtpA-2 was edited at 35% in immature and 52% in mature leaves, and ndhA-2 was edited 63% in immature versus 88% in mature leaves. The newly discovered site ndhD-3 is edited 75% in immature leaves and 94% in mature leaves.
The editing efficiency of ndhD-1 was found to be higher when we used a PCR primer located just before the C target of editing (F1ndhD1: –22 to –3) instead of one located further upstream (F2ndhD1: –50 to –28). Using the more proximal primer changes the edited percentage of ndhD-1 from 34 to 45%. According to Hirose and Sugiura (25), the monocistronic ndhD transcripts in tobacco exhibit different 5′ ends. Our results suggest that longer 5′ end ndhD transcripts might be less edited than shorter ones. There is evidence also in leek that different ndhD transcripts may be differentially edited. Del Campo et al. (31) found all full-length monocistronic ndhD transcripts to be edited at the leek ndhD-1 site by sequencing RT–PCR products made from gel-isolated RNAs, even though they failed to detect editing of this site from total RNA. They proposed that differential cleavage and editing regulate the production of NDH-D protein from an abundant polycistronic transcript that also contains psaC, which encodes a protein of photosystem I that is far more abundant than the NADH dehydrogenase.
Transcripts of all genes carrying editing sites were detected in tobacco root plastids by RT–PCR, even those encoding photosynthesis-associated polypeptides. We previously reported that maize roots also contain transcripts of all plastid genes carrying edited sites, though in every case the relative abundance of the transcripts was reduced compared with leaves (9).
In leaves, 30 of 34 C targets of editing exhibit >80% conversion to U, while in roots, only 17 sites exhibit at least 80% editing (Fig. 2). There are 12 editing sites that exhibit no significant difference in editing between leaves and roots (Fig. 2). The discrepancy between editing extent in leaves and roots is due largely to a reduction in the editing extent of nearly all sites of ndh transcripts in root, the only exception being ndhB-6. Five of these sites, ndhB-3, ndhD-1, ndhD-2, ndhD-3 and ndhF-2, remain nearly unedited in root plastids, although they are almost fully edited in green leaves except for ndhD-1.
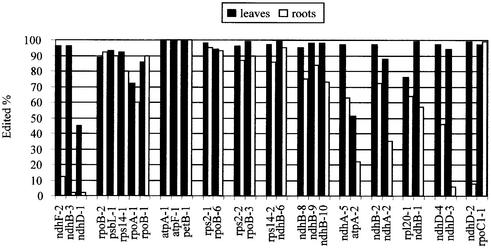
Similar editing efficiency ratios between sites within cis-element clusters
Of the 34 sites, 30 can be grouped in clusters of 2–5 members that have putative conserved cis-elements (22). Additional clusters may be detected in the future as additional C-to-U editing events are discovered. Two clusters have been shown to exhibit cross-competition when one cluster member is overexpressed. All other clusters have been assembled solely by sequence inspection. The extent of editing in leaves versus roots in the 12 observed editing site clusters is shown in Figure 4. Eleven of the clusters exhibit similar ratios of leaf/root editing extent. The largest discrepancy between sites grouped into a cluster is the editing of ndhD-2 and rpoC1-1; both are highly edited in leaves, but editing of ndhD-2 is greatly reduced in roots, unlike rpoC1-1 (Fig. 4).
Figure 4.
Editing efficiencies of 30 editing sites in tobacco leaves and roots that can be grouped according to cis-element cluster. The first two clusters were shown to cross-compete in transgenic chloroplasts (22).
Different editing extent of a cluster member within the same organ
Inspection of Figure 4 reveals that most members of a cluster are edited to the same extent in the same organ. There are a few exceptions, most notably ndhD-1, which is much less edited in leaves than its partners, ndhF-2 and ndhB-3. When ndhF-2 was overexpressed in transgenic chloroplasts, editing of ndhD-1 and ndhB-3 was reduced, providing strong evidence for a shared trans-factor. One possible explanation for the lower leaf editing of ndhD-1 relative to the other two sites would be a lower affinity of the trans-acting factor for the sequences surrounding ndhD-1 versus the other two sites. Inspection of the sequences at these sites reveals that ndhD-1 exhibits two single nucleotide polymorphisms in the putative cis-element (Fig. 5), which possibly could affect binding of an editing factor.
Figure 5.
Putative cis-elements conserved in the upstream sequences of sites in ndhF-2 and atpA-2 clusters. Bold letters represent conserved nucleotides between members of the cluster, and lower case letters indicate nucleotide polymorphism within a putative cis-element. Gaps (–) were introduced to show similarities. C, C target of editing.
The editing extent of atpA-2 is also low in both leaves and roots relative to its partner, ndhA-5 (Fig. 4). AtpA-2 is a silent site; editing does not affect the amino acid sequence of the encoded protein. Therefore, selection pressure for efficient editing would not be expected, and the cis-elements near atpA-2 may not be as efficient as those of ndhA-5 in their interaction with a required editing trans-factor (Fig. 5).
The editing defect of the ndhF-2 cluster in roots is conserved among different plant species
Because the ndhF-2 cluster showed the strongest developmental regulation, we investigated whether members of this cluster also co-varied in editing extent in other species. The C target of editing of members of the tobacco ndhF-2 cluster (ndhF-2, ndhB-3 and ndhD-1) is conserved in several species such as Atropa belladonna (6) and spinach (5). It is the only cluster that is present in both N.tabacum and A.thaliana. We analyzed the editing extent of these sites in spinach and Arabidopsis leaves and roots. In Arabidopsis, we found that none of the three sites are edited in roots while they are edited in leaves (Table 3). In spinach roots, they are edited, but to a much lower extent than in leaves (Table 3). These data suggest that a cluster-specific editing factor also operates on these three sites in Arabidopsis and spinach.
Table 3. Reduced editing in roots of the tobacco ndhF-2 cluster is conserved in Arabidopsis and spinach.
| Tobacco |
Arabidopsis |
Spinach |
||||
|---|---|---|---|---|---|---|
| Leaf | Root | Leaf | Root | Leaf | Root | |
| ndhF-2 | 91% | 11% | 70% | 0% | 90% | 34% |
| ndhB-3 | 97% | 2% | 90% | 0% | 99% | 24% |
| ndhD-1 | 45% | 2% | 61% | 0% | 41% | 16% |
| atpF-1a | 100% | 100% | 100% | 100% | Genomic T | |
aatpF-1 is not part of the tobacco ndhF-2 cluster. It is shown as a control for the Arabidopsis root cDNA. In spinach, a T is present at the genomic level at the location of atpF-1.
Editing efficiency of maize ndh transcript editing sites with 5′ elements similar to those in non-ndh genes
Though the role of the chloroplast NADH dehydrogenase is not entirely understood, it is thought to function in cyclic electron flow around photosystem I (34) and therefore would not be expected to be needed in roots. The most highly edited NADH dehydrogenase subunit editing site in tobacco roots is ndhB-6, which exhibits cis-sequence similarity to rps14-2, an editing site in a ribosomal protein. Possibly the necessity for editing of rps14-2 in tobacco roots has resulted in incidental editing of ndhB-6. To test the hypothesis that ndh editing sites could be affected by clustering with the gene regulatory subunits, we considered relevant data from our previous study of maize editing efficiencies (9). Before discovering conserved cis-elements, we surveyed the editing extent of all 27 known editing sites in maize plastids in a number of different tissues, including leaf and root. As in tobacco, we observed significant reduction in editing in roots versus leaves for many transcripts of NADH dehydrogenase subunits. However, if the data are examined in light of the clusters that can be assembled by inspection of maize sequences, we can observe that, like ndhB-6 in tobacco, relatively high editing occurs at ndh transcript editing sites that cluster with editing sites present in transcripts for components of the gene regulatory machinery (Table 4). Editing of RNA polymerase and ribosomal protein transcripts is likely to be necessary to produce functional transcriptional and translational apparatus for expression of plastid genes involved in non-photosynthetic functions of the plastids, which are the site of a number of metabolic processes. On the other hand, editing of ycf3-2 and ndhF-1, whose transcripts encode genes not useful in non-photosynthesizing tissue, is reduced in roots relative to leaves (Table 4). The protein encoded by ycf3 is involved in assembly of the photosystem I complex (35).
Table 4. Leaf and root editing extents of maize NADH dehydrogenase subunit editing sites that exhibit sequence similarities to non-ndh transcripts.
| Cluster no. | Editing site | Leaf | Root |
|---|---|---|---|
| 1 | ndhB-6 | 99 | 71 |
| 1 | rpoB-5 | 81 | 72 |
| 2 | ndhB-8 | 100 | 98 |
| 2 | atpA-3 | 100 | 100 |
| 2 | rpl20-1 | 95 | 98 |
| 3 | ycf3-2 | 92 | 32 |
| 3 | ndhF-1 | 100 | 50 |
| 4 | ndhB-3 | 100 | 49 |
| 4 | rpoC2-1 | 90 | 90 |
| 4 | ndhA-3 | 100 | 100 |
Editing extents are taken from Peeters and Hanson (9) and are reproducible within ±5%, usually ±2%. The four clusters shown are numbered arbitrarily to show grouping of sites.
The editing defect in roots is not due to a lack of photosynthesis.
Karcher and Bock (26) proposed that ndhB-4 (site III according to their nomenlature) is edited neither in leaf of tobacco non-photosynthetic mutants nor in etiolated seedlings of maize because of a lack of active photosynthesis. To determine whether the absence of photosynthesis is important in the editing extent of the ndhF-2 cluster, we examined a second non-photosynthetic tissue in addition to roots. We found that all three members of ndhF-2 were highly edited in non-photosynthetic etiolated seedlings (Table 5). Because of this discrepancy with Karcher and Bock’s hypothesis of a relationship between photosynthesis and editing, we analyzed the editing extent of ndhB-4 in these etiolated seedlings. To our surprise, we found that, in contrast to the Karcher and Bock (26) report, ndhB-4 transcripts were 86% edited in etiolated seedlings. We also previously found a high degree of editing of ndhB-4 in leaves of maize etiolated seedlings, though this site is edited at only 9% in maize roots (9). Our data do not support a correlation between the photosynthetic capacity of plastids and editing extent of plastid transcripts. Instead, we suggest that the editing extent is affected by the abundance of trans-factors needed for editing, and such trans-factors may be expressed in etiolated leaf tissue in order to produce functional transcripts encoding NDH subunits that will then be available to the chloroplast upon light exposure and greening.
Table 5. Editing extent of the tobacco ndhF-2 cluster in whole green or etiolated seedlings (15 days old).
| Green seedlings | Etiolated seedlings | |
|---|---|---|
| ndhF-2 | 92% | 87% |
| ndhB-3 | 97% | 90% |
| ndhD-1 | 44% | 25% |
| ndhB-4a | 98% | 86% |
andhB-4 is not part of the tobacco ndhF-2 cluster but, in contrast to our data, it was described as not edited at all in non-photosynthetic tissues (26).
The NDH-D subunit is not detectable in root plastids
The C target of editing at the ndhD-1 site is within the ACG codon, and editing presumably creates the AUG translational start codon. Thus, low efficiency of ndhD-1 editing, as occurs in roots, could affect translation of the ndhD transcript. The editing extent of ndhD-1 is <50% in leaves. We used a barley anti-NDH-D antibody (31) to examine leaf and root proteins. The NDH-D subunit was detected in tobacco leaf tissue but not in root (Fig. 6). This result is consistent with a requirement for an editing-generated start codon. However, we have observed from RT–PCR experiments that less ndhD transcript is present in tobacco roots versus leaves (data not shown), so that a reduced abundance of transcript presumably is another factor leading to the lack of detectable NDH-D subunit in roots. In maize, we previously reported that, according to quantitative RT–PCR, transcripts of five different ndh genes exhibited a 50- to 200-fold reduction in abundance in root tissue compared with leaf tissue (9).
Figure 6.
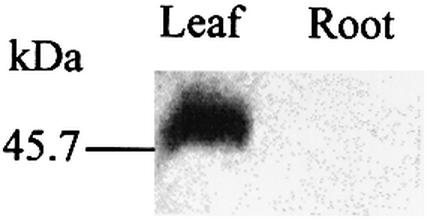
The NDH-D polypeptide is not detected in tobacco root plastids. Immunoblot analysis of total protein from leaf (10 µg) and root (40 µg) using an anti-NDH-D antibody. The calculated molecular mass of NDH-D is 56.3 kDa (MWCALC, Infobiogen). The migration of BioRad pre-stained standard carbonic anhydrase is shown on the left.
DISCUSSION
The two Cs in ndhA transcripts, the eight Cs in ndhB transcripts and the C in ndhF transcripts that exhibit incomplete editing in roots would be predicted to give rise to a large number of proteins carrying amino acid polymorphisms, if both edited and unedited transcripts are translated. At present, it is not known whether unedited transcripts are translated in chloroplasts in vivo. However, both unedited and edited transcripts functioned equally efficiently during in vitro translation, so it is likely that both are translated in vivo (36). In plant mitochondria, both unedited and edited transcripts are present on mitochondrial polysomes, and whether the gene product from unedited transcripts accumulates appears to depend on post-translational factors (37–40). The absence of one subunit of a chloroplast complex usually disrupts assembly of the entire complex (41). If this is true for the NADH dehydrogenase complex, then the fact that the other editing sites within ndh transcripts are less edited in root than in leaf chloroplasts would have no functional importance; the absence of NDH-D subunit would prevent assembly of any complex.
Our finding that overexpression of an editing site in transgenic plants causes reduction in editing at other sites led us to the hypothesis that two or more C targets of editing are operated on by the same trans-factor (22). Similarities in sequences immediately 5′ to the Cs exhibiting cross-competition further suggested that the similar cis-elements could comprise recognition signals for a trans-factor. Alternatively, the similar cis-elements could be recognized by separate, though probably related, trans-factors that operate on the members of a cross-competing cluster. Current data from studies of chloroplast editing in vitro implicate protein rather than RNA trans-factors in the editing machinery (20). Because editing occurs in mutants lacking chloroplast translation (18), protein components of the editing apparatus are thought to be nuclear encoded. If the regulatory sequences on a gene encoding an editing factor result in some tissue specificity of expression, then the abundance of the factor, and the extent of editing of any site on which it operates, could be expected to differ between tissues.
Our data are consistent with the hypothesis that trans-factor expression varies, that the abundance of trans-factors limits editing efficiency and, therefore, that the extent of editing of members of the editing site cluster tends to co-vary. In addition to our data reported here, Schmitz-Linneweber et al. (6) observed in A.belladonna, which shares 29 sites with tobacco, that rpoA-1, rps14-1 and rpoB-2 are partially edited in leaf. As in tobacco, there are putative shared cis-elements upstream of the three edited Atropa Cs, and the developmental co-variation can be explained by a shared developmentally regulated trans-factor. We also found a few exceptions to editing extent co-variation. An occasional site within a cluster was edited more or less than other members, which could be explained by differential affinity of a trans-factor for slightly polymorphic cis-elements found in different members of the same cluster. Because of these possible differences in affinity, a more appropriate comparison of cluster members comes from comparing the ratio of editing extent of each cluster member in one tissue versus another. We found that editing of the members of the same cluster either all did not change between leaf and roots or all were more highly edited in leaves than in roots, with the ratio of leaf/root editing approximately equal within cluster members (Fig. 4). The editing of the ndhD-2/rpoC1-1 cluster is a striking exception; the upstream sequences of these two sites are very similar, but editing of ndhD-2 is far more reduced in roots than is rpoC1-1. This finding could be explained by the presence of two different trans-factors that operate on these two sites, despite their sequence similarity.
An intriguing example of similar editing extent within two members of a cluster is provided by the tobacco ndhB-6/rps14-2 cluster. Though most sites within ndh gene transcripts are quite reduced in editing extent in roots, ndhB-6 is nearly completely edited, as is rps14-2. Possibly the requirement for functional plastid ribosomes in roots has selected for abundant trans-factor in roots, which then coincidentally results in high ndhB-6 editing though the NDH-B subunit would not be needed in roots, which evidently lack NADH dehydrogenase (Fig. 6). Data from our previous maize study (Table 4) support the hypothesis that the requirement for editing of transcripts encoding the gene expression machinery results in high editing extent in transcripts not needed in the root. The cluster concept of editing sites thus explains what otherwise would be a puzzling finding—efficient editing of transcripts that encode proteins that are non-essential in roots.
By assaying editing efficiency in leaf versus root tissue, we are determining the steady-state editing efficiencies in these tissues, which therefore may reflect the tissue-specific abundance of a trans-factor specific to a particular cluster. Under rapidly changing conditions, the abundance of particular chloroplast transcripts as well as the amount of trans-factors may become important in determining editing extent. For example, when maize plants were shifted to 37°C, chloroplasts became 5–10 times more transcriptionally active than at 20°C, and the editing extent of transcripts of rps14 and rpl20 decreased from nearly 100% to 30%. After rapid change in environmental conditions, editing extents undergo a decrease if the rate of transcription exceeds the rate of editing (42).
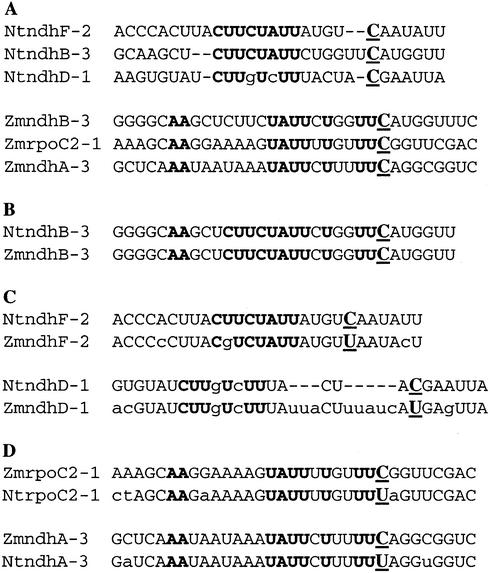
Developmental co-variation of the ndhF-2 editing cluster occurs not only in tobacco, but also in spinach and Arabidopsis. Most editing site clusters cannot be compared between species, because editing at a particular site is species specific. The plastid genome of one species often carries a genomically encoded T where another species must convert a C to U by RNA editing in order to encode the conserved amino acid residue. The existence of 5′ cis-elements near C targets of editing, along with the evidence for shared trans-factors, has interesting implications for the evolution of new editing sites and the loss of existing sites. Consider the tobacco ndhF-2 cluster, which is not conserved in maize (Fig. 7A). Of the three tobacco cluster members, only the ndhB-3 site in maize contains a C that is edited. The Cs at the comparable sites of the two other members of the ndhF-2 cluster are genomically encoded as T, thus requiring no editing (Fig. 7C). The sequence 5′ to the unedited maize ndhD-1 region exhibits considerable differences from the edited tobacco sequence (Fig. 7C). The sequence present in the maize region homologous to the edited tobacco ndhF-2 has a polymorphism that disrupts a conserved 8 nt element present in the tobacco ndhF-2 cluster (Fig. 7C).
Figure 7.
NdhB-3 shares sequence similarities in its upstream region with other editing sites in maize (Zm, Zea mays; Nt, Nicotiana tabacum). (A) Sequence alignment of the ndhF-2 cluster in tobacco and the putative ndhB-3 cluster in maize. (B) Comparison of sequences in maize and tobacco ndhB-3, edited in both species, illustrating that the two sites contain both the tobacco and maize conserved elements. (C) Sequence comparison between sites edited in tobacco and the homologous sites in maize, which carry a genomic T. (D) Sequence comparison between sites edited in maize and the homologous sites in tobacco, which carry a genomic T. Bold letters represent conserved nucleotides between members of the cluster, and lower case letters indicate nucleotide polymorphisms. Gaps (–) were introduced to show similarities. C, C target of editing; U, location of the editing sites in the other species.
In maize, the ndhB-3 site has become grouped with two other C targets of editing, maize rpoC2-1 and ndhA-3 (Fig. 7A). In tobacco, these sites are not edited because a genomic T is present at the corresponding location (Fig. 7D). Though maize ndhB-3 retains the conserved cis-element found in tobacco ndhB-3 (Fig. 7B), maize ndhB-3, maize rpoC2-2 and ndhA-3 share a different cis-element (Fig. 7A) not shared by the tobacco ndhB-3 cluster (Fig. 7A). Sequence inspection suggests that the trans-factor operating on the maize cluster should recognize a cis-element different from that which acts on the tobacco cluster. However, it appears that the maize ndhB-3 site could possibly be editable if it were present in tobacco, as it has retained the tobacco element (Fig. 7B). In contrast, the maize ndhD-1 site, which carries a genomic T (Fig. 7C), has an insertion relative to tobacco that might render it uneditable by the tobacco factor if the maize ndhD-1 sequence carried a genomic C. These examples provide a fascinating glimpse into the evolutionary changes that occur as editing sites are acquired and lost.
Our developmental study suggests that another constraint on acquisition of a new site could be the gene regulatory sequences on a nuclear-encoded trans-factor. If a T to C mutation arises in a gene that must be expressed properly in a particular tissue for optimal function of plastids, then the nuclear factor presumably must be synthesized at a sufficient level to correct the defect in the new site as well as other sites already operated upon by the factor. Thus selection would operate on the nuclear trans-factor’s gene regulatory sequences as well as on the sequences immediately 5′ to the new T to C mutation in order to optimize editing efficiency in those tissues where functional transcripts and proteins are needed.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mercedes Martin for a gift of anti-NDH-D antibody. This work was supported by NIH 2 RO1 GM50723 grant to M.R.H.
REFERENCES
- 1.Smith H.C., Gott,J.M. and Hanson,M.R. (1997) A guide to RNA editing. RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 2.Brennicke A., Marchfelder,A. and Binder,S. (1999) RNA editing. FEMS Microbiol. Rev., 23, 297–316. [DOI] [PubMed] [Google Scholar]
- 3.Maier R.M., Zeltz,P., Kossel,H., Bonnard,G., Gualberto,J.M. and Grienenberger,J.M. (1996) RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol., 32, 343–365. [DOI] [PubMed] [Google Scholar]
- 4.Bock R. (2001) RNA editing in plant mitochondria and chloroplasts. In Bass,B.L. (ed.), RNA Editing. Oxford University Press, pp. 38–60.
- 5.Tsudzuki T., Wakasugi,T. and Sugiura,M. (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol., 53, 327–332. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz-Linneweber C., Regel,R., Du,T.G., Hupfer,H., Herrmann,R.G. and Maier,R.M. (2002) The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol., 19, 1602–1612. [DOI] [PubMed] [Google Scholar]
- 7.Giege P. and Brennicke,A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA, 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notsu Y., Masood,S., Nishikawa,T., Kubo,N., Akiduki,G., Nakazono,M., Hirai,A. and Kadowaki,K. (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Gen. Genet., 268, 434–445. [DOI] [PubMed] [Google Scholar]
- 9.Peeters N.M. and Hanson,M.R. (2002) Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA, 8, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki Y., Kozaki,A., Ohmori,A., Iguchi,H. and Nagano,Y. (2001) Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem., 276, 3937–3940. [DOI] [PubMed] [Google Scholar]
- 11.Zito F., Kuras,R., Choquet,Y., Kossel,H. and Wollman,F.A. (1997) Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol., 33, 79–86. [DOI] [PubMed] [Google Scholar]
- 12.Carrillo C. and Bonen,L. (1997) RNA editing status of nad7 intron domains in wheat mitochondria. Nucleic Acids Res., 25, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Campo E.M., Sabater,B. and Martin,M. (2000) Transcripts of the ndhH-D operon of barley plastids: possible role of unedited site III in splicing of the ndhA intron. Nucleic Acids Res., 28, 1092–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanlungo S., Quinones,V., Moenne,A., Holuigue,L. and Jordana,X. (1995) Splicing and editing of rps10 transcripts in potato mitochondria. Curr. Genet., 27, 565–571. [DOI] [PubMed] [Google Scholar]
- 15.Hirose T., Fan,H., Suzuki,J.Y., Wakasugi,T., Tsudzuki,T., Kossel,H. and Sugiura,M. (1996) Occurrence of silent RNA editing in chloroplasts: its species specificity and the influence of environmental and developmental conditions. Plant Mol. Biol., 30, 667–672. [DOI] [PubMed] [Google Scholar]
- 16.Wilson R.K. and Hanson,M.R. (1996) Preferential RNA editing at specific sites within transcripts of two plant mitochondrial genes does not depend on transcriptional context or nuclear genotype. Curr. Genet., 30, 502–508. [DOI] [PubMed] [Google Scholar]
- 17.Drescher A., Hupfer,H., Nickel,C., Albertazzi,F., Hohmann,U., Herrmann,R.G. and Maier,R.M. (2002) C-to-U conversion in the intercistronic ndhI/ndhG RNA of plastids from monocot plants: conventional editing in an unconventional small reading frame? Mol. Gen. Genet., 267, 262–269. [DOI] [PubMed] [Google Scholar]
- 18.Zeltz P., Hess,W.R., Neckermann,K., Borner,T. and Kossel,H. (1993) Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J., 12, 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhuri S., Carrer,H. and Maliga,P. (1995) Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J., 14, 2951–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose T. and Sugiura,M. (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J., 20, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto T., Obokata,J. and Sugiura,M. (2002) Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol., 22, 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chateigner-Boutin A.L. and Hanson,M.R. (2002) Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol., 22, 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison L.A. (2000) The role of sigma factors in plastid transcription. Biochimie, 82, 537–548. [DOI] [PubMed] [Google Scholar]
- 24.Hirose T., Kusumegi,T., Tsudzuki,T. and Sugiura,M. (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet., 262, 462–467. [DOI] [PubMed] [Google Scholar]
- 25.Hirose T. and Sugiura,M. (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J., 16, 6804–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karcher D. and Bock,R. (2002) The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J. Biol. Chem., 277, 5570–5574. [DOI] [PubMed] [Google Scholar]
- 27.Karcher D. and Bock,R. (1998) Site-selective inhibition of plastid RNA editing by heat shock and antibiotics: a role for plastid translation in RNA editing. Nucleic Acids Res., 26, 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karcher D. and Bock,R. (2002) Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr. Genet., 41, 48–52. [DOI] [PubMed] [Google Scholar]
- 29.Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant., 15, 473–479. [Google Scholar]
- 30.Reed M.L., Wilson,S.K., Sutton,C.A. and Hanson,M.R. (2001) High-level expression of a synthetic red-shifted GFP coding region incorporated into transgenic chloroplasts. Plant J., 27, 257–265. [DOI] [PubMed] [Google Scholar]
- 31.Del Campo E.M., Sabater,B. and Martin,M. (2002) Post-transcriptional control of chloroplast gene expression. Accumulation of stable psaC mRNA is due to downstream RNA cleavages in the ndhD gene. J. Biol. Chem., 277, 36457–36464. [DOI] [PubMed] [Google Scholar]
- 32.Neckermann K., Zeltz,P., Igloi,G.L., Kossel,H. and Maier,R.M. (1994) The role of RNA editing in conservation of start codons in chloroplast genomes. Gene, 146, 177–182. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Serrano M., Del Campo,E.M., Sabater,B. and Martin,M. (2001) Primary transcripts of ndhD of Liliaceae and Aloaceae require editing of the start and 20th codons. J. Exp. Bot., 52, 179–180. [PubMed] [Google Scholar]
- 34.Burrows P.A., Sazanov,L.A., Svab,Z., Maliga,P. and Nixon,P.J. (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J., 17, 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naver H., Boudreau,E. and Rochaix,J.D. (2001) Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell, 13, 2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirose T., Kusumegi,T. and Sugiura,M. (1998) Translation of tobacco chloroplast rps14 mRNA depends on a Shine–Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett., 430, 257–260. [DOI] [PubMed] [Google Scholar]
- 37.Lu B., Wilson,R.K., Phreaner,C.G., Mulligan,R.M. and Hanson,M.R. (1996) Protein polymorphism generated by differential RNA editing of a plant mitochondrial rps12 gene. Mol. Cell. Biol., 16, 1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu B. and Hanson,M.R. (1994) A single homogeneous form of ATP6 protein accumulates in petunia mitochondria despite the presence of differentially edited atp6 transcripts. Plant Cell, 6, 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams M.A., Tallakson,W.A., Phreaner,C.G. and Mulligan,R.M. (1998) Editing and translation of ribosomal protein S13 transcripts: unedited translation products are not detectable in maize mitochondria. Curr. Genet., 34, 221–226. [DOI] [PubMed] [Google Scholar]
- 40.Phreaner C.G., Williams,M.A. and Mulligan,R.M. (1996) Incomplete editing of rps12 transcripts results in the synthesis of polymorphic polypeptides in plant mitochondria. Plant Cell, 8, 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor W.C., Barkan,A. and Martienssen,R.A. (1987) Use of nuclear mutants in the analysis of chloroplast development. Dev. Genet., 8, 305–320. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima Y. and Mulligan,R.M. (2001) Heat stress results in incomplete C-to-U editing of maize chloroplast mRNAs and correlates with changes in chloroplast transcription rate. Curr. Genet., 40, 209–213. [DOI] [PubMed] [Google Scholar]