Abstract
Malonyl-CoA is an essential intermediate in fatty acid synthesis in all living cells. Here we demonstrate a new role for this molecule as a global regulator of lipid homeostasis in Gram-positive bacteria. Using in vitro transcription and binding studies, we demonstrate that malonyl-CoA is a direct and specific inducer of Bacillus subtilis FapR, a conserved transcriptional repressor that regulates the expression of several genes involved in bacterial fatty acid and phospholipid synthesis. The crystal structure of the effector-binding domain of FapR reveals a homodimeric protein with a thioesterase-like ‘hot-dog' fold. Binding of malonyl-CoA promotes a disorder-to-order transition, which transforms an open ligand-binding groove into a long tunnel occupied by the effector molecule in the complex. This ligand-induced modification propagates to the helix-turn-helix motifs, impairing their productive association for DNA binding. Structure-based mutations that disrupt the FapR–malonyl-CoA interaction prevent DNA-binding regulation and result in a lethal phenotype in B. subtilis, suggesting this homeostatic signaling pathway as a promising target for novel chemotherapeutic agents against Gram-positive pathogens.
Keywords: fatty acid synthesis, Gram-positive bacteria, lipid homeostasis, malonyl-CoA, X-ray crystallography
Introduction
Our understanding of the relationship between metabolic signals and global changes in gene expression is limited by the difficulty in identifying intracellular signaling molecules that interact with key regulatory proteins. This gap is particularly apparent in the control of membrane lipid homeostasis. Fatty acids and their derivatives play essential roles in all living organisms as components of membranes and source of metabolic energy. Biosynthesis of these compounds involves repeated cycles of condensation, reduction and dehydration of carbon–carbon linkages, which are carried out by a single multifunctional polypeptide (type I systems) in higher eukaryotes (Smith et al, 2003) and by discrete proteins (type II systems) in bacterial cells, plant chloroplasts and malaria parasites (White et al, 2005). Despite the complexity of these biosynthetic pathways, biological membranes maintain stable compositions that are characteristic for different organisms, tissues and intracellular organelles. However, the precise homeostatic mechanisms maintaining the concentration of lipids at particular levels are largely unknown.
Transcriptional regulation of bacterial lipid biosynthetic genes is poorly understood at the molecular level. Indeed, the only well-documented example is probably that of fabA and fabB, two essential genes for the synthesis of unsaturated fatty acids in Escherichia coli (Lu et al, 2004; Schujman and de Mendoza, 2005). Two transcriptional regulators belonging to the TetR superfamily, the activator FadR and the repressor FabR, were shown to regulate the expression of the fabA and fabB genes. FadR was discovered as a repressor of the β-oxidation regulon (Overath et al, 1969; Dirusso and Nunn, 1985), and subsequently found to also activate fabA and fabB transcription (Henry and Cronan Jr, 1991, 1992; Lu et al, 2004; Schujman and de Mendoza, 2005). Binding of FadR to its DNA operator is antagonized by different long-chain acyl-CoAs (Dirusso et al, 1992; Raman and Dirusso, 1995; Cronan Jr and Subrahmanyam, 1998; Dirusso et al, 1998), in agreement with the structural model (van Aalten et al, 2000, 2001; Xu et al, 2001). The second E. coli regulator, FabR, acts as a repressor in the regulation of fabA and fabB (Zhang et al, 2002), although it is still unclear which signal(s) modulates its DNA binding activity.
A major advance in our understanding of the transcriptional control of bacterial lipid synthesis was achieved through the identification of FapR, a global transcriptional repressor that controls the expression of many genes involved in the biosynthesis of fatty acids and phospholipids (the fap regulon) in Bacillus subtilis (Schujman et al, 2003). FapR has highly conserved homologs in many Gram-positive bacteria, including several human pathogens (Schujman et al, 2003). As well, in all these organisms the consensus binding sequence of FapR is largely invariant in the putative promoter regions of the fapR gene, indicating that the regulation mechanism observed in B. subtilis is conserved in many other organisms.
We demonstrate here that FapR belongs to a new class of bacterial repressors and that malonyl-CoA, an essential intermediate in fatty acid synthesis, operates as the direct and specific inducer of FapR-regulated promoters. The effector-binding domain of FapR was found to display a ‘hot-dog' fold, similar to that of several thioesterases known to process acyl-CoA substrates but different from other known bacterial transcriptional regulators. Binding of malonyl-CoA promotes a disorder-to-order transition in the protein that causes the FapR-DNA complex to dissociate or prevents its formation. Finally, we show that site-directed mutations disrupting the FapR–malonyl-CoA interaction result in a lethal phenotype in B. subtilis, validating the structural model and suggesting that this homeostatic signaling pathway could be a target for novel chemotherapeutic agents against Gram-positive pathogens.
Results and discussion
Malonyl-CoA is a direct and specific regulator of FapR activity
Two observations suggested that the intracellular pool of malonyl-CoA might be associated with the regulation of FapR activity. First, expression of the fap regulon is derepressed by antibiotics that inhibit fatty acid biosynthesis (with the concomitant increase in the intracellular levels of malonyl-CoA; Schujman et al, 2001). Second, this upregulation is abolished by precluding the transcription of genes encoding the subunits of the acetyl-CoA carboxylase (ACC), which catalyzes the synthesis of malonyl-CoA (Schujman et al, 2003). A major question is whether malonyl-CoA directly regulates FapR activity or acts as a building block, converted into another product that is the signaling molecule. For the malonate group to be used in fatty acid synthesis, it must be transferred from malonyl-CoA to acyl-carrier-protein by malonyl-CoA:ACP transacylase, the product of the fabD gene (de Mendoza et al, 2002). After conditional inhibition of the B. subtilis fabD gene (Morbidoni et al, 1996), we still observed transcriptional induction of the fap regulon by antibiotics that inhibit the biosynthetic pathway (data not shown), suggesting that malonyl-CoA could be the direct effector of FapR. To prove this hypothesis, we performed in vitro transcription experiments, initiating at fapR (see Materials and methods). A transcript of the expected size (93 nt) was obtained in the absence of FapR, whereas fapR transcription was inhibited in reactions containing the repressor (Figure 1), confirming that FapR represses transcription. To test whether malonyl-CoA relieves FapR-mediated repression, we repeated the experiments in the presence of FapR and malonyl-CoA at concentrations ranging from 15 to 500 μM. As shown in Figure 1, increasing concentrations of malonyl-CoA gradually induced in vitro fapR transcription. Furthermore, this effect is highly specific since several acyl-CoA derivatives related to malonyl-CoA (such as acetyl-CoA, propionyl-CoA, succinyl-CoA and butyryl-CoA), were not able to relieve the inhibitory action of the repressor on fapR transcription (Figure 1). Similar results were obtained by in vitro transcription analysis using the promoters of other genes belonging to the fap regulon, such as fabI and yhdO, which were repressed by FapR and specifically induced by malonyl-CoA (data not shown). Thus, all these experiments prove that FapR is unable to repress transcription of the fap regulon in the presence of malonyl-CoA, and that this molecule operates as a direct and specific inducer of the fap promoters.
Figure 1.
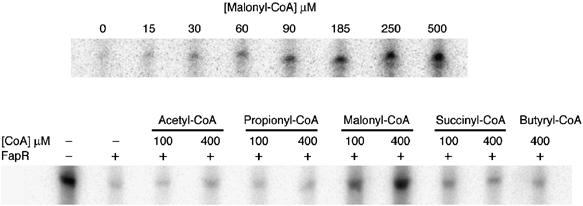
Effect of malonyl-CoA and short-chain acyl-CoA thioesters on FapR-mediated repression. In vitro transcription was performed with a PfapR promoter fragment (6.4 nM) as the template in the presence of FapR (120 nM) and different concentrations of malonyl-CoA (upper panel) or malonyl-CoA analogs (lower panel).
Malonyl-CoA binds FapR and modulates the repressor-operator interaction
Sequence analysis suggests that FapR, like many bacterial repressors, is a two-domain protein with an N-terminal DNA-binding domain (DBD) connected through a helical linker to a larger C-terminal domain (Figure 2). This second domain has weak sequence identity with thioesterases, a family of enzymes known to bind and process acyl-CoA substrates (Dillon and Bateman, 2004), suggesting that the thioesterase-like domain (TLD) of FapR might bear the effector-binding function. To prove this hypothesis, we produced two deletion mutants of FapR (FapRΔ43 and FapRΔ67) that contain the TLD domain but lack the DBD (Figure 2). One of them (FapRΔ43) still harbors the linker helix αL. Isothermal titration calorimetric (ITC) measurements demonstrate that malonyl-CoA binds full-length FapR with an affinity constant in the micromolar range (Figure 3A), which is within a physiologically relevant range (Heath et al, 2002). The malonyl-CoA binding site is entirely defined by the C-terminal domain of the protein, since malonyl-CoA also binds FapRΔ43 with a similar affinity constant. In each case, two molecules of inducer bind the protein dimer in independent non-cooperative events, as indicated by a Scatchard plot derived from the thermodynamic data (Supplementary Figure S1). However, the binding of malonyl-CoA to FapR and FapRΔ43 display some significant differences. While effector binding to FapR was largely entropy driven (ΔH°/ΔG°=24%), binding to FapRΔ43 was enthalpy driven (ΔH°/ΔG°=67%) and displayed a three-fold decrease of the heat capacity change (ΔCp) on binding (see Figure 3A, legend). Both the different nature of the binding driving force and the change of ΔCp might be attributed to a different environment of the malonyl-CoA binding-site in the absence of the HTH motif. On the other hand, under the same experimental conditions the shorter construct FapRΔ67 presented a flat binding isotherm at three different temperatures (Figure 3A). A null enthalpy value over a 20° temperature interval strongly argues for a severe decrease of the malonyl-CoA binding affinity upon deletion of the linker helix αL, indicating that this helix must play an important role in the formation of a competent effector-binding site in FapR.
Figure 2.
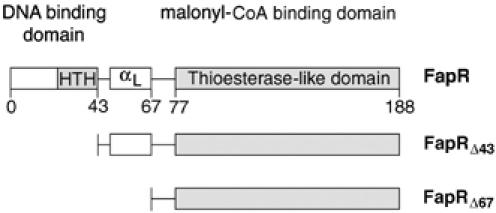
Domain organization of B. subtilis FapR and the two deletion mutants used in this work.
Figure 3.
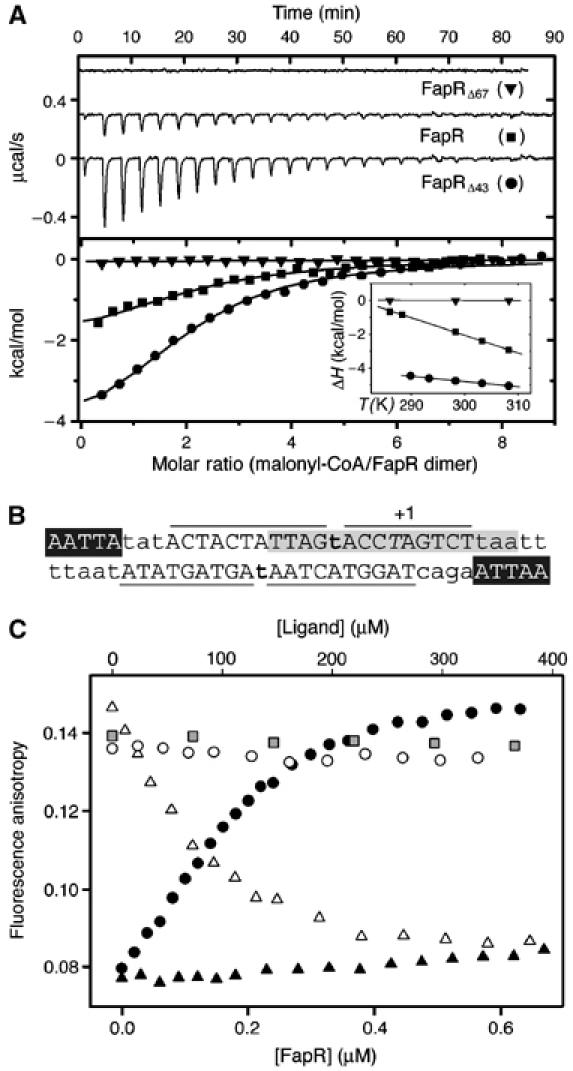
Malonyl-CoA binds to the C-terminal domain of FapR and modulates DNA-binding activity. (A) Isothermal calorimetric titrations of FapR with malonyl-CoA. The top panel shows the raw heat signal for 10 μl injections of 240 μM malonyl-CoA into solutions of 16 μM FapR, 20 μM FapRΔ43, and 20 μM FapRΔ67, respectively, at 298 K (curves have been offset by 0.3 μcal/s for clarity). The bottom panel shows the transition curves of the three proteins (FapR, Kd=2.4±0.2 μM; FapRΔ43, Kd=7.1±0.9 μM). The inset shows the enthalpy changes for the above reactions as a function of temperature (FapR, ΔH°=−1.8±0.2 kcal mol−1, ΔCp=−103.0±0.3 cal mol−1 K−1; FapRΔ43, ΔH°=−4.8±0.3 kcal mol−1, ΔCp=−31.9±1.1 cal mol−1 K−1). FapRΔ67 displays a flat binding isotherm at all tested temperatures between 288 and 308 K. (B) PfapR operator region. Capital letters indicate bases protected by FapR from DNAseI digestion; lines delimitate protected regions. Bold letters show hypersensitive spots. Italic T indicates the transcription start base. The 17 bp inverted repeat conserved in all operators of the fap regulon (Schujman et al, 2003) is shown in gray and white letters indicate a 5 bp inverted repeat separated by 23 nt. (C) Fluorescence anisotropy changes on addition of FapRWT to 9.5 nM 34 bp F-dsDNA (black circles); on addition of FapRWT to a mixture of 9.5 nM 34 bp F-dsDNA and 307.7 μM malonyl-CoA (black triangles); and on addition of malonyl-CoA (white triangles), acetyl-CoA (white circles) or malonic acid (gray squares) to the previously formed F-dsDNA/FapRWT complex.
The DBD of FapR is predicted to contain a typical HTH domain, which displays a conserved sequence motif in the putative DNA-recognition helix. This motif is similar to that of the deoxyribose repressor DeoR family (Aravind et al, 2005), although the FapR family of repressors lacks the β-hairpin characteristic of the DeoR winged domains (Ni et al, 1999). DNAse I footprinting analyses on both strands of the fapR promoter (Supplementary Figure S2) revealed that the repressor protects a 34 bp DNA region composed by the dyad symmetric consensus element flanked by 5′-AATTA-3′ inverted repeats (Figure 3B). ITC measurements at 25°C show that wild-type FapR binds the 34 bp dsDNA with a dissociation constant of 0.2 μM in an enthalpy-driven event (data not shown). To investigate the effect of malonyl-CoA on FapR–DNA interactions, we performed titration experiments adding FapR to fluorescein-labeled 34 bp dsDNA. Direct binding of DNA to FapR is evidenced by an increase of the fluorescence anisotropy (Figure 3C). Addition of malonyl-CoA to the FapR-DNA adduct results in a significant decrease of the fluorescence anisotropy, whereas acetyl-CoA or malonic acid are unable to induce the same effect. Furthermore, attempts to bind DNA in the presence of saturating concentrations of malonyl-CoA result in a modest increase in the fluorescence anisotropy, indicating that DNA binding barely takes place under these conditions (Figure 3C). These experiments confirm that malonyl-CoA is a specific effector molecule that releases FapR from (or prevents binding to) its DNA operator.
Structural basis of malonyl-CoA-binding specificity
The crystal structure of the C-terminal domain of FapR (FapRΔ67) was determined at 2.5 Å resolution using a combination of MAD and molecular replacement techniques (Table I). The protein is a homodimer that displays the typical ‘hot-dog' fold characteristic of the thioesterase enzyme family (Leesong et al, 1996; Li et al, 2000). Each monomer folds into an α/β globular domain with a central α-helix (αC) surrounded by a six-stranded antiparallel β-sheet ‘bun'. Topologically, αC is inserted between strands β2 and β3, with the last four β-strands (β3–β6) arranged in a Greek key motif. The dimer is formed by extensive interaction between the β3 strands of both monomeric partners, giving rise to a single inter-monomeric antiparallel β-sheet, which wraps along its concave side both αC helices (Figure 4). In each monomer, the N-terminal residues of FapRΔ67 (up to position 73) are present but disordered in the crystal structure.
Table 1.
Data collection, phasing and refinement statistics
| Data set | SeMet-labeled FapR | FapRΔ67 | FapRΔ43–malonyl-CoA | ||
|---|---|---|---|---|---|
| Data collection | |||||
| Resolution (Å)a | 40–3.5 (3.69–3.5) | 60–2.5 (2.64–2.5) | 63.2–3.1 (3.27–3.1) | ||
| Wavelength (Å) | 0.9791 | 0.9793 | 0.9755 | 1.072 | 0.9794 |
| Measured reflections | 22 839 | 22 837 | 23 045 | 74 467 | 87 334 |
| Multiplicitya | 5.8 (5.8) | 5.8 (5.7) | 5.8 (5.8) | 5.0 (4.3) | 6.9 (7.2) |
| Completeness (%)a | 99.4 (99.4) | 99.5 (99.7) | 99.3 (99.2) | 80.5 (50.0) | 100 (100) |
| Rsym (%)a,b | 9.3 (27.5) | 9.7 (32.6) | 11.5 (40.3) | 8.3 (28.7) | 7.9 (31.0) |
| 〈I/σ〉a | 13.5 (4.9) | 12.8 (4.2) | 11.3 (3.3) | 13.7 (4.8) | 19.3 (6.0) |
| Refinement | |||||
| Resolution (Å) | 30–2.5 | 63.2–3.1 | |||
| Rcryst c (No. refs) | 0.221 (14035) | 0.187 (11604) | |||
| Rfree c (No. refs) | 0.267 (790) | 0.227 (941) | |||
| R.m.s. bonds (Å) | 0.019 | 0.02 | |||
| R.m.s. angles (deg) | 1.72 | 2.14 | |||
| Protein atoms | 2964 | 2238 | |||
| Water molecules | 8 | 10 | |||
| Ligand atoms | — | 64 | |||
| aValues in parentheses apply to the high resolution shell. | |||||
b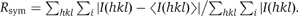
| |||||
c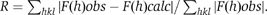
| |||||
| Rcryst and Rfree were calculated from the working and test reflection sets, respectively. | |||||
Figure 4.
Overall structure of the FapRΔ43–malonyl-CoA complex. Ribbon model with secondary structure elements (β-strands 1–6, the linker helix αL and central helix αC) indicated for one monomer.
Formation of the FapR dimer buries 1150 Å2 from each monomer surface, which represents 13.5% of the total accessible surface. The protein presents this same oligomeric arrangement in solution, as indicated by studies in solution of full-length FapR and the two deletion mutants (FapRΔ43 and FapRΔ67) using gel filtration (Supplementary Figure S3), dynamic light scattering and glutaraldehyde cross-linking (data not shown) experiments.
To further investigate the FapR–malonyl-CoA interactions, we crystallized FapRΔ43 in complex with the effector molecule and determined the structure of the complex at 3.1 Å resolution (Table I). Crystals of this complex belong to a different space group than those of the unliganded proteins FapRΔ43 or FapRΔ67 (see Materials and methods), further suggesting that malonyl-CoA binding induced a conformational change in the protein. Two malonyl-CoA molecules are deeply buried into the globular core of the dimer (Figure 4). A large fraction of the ligand molecules is positioned within equivalent tunnels that run perpendicular to both the extended β-sheet and the central αC helices on either side of the dimer. Instead, the 3′P-nucleoside moiety of the ligand (disordered in the crystal structure) sticks out from the globular core and is exposed to the bulk solvent, as observed in other acyl-CoA-thioesterase complexes (Thoden et al, 2003). The linker helices αL, which were missing in FapRΔ67, are now visible in the structure of the FapRΔ43–malonyl-CoA complex (Figure 4), clearly detached from the TLD core.
The malonyl moiety of the ligand is critical for specificity, since analogous acyl-CoA derivatives are significantly less active in transcription experiments (Figure 1) and do not interfere with FapR–DNA binding (Figure 3C). Both the size of the malonyl moiety (to fit within the binding cavity) and the presence of a carboxylate group at position 1 are primary factors influencing effector-binding specificity. The malonyl carboxylate sits on top of the N-terminal end of helix αC, within a hydrophobic pocket defined by residues Leu69′, Val119′, Asn115′, Ser116′, Phe99 and His108 (primed/unprimed numbers indicate residues from each monomer in the homodimer). Upon malonyl-CoA-binding, the guanidinium group of Arg106 moves from its outer position in free FapR towards the hydrophobic pocket, where it stacks against the aromatic ring of Phe99 and forms a salt bridge with the malonyl CO2− group. Interestingly, Phe99 and Asn115′ (H-bonded to the thioester carbonyl oxygen of the malonyl moiety) are largely conserved in the FapR family of bacterial repressors and occupy the equivalent positions of the two catalytic residues in homologous thioesterases (Thoden et al, 2002).
Malonyl-CoA binding to FapR induces significant conformational changes
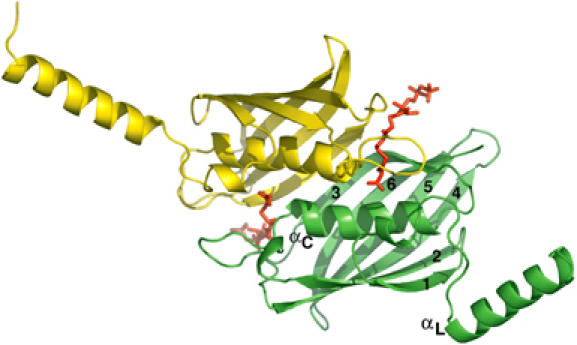
The structural comparison of malonyl-CoA-bound and free FapR reveals that an open groove in the ligand-free protein becomes a tunnel enclosing the effector molecule in the complex (Figure 5A). These changes are due to the structuring of three protein loops, which are partially or totally disordered in ligand-free FapR but display a well-defined conformation in the complex (Figure 5B), with no obvious crystal packing interactions that could account for these differences. The open cleft in free FapR facilitates ligand binding, avoiding the costly path of introducing the charged malonate moiety through a pre-existent hydrophobic tunnel (Benning et al, 1998).
Figure 5.
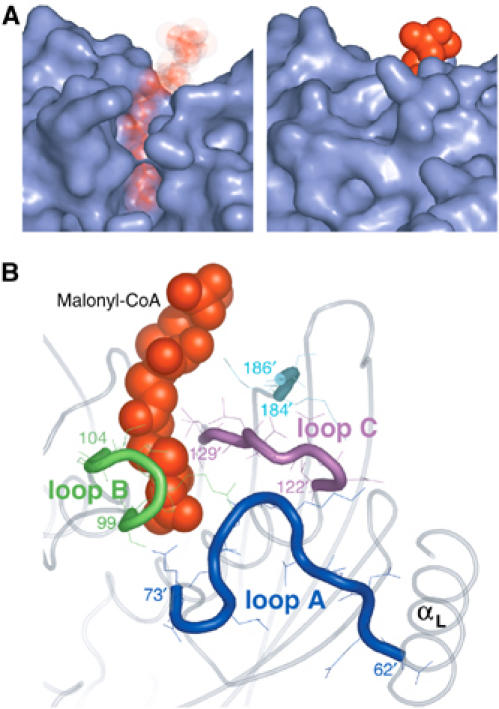
(A) Molecular surface of the binding site in the ligand-free (left) and ligand-bound (right) FapR structures. Malonyl-CoA (in red) is shown for reference in the ligand-free structure. (B) Formation of the tunnel involves the structuring of three protein loops (A–C) and the C-terminal end of the protein, which are highly mobile or disordered in ligand-free FapR.
The critical interaction between the malonyl carboxylate and Arg106 side-chain at the end of loop B triggers the formation of several H-bonding interactions that ultimately stabilize the conformation of loops A–C in the complex (Figure 6). Upon malonyl-CoA binding, the Arg106 guanidinium group gets engaged in electrostatic interactions with both the malonyl CO2− and the main-chain carbonyl of Glu73′ in loop A. The movement of Arg106 allows the rearrangement of the Glu73′ side-chain, which now forms a strong H-bond with Ser100 on loop B. In turn, the new conformation of loop B facilitates the interaction of the Arg101 guanidinium with the main-chain carbonyls of Lys67′ (loop A) and Glu125′ (loop C), further stabilized by the formation of a strong salt bridge between Lys67′ (loop A) and Asp123′ (loop C). This closure event extends like a ‘zipper' towards the N-terminus of loop A, involving the formation of additional H-bonds, up to the linker helix αL connecting the TLD and DBD domains. Additional interactions of the ligand phosphates with Tyr183 and Lys186 contribute to stabilize the C-terminal segment of the protein, which is also disordered in ligand-free FapR but makes contacts with loop C residues in the complex.
Figure 6.
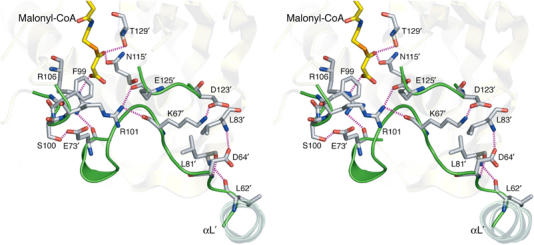
Stereoview of the FapRΔ43–malonyl-CoA complex showing the main hydrogen-bonding interactions (in magenta) involved in the ‘zipper' effect, which are absent in the ligand-free protein (see text). Loops A–C, which are disordered in ligand-free FapR, are shown in green.
A model of action for FapR
The malonyl-CoA-induced structural rearrangements described above severely constrain the spatial mobility of the linker helix αL with respect to the TLD core, supporting a mechanism for the regulation of transcription (Figure 7). According to this model, the flexibility of loop A in the repressed state allows the two DBDs of FapR to adopt a competent operator-binding conformation. When malonyl-CoA binds to FapR, a disorder-to-order transition of loop A during formation of the ligand-binding tunnel modifies the orientation of helix αL, pulling apart the DBDs from each other and disrupting their productive association for operator binding. This allosteric mechanism differs from that of other bacterial repressors, for which the ability to bind DNA typically involves a moderate hinge-bending motion between domains (Friedman et al, 1995; Orth et al, 2000; van Aalten et al, 2001).
Figure 7.
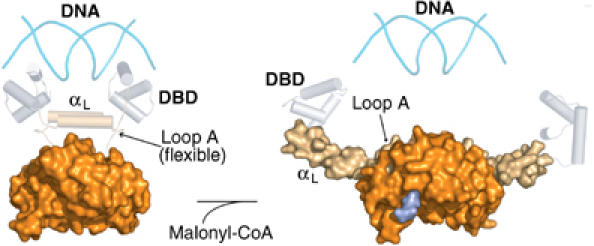
Proposed model of action of FapR. The figure shows the repressed (operator-bound, left) and derepressed (malonyl-CoA-bound, right) states of FapR. In each case, molecular surfaces represent the experimental X-ray structures. The DBDs (gray cylinders) are modeled on the MecI/BlaI repressor family (Garcia-Castellanos et al, 2003). The proposed αL–αL interaction for the repressed state is indeed observed between neighboring dimers in the crystal structure of the FapRΔ43–malonyl-CoA complex.
Several lines of evidence support the above model. First, most residues involved in the ‘zipper' effect are largely conserved in FapR proteins from different species (Schujman et al, 2003). Second, the structure of the complex shows the linker helices (αL) protruding away from the dimeric core in opposite directions (Figure 4), incompatible with an architecture allowing DNA-binding through the two N-terminal domains. In agreement with this observation, residues 68–73 from loop A, which are determinant for αL stiffening in the derepressed state, are present but disordered in two different crystal forms of unbound FapR, suggesting a flexible conformation. Such flexibility is confirmed by proteolysis experiments showing that loop A is accessible to trypsin in the ligand-free protein as well as in the presence of inactive acyl-CoA molecules. Instead, binding of malonyl-CoA to FapR protects this region against trypsin cleavage (Supplementary Figure S4), fully supporting the conclusions derived from the structural model.
In the absence of inducer, the FapR model predicts a direct interaction between the two amphipatic linker helices αL (Figure 7) in order to achieve a functional association of the associated HTH motifs for DNA binding. Although we lack the crystal structure of the FapR–DNA complex, the helix–helix interaction may resemble that observed between neighboring dimers in the FapRΔ43–malonyl-CoA complex (Figure 8A). We therefore produced three point mutants to test this hypothesis. One of them, Ile54-Cys, was designed to crosslink the two helices through a disulfide bridge, without affecting the protein functionality. The other two, Ile54-Asp and Val57-Pro, were intended to interfere with helix dimerization and DNA-binding capacity. As expected, FapRI54C was found to form a covalent dimer (Figure 8B) without losing its DNA-binding capability (Figure 8C), whereas the introduction of an aspartate at position 54 (FapRI54D) or a proline at position 57 (FapRV57P) precluded a functional helix–helix association and rendered the protein unable to bind DNA (Figure 8C). These results strongly support a close spatial proximity of the two linker helices in inducer-free FapR, as predicted by the model, and demonstrate a significant conformational change of the protein upon malonyl-CoA binding, since disulfide bridge formation in FapRI54C would not be possible if the helices had the orientation observed in the crystal structure of the repressor-inducer complex (Figure 4).
Figure 8.
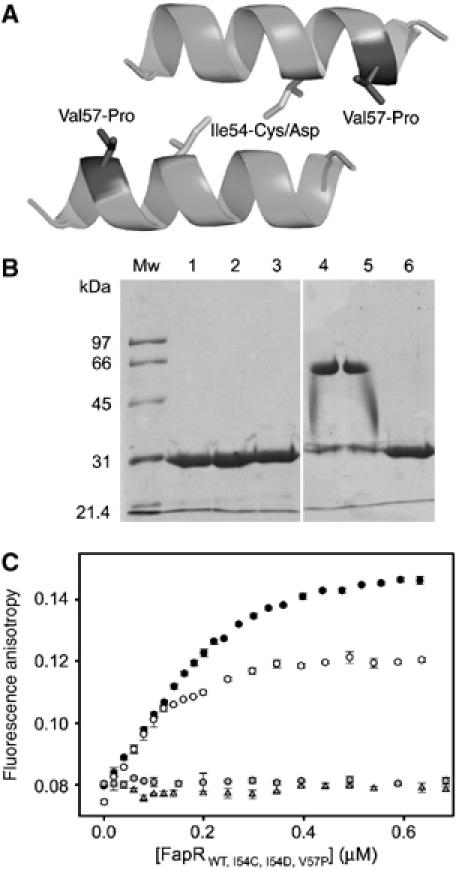
Mutational analysis of linker helix dimerization. (A) Helix–helix interaction as observed in crystal packing contacts of the FapRΔ43–malonyl-CoA complex. The amino-acid positions mutated are indicated. (B) SDS–PAGE of purified FapR (lanes 1–3) and FapRI54C (lanes 4–6) under oxidative (lanes 1 and 4), nonoxidative (lanes 2 and 5) and reducing (lanes 3 and 6) conditions. Both proteins behave as a homodimer in solution, as indicated by gel filtration chromatography (data not shown). (C) Fluorescence anisotropy changes on addition of wild-type FapR (black circles), FapRI54C (white circles), FapRI54D (gray triangles) and FapRV57P (gray circles) to 9.5 nM 34 bp F-dsDNA. As for the wild-type protein, the FapRI54C–DNA complex is dissociated upon addition of malonyl-CoA (data not shown).
Disruption of the malonyl-CoA–FapR interaction is deleterious for cell growth
To further validate the proposed model, we designed site-directed mutations predicted to impair the malonyl-CoA binding properties of FapR. Thus, the substitution Arg106-Ala was aimed at disrupting its interaction with the malonyl, while the double substitution Gly107-Val/Leu128-Trp would block the tunnel region, hindering the formation of the complex. Fluorescence anisotropy experiments confirmed that the mutant proteins are still able to bind the DNA target, whereas DNA dissociation could only be achieved at higher concentrations of malonyl-CoA compared to wild-type FapR (Figure 9A). Consistently, direct ITC measurements of the FapRR106A–malonyl-CoA interaction showed a flat isotherm using different ligand concentrations (20–100 μM FapRR106A, 0.5–2 mM malonyl-CoA) at different temperatures (298 and 308 K), confirming a significant (>1000 fold) decrease in the affinity of the mutant protein for malonyl-CoA (data not shown).
Figure 9.
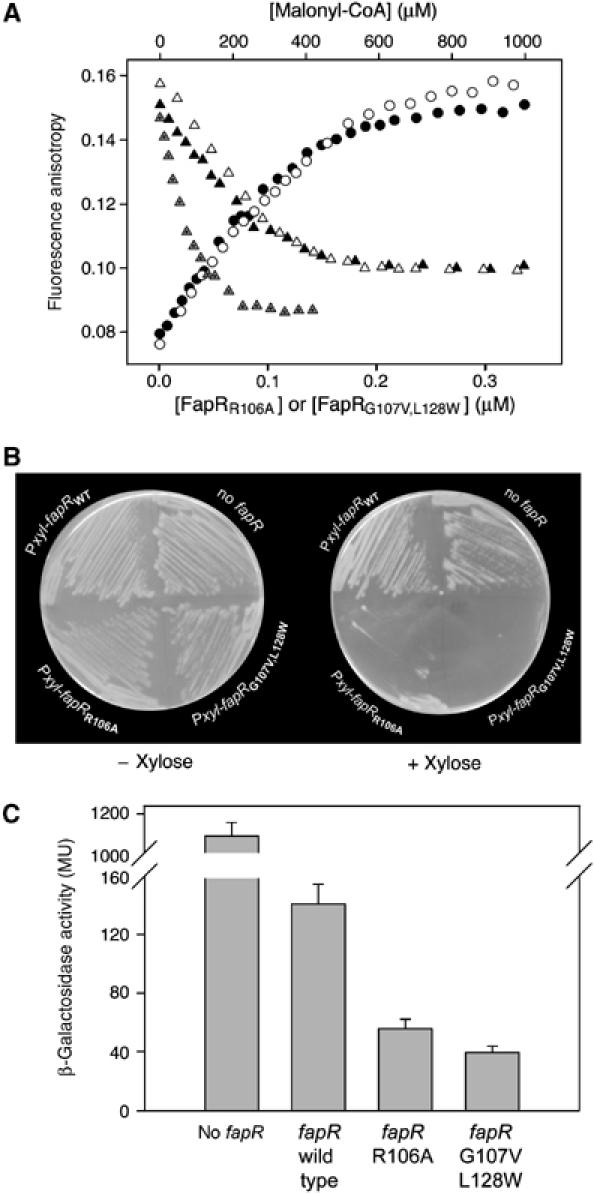
Mutations in the FapR malonyl-CoA binding pocket are deleterious for cell growth. (A) Fluorescence anisotropy changes upon addition of FapRR106A to 9.5 nM 34 bp F-dsDNA (black circles); on addition of FapRG107V,L128W to 9.5 nM 34 bp F-dsDNA (white circles); on addition of malonyl-CoA to the previously formed F-dsDNA/FapRWT complex (gray triangles); on addition of malonyl-CoA to the previously formed F-dsDNA/FapRR106A complex (black triangles); and on addition of malonyl-CoA to the previously formed F-dsDNA/FapRG107V,L128W complex (white triangles). (B) Plasmids were constructed in which fapRWT, fapRR106A or fapRG107V,L128W were under the control of a xylose-inducible promoter (Pxyl) and introduced into a strain, GS364, that was a mutant for fapR and contained a PfabHA-lacZ fusion. The strains were grown on LB solid medium in the absence and in the presence of xylose (1%) (C) β-galactosidase activities from PfabHA-lacZ in strain GS364 expressing fapRWT,fapRR106A or fapRG107V,L128W. To induce expression of the fapR alleles, the different strains previously grown on LB agar solid medium, were resuspended in LB medium supplemented with xylose 1% to an OD525 of 0.1. After two hours of growth at 37°C, samples were collected and β-galactosidase activities determined.
These fapR point mutants were also expressed under the control of the xylose inducible PxylA promoter in a B. subtilis strain containing a fapR deletion. Xylose-induced expression of FapRR106A caused a marked drop in cell viability, while cells expressing FapRG107V,L128W failed to grow (Figure 9B). As expected, transcription of the FapR-regulated promoter fabHA was greatly decreased when the expression of the two FapR mutants was induced with xylose (Figure 9C). These data validate the hypothesis that malonyl-CoA plays a central role as a metabolic signal controlling lipid biosynthesis in B. subtilis and, given the widespread occurrence of this pathway in several Gram-positive pathogens (Schujman et al, 2003), the lethal phenotype associated with the disruption of FapR–malonyl-CoA interactions provides the basis for the development of new antibacterial compounds.
In conclusion, malonyl-CoA was known to serve a dual function as a general intermediate in fatty acid synthesis and as a regulatory effector controlling the import of fatty acids into mitochondria for β-oxidation (Dowell et al, 2005). We now demonstrate a novel role for this compound as a signaling molecule regulating the activity of FapR to control membrane lipid homeostasis. This molecule is ideally suited for sensing the status of fatty acid biosynthesis because the acc genes, responsible for malonyl-CoA synthesis, are not under FapR control and the only known fate of malonyl-CoA is fatty acid synthesis in B. subtilis and most other bacteria. The control of FapR activity provides the first reported example of transcriptional regulation mediated by this biosynthetic intermediate. The possibility of this type of regulation has long been recognized in higher organisms, where malonyl-CoA acts as a mediator linking fatty acid synthesis to the control of food intake and energy expenditure, probably by modulating the activity of a signaling protein that controls the expression of hypothalamic neuropeptides (Dowell et al, 2005). Therefore, malonyl-CoA appears to be a universal signal molecule forming part of a control mechanism that monitors energy balance, conserved from bacteria to humans.
Materials and methods
Plasmids and strains
Plasmids were constructed using standard methods and amplified in E. coli DH5α and XL1-Blue. PCR-fragments were amplified from chromosomal DNA of B. subtilis strain JH642 (Dean et al, 1977). Oligonucleotide primers were purchased from Sigma Genosys.
To obtain a fapR null strain, four DNA fragments were amplified by PCR and fused by overlap extension PCR (Ho et al, 1989). These fragments contained the last 457 bp of recG (gene upstream of fapR), the promoterless and terminatorless cat gene, the promoter region of fapR and the last 150 bp of fapR plus the first 350 bp of the downstream gene plsX. Strain GS273 (amyE∷PfabHA-lacZ Sp) (Schujman et al, 2003) was transformed with the whole PCR fragment to give strain GS364. The fapR deletion was confirmed by PCR and Western blot analyses.
Site-directed mutagenesis was performed by overlap extension PCR (Ho et al, 1989). For ectopical expression of fapR alleles, a 631 bp DNA fragment containing the ribosome binding site sequence and full-length fapR allele was amplified using the primers ylpC5Hind (5′-GTACCCAAGCTTAATTGTCCGGATGGTGTT-3′) and ylpC3Bam (5′-CTACAGGGATCCTCATGCATGCTCCACCTT-3′). For each allele, the PCR fragment was digested with HindIII and BamHI (sites underlined in the primer sequences) and cloned into pGES247 (pDG1730 (Guerout-Fleury et al, 1996) containing xylR-PxylA), downstream of the xylA promoter. EcoRI–BamHI DNA fragments from the resulting plasmids, containing PxylA-fapR alleles, were subcloned into pHPKS (Johansson and Hederstedt, 1999) digested with EcoRI–BamHI and transformed into GS364. Plasmids expressing the histidine tagged fusions of FapR alleles were obtained as described (Schujman et al, 2003).
Limited proteolysis and N-terminal sequence analyses
Recombinant purified FapR or FapR43 (25 μg) were incubated with trypsin (0.05 μg, Sigma) in 250 μl of 25 mM Tris–HCl pH 7.5, 10 mM CaCl2, in the presence of 5 mM malonyl-CoA (or related analogs) for 0–90 min at 37°C. The reactions were stopped by addition of 100 mM Tris–HCl pH 8.8, 2% SDS, 10% glycerol, 100 mM DTT and 0,01% bromophenol blue. Samples were boiled for 3 min at 95°C and analyzed by SDS–PAGE. Proteolytic fragments were transferred onto Hybond-P PVDF (Amersham Biosciences) and the Coomassie stained bands were subjected to N-terminal sequence analysis (ABI494 Protein Sequencer, Applied Biosystems).
In vitro transcription
In vitro transcription assays were performed as described (Opdyke et al, 2001). As templates for the reactions, DNA fragments containing the promoter regions of fapR (192 bp), yhdO (175 bp) and fabI (185 bp) were amplified by PCR using the primers ylpC116U (5′-CGGAGGCATGCACGGGGAAATATTAAG-3′) and ylpC116L (5′-TGCTTGGATCCTCTGCTGAAGTAATTCC-3′), yhdOKpn (5′-GTTTAAGGTACCGCCGACCAGAGAGGAA-3′) and yhdOBam (5′-TATACGGATCCCACTTCCTTTGTCGTAATC-3′), fabI116U (5′-AGCTTAGGTACCAGCCGATGAGATCAC-3′) and fabI116L (5′-ATGTTACGGGATCCAAGTGAAAAAATC-3′), respectively. Gels were developed and digitalized with a Storm 840 scanner (Amersham). Short chain acyl-CoAs were purchased from Sigma, diluted in 10 mM HAc and their concentration calculated from absorbance at 260 and 232 nm.
DNAse I footprinting assay
This assay was performed as described (Aguilar et al, 2001). DNA for the assay was obtained by PCR using primers ylpC116U and ylpC116L.
Protein production
Two fragments of the B. subtilis fapR gene, encoding the amino acids 44–188 (FapR43) and 68–188 (FapR67), and five point mutants of the full-length protein (FapRI54C, FapRI54D, FapRV57P, FapRR106A and FapRG107V,L128W) were generated. All constructs were subcloned in the expression plasmid pQE30 (Qiagen) using standard protocols. Recombinant proteins were separately overexpressed in IPTG-induced E. coli BL21(DE3) pLysS cells, following standard protocols (Coligan et al, 2002). After 16 h at 20°C, cells were harvested and resuspended in 50 ml of BugBuster Protein Extraction Reagent (Novagen) containing protease inhibitors (Complete EDTA-free, Roche). After centrifugation, the proteins were purified in a two-step chromatographic procedure using a HisTrap Chelating column (Amersham Biosciences) followed by a MonoQ HR 5/5 (Amersham Biosciences). Selenomethionine (SeMet) labeled FapR was expressed in E. coli strain B834(DE3) (Novagen) in a DLM medium containing 0.2 g/l SeMet (2), and purified using the same procedure as described for the non-labeled protein.
Protein crystallization
The purified proteins FapR, FapRΔ43 or FapRΔ67 were concentrated to 10 mg/ml in 10 mM Tris–HCl pH 7.5. Initial crystallogenesis screenings were carried out with a nanoliter dispenser robot (Cartesian Technologies) at 18°C, and promising conditions were further refined by hand using the hanging-drop vapor-diffusion method. Both the native and SeMet-labeled FapR crystallized in 50 mM Tris pH 8.5, 10 mM MgCl2 and 15% PEG 4000. Rod-like crystals appeared after 15–30 days, grew to a maximum size of 0.15 × 0.06 × 0.06 mm3 and belong to spacegroup P41212, with cell dimensions a=b=59.1 Å, c=157.8 Å and two monomers of FapR in the asymmetric unit. N-terminal protein sequencing of the crystallized material yielded the sequence SLSLD, indicating that FapR has undergone proteolysis at Lys67–Ser68 during protein purification or storage.
Crystals of FapRΔ67 were grown in 100 mM HEPES pH 7.5, 2 M (NH4)2SO4, 2% PEG 400. They are orthorhombic, P212121, with cell dimensions a=39.2 Å, b=84.3 Å, c=155.4 Å, and have four monomers in the asymmetric unit, though the molecular packing is similar to that of the tetragonal FapR crystals. When soaked with malonyl-CoA, these crystals shattered and eventually dissolved, suggesting a ligand-induced conformational rearrangement of the protein. Diffraction-quality crystals of the complex could be obtained with FapRΔ43 (but not with FapRΔ67) in the presence of malonyl-CoA (5 mM) in 25% ethylenglycol. The crystals appeared after 30 days and belong to the tetragonal space group P41212, with cell dimensions a=b=89.4 Å, c=162.2 Å and two monomers in the asymmetric unit. Crystals of FapRΔ43 could also be grown in the absence of malonyl-CoA, but these crystals, which have trigonal (or hexagonal) point symmetry, diffracted to very low resolution (<6 Å) and were not further characterized.
X-ray structure determination and refinements
All X-ray diffraction data processing and routine crystallographic computational tasks were performed with the CCP4 suite of programs (Collaborative Computational Project, 1994). One flash-frozen crystal of SeMet-labeled FapR was subjected to a three-wavelength MAD experiment to 3.5 Å resolution at the ESRF (Grenoble, beamline ID29). Two Se atoms in the asymmetric unit (one per monomer) were located and their parameters refined with the program SHARP (Bricogne et al, 2003). The good quality of the MAD phases (mean figure-of-merit for acentric reflections=0.584, anomalous phasing power=1.26) resulted in readily interpretable electron density maps. Polypeptide chain tracing was carried out with the program O (Jones et al, 1991) and initial refinement was performed with the program CNS (Brünger et al, 1998) using a conservative strategy with non-crystallographic constraints and simulated annealing protocols.
As FapR was found to be cleaved before Ser68, we continued to work with the FapRΔ67 construction to improve the diffraction quality of the crystals. Using 37% sucrose in mother liquor as cryoprotectant, a diffraction data set was collected at 2.5 Å resolution using synchrotron radiation (ID23-ESRF). The diffraction pattern remains highly anisotropic (2.5 Å along a* and c*, 2.9 Å along b*), accounting for the low completeness in the higher resolution shell (Table I). The structure was solved by molecular replacement methods using the partially refined model at 3.5 Å resolution as the search probe. Model rebuilding with the program O was iterated with restrained refinement as implemented in the program Refmac5 (Murshudov et al, 1997), maintaining noncrystallographic restraints among the four independent monomers and including TLS parameters to model anisotropic displacements (Winn et al, 2003).
A diffraction data set of the FapRΔ43–malonyl-CoA complex was collected using synchrotron radiation (ID14.4-ESRF) to 3.1 Å resolution. The structure was solved by molecular replacement methods using the FapRΔ67 model and refined as above. The two malonyl-CoA molecules are clearly defined in the electron density maps (Supplementary Figure S5), except for the adenine bases and the 3′P-ribose moieties, which were not included in the atomic model. Data collection and refinement statistics for the final atomic models are given in Table I.
Fluorescence anisotropy titrations
The fluorescein-labeled HPLC-purified oligonucleotide (5′-GCCAATTATATACTACTATTAGTACCTAGTC TTAATTCCG-3′) was purchased from Eurogentec (Liege, Belgium) and annealed with 1.1. equivalents of a complementary HPLC-purified oligonucleotide (Eurogentec) to generate the double-stranded (34 bp) F-dsDNA containing the FapR-recognition sequence. Annealing was carried out by heating the reaction mixture to 95°C for 5 min, followed by slow cooling (16 h) to room temperature in 10 mM Tris-acetate, pH 8.0. dsDNA was separated from single-stranded DNA by PAGE and extracted from the acrylamide band by incubation at 42°C for 16 h in 20 mM Tris–HCl, pH 8.0, 200 mM NaCl, 2 mM EDTA. After filtration, the DNA solution was precipitated with 2 volumes of 100% ethanol, and 0.1 volumes of 3 M NaAc by incubation for 2 h at −20°C. The sample was centrifuged at 14000 r.p.m. for 15 min at 4°C, the supernatant was removed and the pellet was resuspended in 100 μl sterile Milli-Q water. dsDNA was quantified by absorbance at 260 nm.
Fluorescence anisotropy measurements were performed with a Varian Cary Eclipse Fluorescence Spectrophotometer, using an excitation wavelength of 494 nm (slit width 10 nm), an emission wavelength of 519 nm (slit width 10 nm), and a 2 s integration time. The temperature was kept constant at 25°C, using a thermally jacketed cell holder. The G-factor was measured for the F-dsDNA solution and was then kept constant for the titration of F-dsDNA with FapR and for the titration of the F-dsDNA/FapR complex with malonyl-CoA, acetyl-CoA or malonate. All titrations were performed adding small amounts of a concentrated solution of the variable ligand (wild-type or mutant FapR, malonyl-CoA, acetyl-CoA or malonate) to a fixed amount of the F-dsDNA or the F-dsDNA/FapR complex solutions. After each addition of ligand the mixture was allowed to equilibrate for 1 min, with slow stirring, and then three measurements were taken and averaged. In all cases, binding was performed in 20 mM Tris–Cl, pH 8.0, 50 mM NaCl and 10 mM MgCl2.
Isothermal titration calorimetry
ITC experiments were performed using the MCS instrument from MicroCal Inc. (Northampton, MA) as described (Schaeffer et al, 2002). Raw calorimetric data were corrected for the heat of dilution, and analyzed using the program ORIGIN™ provided by the manufacturer. The association constant (Ka), molar binding stoichiometry (N) and molar binding enthalpy (ΔH°) were determined by fitting the binding isotherm to a model with one set of sites. Molar changes in heat capacity (ΔCp) were determined using a linear regression analysis of the binding enthalpy as a function of temperature. The Gibbs free energy (ΔG°) was calculated from the expression: 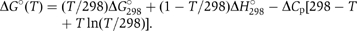 using the calorimetrically determined binding constant at 298K as a reference point.
using the calorimetrically determined binding constant at 298K as a reference point.
General methods
Assays of β-galactosidase activity were performed as described previously and presented as Miller units (Harwood and Cutting, 1990). Preparation of B. subtilis competent cells was carried out by the two-step method (Harwood and Cutting, 1990). Antibiotics were used at the following concentrations: chloramphenicol, 5 μg/ml; erythromycin plus lincomycin, 0.5 μg/ml and 12.5 μg/ml; spectinomycin, 100 μg/ml. X-gal was used at 40 μg/ml.
Supplementary Material
Supplementary Figures
Acknowledgments
We thank JA Opdyke and CP Moran for advice with in vitro transcription experiments, and A Haouz (PF6, Institut Pasteur) for assistance with robotic crystallization. This study was supported by grants from the Institut Pasteur (France), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina) and Agencia de Promoción Científica y Tecnológica (FONCYT, Argentina). AJV and D de M are International Research Scholars from Howard Hughes Medical Institute. Atomic coordinates and structures factors have been deposited with the Protein Data Bank, accession codes 2F41 (FapR) and 2F3X (FapR–malonyl-CoA complex).
References
- Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D (2001) Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J 20: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Benning MM, Wesenberg G, Liu R, Taylor KL, Dunaway-Mariano D, Holden HM (1998) The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J Biol Chem 273: 33572–33579 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT (2002) In Current Protocols in Molecular Biology, pp 3.12–3.13. New York: John Wiley & Sons, Inc. [Google Scholar]
- Collaborative Computational Project n.4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cronan JE Jr, Subrahmanyam S (1998) FadR, transcriptional co-ordination of metabolic expediency. Mol Microbiol 29: 937–943 [DOI] [PubMed] [Google Scholar]
- de Mendoza D, Schujman GE, Aguilar PS (2002) Biosynthesis and function of membrane lipids. In Bacillus subtilis and its Closest Relatives: from Genes to Cells, Sonenshein AL, Hoch JA, Losick R (eds), pp 43–55. Washington, DC: ASM Press [Google Scholar]
- Dean DR, Hoch JA, Aronson AI (1977) Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol 131: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Bateman A (2004) The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics 5: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirusso CC, Heimert TL, Metzger AK (1992) Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J Biol Chem 267: 8685–8691 [PubMed] [Google Scholar]
- Dirusso CC, Nunn WD (1985) Cloning and characterization of a gene (fadR) involved in regulation of fatty acid metabolism in Escherichia coli. J Bacteriol 161: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirusso CC, Tsvetnitsky V, Hojrup P, Knudsen J (1998) Fatty acyl-CoA binding domain of the transcription factor FadR. Characterization by deletion, affinity labeling, and isothermal titration calorimetry. J Biol Chem 273: 33652–33659 [DOI] [PubMed] [Google Scholar]
- Dowell P, Hu Z, Lane MD (2005) Monitoring energy balance: metabolites of fatty acid synthesis as hypothalamic sensors. Annu Rev Biochem 74: 515–534 [DOI] [PubMed] [Google Scholar]
- Friedman AM, Fischmann TO, Steitz TA (1995) Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science 268: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Garcia-Castellanos R, Marrero A, Mallorqui-Fernandez G, Potempa J, Coll M, Gomis-Ruth FX (2003) Three-dimensional structure of MecI. Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J Biol Chem 278: 39897–39905 [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61 [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM (1990) Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons Ltd [Google Scholar]
- Heath RJ, Jackowski S, Rock CO (2002) Fatty acid and phospholipid metabolism in prokaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes, Vance DE, Vance JE (eds), 4th edn, pp 55–92. Amsterdam: Elsevier [Google Scholar]
- Henry MF, Cronan JE Jr (1991) Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J Mol Biol 222: 843–849 [DOI] [PubMed] [Google Scholar]
- Henry MF, Cronan JE Jr (1992) A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell 70: 671–679 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Johansson P, Hederstedt L (1999) Organization of genes for tetrapyrrole biosynthesis in Gram-positive bacteria. Microbiology 145: 529–538 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL (1996) Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure 4: 253–264 [DOI] [PubMed] [Google Scholar]
- Li J, Derewenda U, Dauter Z, Smith S, Derewenda ZS (2000) Crystal structure of the Escherichia coli thioesterase II, a homolog of the human Nef binding enzyme. Nat Struct Biol 7: 555–559 [DOI] [PubMed] [Google Scholar]
- Lu YJ, Zhang YM, Rock CO (2004) Product diversity and regulation of type II fatty acid synthases. Biochem Cell Biol 82: 145–155 [DOI] [PubMed] [Google Scholar]
- Morbidoni HR, de Mendoza D, Cronan JE Jr (1996) Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol 178: 4794–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ni J, Sakanyan V, Charlier D, Glansdorff N, Van Duyne GD (1999) Structure of the arginine repressor from Bacillus stearothermophilus. Nat Struct Biol 6: 427–432 [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Scott JR, Moran CP (2001) A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol Microbiol 42: 495–502 [DOI] [PubMed] [Google Scholar]
- Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol 7: 215–219 [DOI] [PubMed] [Google Scholar]
- Overath P, Pauli G, Schairer HU (1969) Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem 7: 559–574 [PubMed] [Google Scholar]
- Raman N, Dirusso CC (1995) Analysis of acyl coenzyme A binding to the transcription factor FadR and identification of amino acid residues in the carboxyl terminus required for ligand binding. J Biol Chem 270: 1092–1097 [DOI] [PubMed] [Google Scholar]
- Schaeffer F, Matuschek M, Guglielmi G, Miras I, Alzari PM, Beguin P (2002) Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase CelD bind to a cohesin domain of the scaffolding protein CipA with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41: 2106–2114 [DOI] [PubMed] [Google Scholar]
- Schujman GE, Choi KH, Altabe S, Rock CO, de Mendoza D (2001) Response of Bacillus subtilis to cerulenin and acquisition of resistance. J Bacteriol 183: 3032–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, de Mendoza D (2005) Transcriptional control of membrane lipid synthesis in bacteria. Curr Opin Microbiol 8: 149–153 [DOI] [PubMed] [Google Scholar]
- Schujman GE, Paoletti L, Grossman AD, de Mendoza D (2003) FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell 4: 663–672 [DOI] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Joshi AK (2003) Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res 42: 289–317 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Holden HM, Zhuang Z, Dunaway-Mariano D (2002) X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J Biol Chem 277: 27468–27476 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM (2003) The structure of 4-hydroxybenzoyl-CoA thioesterase from arthrobacter sp. strain SU. J Biol Chem 278: 43709–43716 [DOI] [PubMed] [Google Scholar]
- van Aalten DM, Dirusso CC, Knudsen J (2001) The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J 20: 2041–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten DM, Dirusso CC, Knudsen J, Wierenga RK (2000) Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J 19: 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SW, Zheng J, Zhang YM, Rock CO (2005) The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74: 791–831 [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374: 300–321 [DOI] [PubMed] [Google Scholar]
- Xu Y, Heath RJ, Li Z, Rock CO, White SW (2001) The FadR. DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J Biol Chem 276: 17373–17379 [DOI] [PubMed] [Google Scholar]
- Zhang YM, Marrakchi H, Rock CO (2002) The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J Biol Chem 277: 15558–15565 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


