Abstract
Kinetochores (KTs) assemble on centromeric DNA, bi-orient paired sister chromatids on spindle microtubules (MTs) and control cell-cycle progression via the spindle assembly checkpoint. Genetic and biochemical studies in budding yeast have established that three ‘linker' complexes, MIND, COMA and NDC80, play essential but distinct roles in KT assembly and chromosome segregation. To determine whether similar linker activities are present at human KTs, we have compared the functions of Nnf1R and Mcm21R, recently identified MIND and COMA subunits, and Nuf2R, a well-characterized NDC80 subunit. We find that the three proteins bind to KTs independent of each other and with distinct cell-cycle profiles. MT–KT attachment is aberrant in Nnf1R- and Mcm21R-depleted cells, whereas it is lost in the absence of Nuf2R. Defective attachments in Nnf1R-depleted cells prevent chromosome congression, whereas those in Mcm21R-depleted cells interfere with spindle assembly. All three human KT proteins are necessary for correct binding of spindle checkpoint proteins to KTs. The differing functions and KT-binding properties of Nnf1R, Mcm21R and Nuf2R suggest that, like their yeast counterparts, the proteins act independent of each other in KT assembly, but that their combined activities are required for checkpoint signaling.
Keywords: kinetochore, mitosis, spindle checkpoint
Introduction
Kinetochores (KTs) are multiprotein complexes that assemble on centromeric (CEN) DNA, bind chromosomes to microtubules (MTs) of the mitotic spindle and mediate the bi-orientation, congression and disjunction of sister chromatids. Unattached KTs also generate checkpoint signals that block cells prior to the initiation of anaphase (Cleveland et al, 2003). The checkpoint is silenced only when all sister chromatid pairs have achieved bi-orientation, a configuration that is uniquely compatible with accurate segregation at anaphase. Because they are based on unusually simple 125 bp CEN sequences, Saccharomyces cerevisiae KTs are among the best understood (Meraldi et al, 2006). Budding yeast KTs contain >70 components organized into ∼14 multiprotein complexes (McAinsh et al, 2003) and the steps involved in assembly of their KTs are currently the subject of detailed analysis. Despite the fact that human CENs differ radically from those in S. cerevisiae, spanning millions rather than hundreds of base pairs, many yeast KT proteins have human orthologs (Fukagawa, 2004; Chan et al, 2005). However, the organization and structure of human KTs remain largely unexplored and it is unclear whether simple and complex KTs assemble around similar complexes.
When visualized by electron microscopy (EM), human KTs have a trilaminar structure. Although S. cerevisiae KTs are not visible in the EM, functional experiments are suggestive of a trilaminar organization comprising DNA-binding, linker, and MT-binding activities (McAinsh et al, 2003). The term ‘linker' is used rather loosely in this context, reflecting a paucity of mechanistic detail, but assembly of linker proteins onto CEN DNA is characterized by a shared hierarchy with respect to other KT subunits. CEN association by linker proteins requires DNA but not MT-associated proteins (MAPs); MAPs and motors require both DNA-binding and linker proteins, whereas DNA-binding subunits associate with CEN DNA independent of linkers, MAPs and motors (De Wulf et al, 2003). This hierarchy implies that S. cerevisiae linkers serve as bridges between proteins in direct contact with DNA and those in contact with MTs, but linkers are likely to have other important activities. Four S. cerevisiae linker complexes have been identified to date: the heterotetrameric NDC80, MIND and COMA complexes and the heterodimeric SPC105 complex (Janke et al, 2001a; Wigge and Kilmartin, 2001; De Wulf et al, 2003; Nekrasov et al, 2003). Recent biochemical and computational analysis has identified potential human orthologs of several budding yeast linker proteins (Cheeseman et al, 2004; Obuse et al, 2004; Meraldi et al, 2006), raising the possibility that linkers form an evolutionarily conserved KT core.
The best-characterized linker is the NDC80 complex, which is composed in both yeast and humans of Ndc80Hec1, Nuf2, Spc24 and Spc25. These four proteins assemble with a 1:1:1:1 stoichiometry into a 57-nm rod with globular domains at both ends whose probable function is binding to other KT proteins (Ciferri et al, 2005; Wei et al, 2005). Association of S. cerevisiae Ndc80p, Nuf2p, Spc24p and Spc25p with CEN DNA in vivo requires the CBF3 DNA-binding complex. CEN association by Stu2p and DASH complex (both of which are MAPs), and Cin8p-Kip1p kinesins is dependent, in turn, on the NDC80 complex (He et al, 2001; Janke et al, 2001a; Wigge and Kilmartin, 2001; De Wulf et al, 2003). Loss-of-function mutations in any of the four essential subunits of the yeast NDC80 complex detach chromosomes from MTs and, by preventing KT recruitment of Mad1 and Mad2, inactivate the spindle checkpoint (He et al, 2001; Janke et al, 2001b; Wigge and Kilmartin, 2001; McCleland et al, 2003). RNAi, antibody injection and immunodepletion generate similar loss-of-function phenotypes for all NDC80 complex subunits tested to date in Xenopus laevis, Caenorhabditis elegans and human cells (DeLuca et al, 2002; Martin-Lluesma et al, 2002; McCleland et al, 2003; Cheeseman et al, 2004; Meraldi et al, 2004). Together, these data suggest that the NDC80 complex has been highly conserved in structure and function from yeast to man.
In contrast to the NDC80 complex, less is known about the functions of MIND and COMA. The S. cerevisiae MIND complex is composed of four proteins, Mtw1p, Nnf1p, Nsl1p and Dsn1p, that assemble into a 215 kDa heterotetramer (De Wulf et al, 2003; Nekrasov et al, 2003; Scharfenberger et al, 2003). Temperature-sensitive mutants in MIND subunits lead to defects in KT-based force generation and provoke the spindle checkpoint (Goshima and Yanagida, 2000; De Wulf et al, 2003), although inactivation of Nsl1p has been reported to lead to a failure of checkpoint signaling under some circumstances (Scharfenberger et al, 2003). Potential orthologs of S. cerevisiae MIND subunits have been identified in human cells (Nnf1R (also known as PMF1), Nsl1RDC8, Dsn1Rc20orf172 and Mis12; Goshima et al, 2003; Cheeseman et al, 2004; Obuse et al, 2004), but the functions of these proteins are not well established. Depletion of Dsn1R has been reported to cause defects in chromosome congression and provoke the checkpoint, whereas depletion of Mis12 abrogates the checkpoint and allows cells to enter anaphase with unaligned and lagging chromosomes (Goshima et al, 2003; Obuse et al, 2004). Whether these differences represent differences in biological function or simply variable degrees of protein depletion has not yet been determined.
COMA is the most highly diverged linker complex. Our recent analysis of KT proteins across multiple species suggests that only the Mcm21p subunit of S. cerevisiae COMA is conserved from budding yeast to humans (Meraldi et al, 2006). However, a number of human homologs can be found of Schizosaccharomyces pombe COMA proteins that lack counterparts in S. cerevisiae (Meraldi et al, 2006). It is tempting to speculate that COMA is involved in linking conserved KT proteins to proteins that are specific to different CEN architectures.
In this paper, we explore the extent to which KTs structure has been conserved through evolution by comparing directly the functions of three potential human linkers: Nuf2R, Nnf1R and Mcm21R. We establish that human Nuf2R, Nnf1R and Mcm21R, like their yeast counterparts, are recruited to KTs independently of each other. Using a combination of live-cell imaging and small inhibitory RNA (siRNA)-based gene silencing, we examine the functions of Nnf1R and Mcm21R in detail. Both proteins are required for chromosome congression and for correct operation of the spindle checkpoint. These and other data lead us to speculate that human NDC80, MIND and COMA probably represent distinct functional structures that cooperate in the assembly of KTs by recruiting MT-binding and checkpoint proteins. In this respect, the human proteins resemble yeast counterparts.
Results
KT localization of Nnf1R and Mcm21R
As a first step in distinguishing the functions of Nuf2R, Nnf1R and Mcm21R, we compared their patterns of localization in mitotic cells. Immunofluorescence microscopy and immuno-EM have previously shown that Nuf2R is present on KTs and spindle poles (Wigge and Kilmartin, 2001; DeLuca et al, 2005), but Nnf1R has been examined only as a YFP-tagged fusion protein (Cheeseman et al, 2004). No data are available for Mcm21R. Rabbit polyclonal antisera were therefore raised against recombinant Nnf1R and Mcm21R. By Western blotting, three independent rabbit anti-Mcm21R antisera recognized a single protein band of 33 kDa in HeLa whole-cell extracts and three independent rabbit anti-Nnf1R antisera recognized a 24 kDa band, consistent with the predicted molecular weights of the two proteins. Depletion of Mcm21R or Nnf1R with siRNA oligonucleotides (siMcm21R-1 or siNnf1R-3; see Supplementary data for details) reduced the intensity of the 33 or 24 kDa bands by >90% confirming the specificity of the antibodies (Figure 1A).
Figure 1.

Localization of Nnf1R and Mcm21R in mitosis. (A) Immunoblots of lysates from cells transfected with siLaminA, siNnf1R-3 or siMcm21R-1 and probed with antibodies as indicated. (B) HeLa cells were stained with DAPI, γ-tubulin for spindle poles (blue), CREST antisera for KTs (red) and Nnf1R or Mcm21R antisera (green). As a control, localization of Nnf1R and Mcm21R was determined following MTs depolymerization with nocodazole for 1 h. (scale for insets). (C) Immunofluorescence images of prometaphase cells following transfection with siLaminA (negative control), siNnf1R-3 or siMcm21R-1 and stained with CREST (red) and Nnf1R or Mcm21R antisera (green) as indicated. Scale bars=10 μm.
To localize Nnf1R and Mcm21R, polyclonal antisera were next used to stain normally growing HeLa cells in conjunction with human anti-KT (CREST) antibodies and DAPI. In cells undergoing mitosis, anti-Nnf1R antisera (but not preimmune control sera) recognized puncta that colocalized precisely with CREST staining (Figure 1B). The specificity of the punctate staining was established by immunofluorescence microscopy of cells depleted of Nnf1R. When HeLa cells were transfected with siNnf1R-3, Nnf1R staining was reduced to background levels, whereas transfection with control siRNA had no effect; CREST staining was equally intense in both (Figure 1C). We therefore conclude that Nnf1R localizes primarily to KTs in mitotic cells. Anti-Mcm21R antibodies (but not preimmune sera) stained KTs and centrosomes brightly and gave rise to diffuse, variable staining in the cytoplasm (Figure 1B). Depletion of Mcm21R (siMcm21R-1) reduced KT staining to very low levels, but had little effect on diffuse cytosolic and centrosomal staining (Figure 1C). From these data, we conclude that Mcm21R localizes to KTs. Centrosomal staining may represent nonspecific binding to an unrelated protein or to Mcm21R that is highly resistant to RNAi-based depletion due to very low protein turnover (relative to KT-bound Mcm21R). Further analysis with additional antibodies or with GFP fusion proteins will be required to fully resolve this issue. In any case, we conclude that a significant fraction of human Mcm21R is KT-bound, as is the case with its yeast ortholog.
The recruitment of proteins to KTs can either be direct, arising from association with core KT factors, or indirect, arising from association with the plus-ends of MT, which themselves become KT-bound during mitosis. To distinguish between direct and indirect KT association by Nnf1R and Mcm21R, cells treated with sufficient nocodazole to fully depolymerize MTs and the proteins localized by immunofluorescence. Both Nnf1R and Mcm21R were observed to retain KT association in the absence of MTs (Figure 1B). Thus, Nnf1R and Mcm21R do not require MTs to localize to KTs, like Nuf2R (DeLuca et al, 2005), but unlike EB1 (Tirnauer et al, 2002).
To determine when in the cell cycle Nuf2R, Nnf1R and Mcm21R are recruited to KTs, HeLa cells were costained with DAPI, with antibodies against CREST and α-tubulin and with anti-Nnf1R, Mcm21R or Nuf2R antisera (Figure 2A). At least 40 KTs in eight or more cells were examined by 3D deconvolution microscopy. The intensity of CREST staining was constant throughout the cell cycle, as expected (Hoffman et al, 2001), and served as an internal reference by which to compare signals on different KTs. The association of Nuf2R with KTs was first detected during prophase, peaked during metaphase and disappeared during anaphase B (Figure 2B; Hori et al, 2003). Nuf2R also accumulated on spindle poles in mitosis and could be found at the spindle mid-zone during anaphase B (data not shown). Nnf1R was found at peak levels on KTs during prometaphase and metaphase and was absent from KTs only during a brief period in telophase/early interphase during which cells contained a postmitotic bridge (Figure 2B). Finally, Mcm21R was found on KTs throughout the cell cycle (Figure 2B) with no significant difference between G1, S and G2. Levels of KT-bound Mcm21R did not rise during mitosis and, if anything, were slightly higher during interphase. Differences in the timing of Nuf2R, Nnf1R and Mcm21R recruitment to KTs support the idea that, in human cells, as in yeast, the three proteins bind KTs independent of each other.
Figure 2.
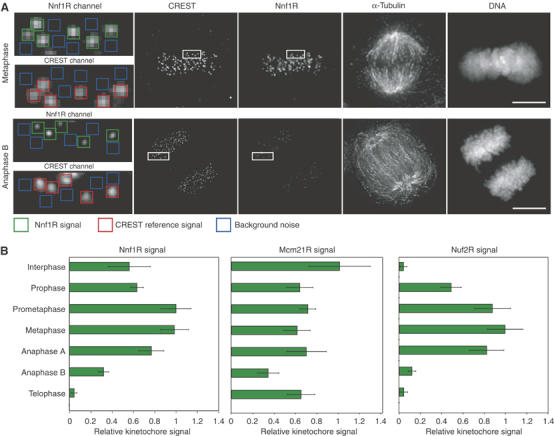
Cell-cycle localization of Nnf1R, Mcm21R and Nuf2R during the mitotic cell cycle. (A) Levels of Nnf1R, Nuf2R and Mcm21R on KTs were determined from deconvolved 3D reconstructions of cells individually stained with DAPI, CREST antisera (KTs) and Nnf1R, Nuf2R or Mcm21R antisera. The intensity of KT signal (green) was determined relative to a CREST reference (red) after background correction (blue) (see Supplementary data for details). Box: 0.5 × 0.5 μm. Scale bar=10 μm. (B) Quantification of Nnf1R, Mcm21R and Nuf2R protein levels at KTs during different cell-cycle stages. Error bars show s.e.m. from repeat determinations in multiple cells.
Depletion of Nnf1R disrupts chromosome segregation
To study the functions of Nnf1R during mitosis, HeLa cells expressing Histone-H2B-GFP were transfected with one of four different siRNA oligonucleotides directed against Nnf1R (siNnf1R1–4), chromosome movement was examined by imaging living cells every 3 min for a period of 6 h and the key events of mitosis were scored morphologically. Nuclear envelope breakdown (NBD) was set as T=0 in each cell and the times of chromosome congression and anaphase onset recorded relative to NBD. Cells were followed into telophase to determine the accuracy of anaphase chromosome disjunction. When control transfected cells were monitored by live-cell imaging (n=127), anaphase onset followed a previously described skew-normal distribution (Meraldi et al, 2004), with ∼90% of cells completing chromosome congression by T=24 min, 80% initiating anaphase by T=36 min and >95% of cells faithfully completed disjunction, as judged by segregation of H2B-GFP-labelled chromatin into two equal masses without evidence of lagging or unaligned chromosomes (Figure 3A–E). In contrast, in >80% HeLa cells transfected with one of four Nnf1R oligonucleotides, chromosomes failed to congress correctly to the metaphase plate by T=24 min (Figure 3A and C). Mitotic outcome, however, was surprisingly different, depending on which siRNA oligo was used: transfection with siNnf1R-1 or 2 caused cells to arrest in mitosis (>70% were mitotic by T=120 min; Figure 3A and D), whereas transfection with siNnf1R-3 or 4 caused cells to exit mitosis with kinetics similar to or slightly faster than control cells (88% had initiated anaphase by T=36 min; Figure 3A and D), despite the presence of mal-oriented chromosomes. As a consequence, siNnf1R-3- or 4-transfected cells experienced massive chromosome loss.
Figure 3.

Chromosome misalignment and missegregation in cells depleted of Nnf1R. (A) Successive frames every 3 or 4 min from live-cell movies of H2B-GFP-expressing HeLa cells transfected with either siLaminA, siNnf1R-1 or siNnf1R-3. Scale bar indicates 10 μm. (B) Cumulative frequency plots of anaphase onset times in siRNA-treated cells, as indicated, with NBD=T0, as determined from live-cell movies. (C) Fraction of cells, treated with siRNAs as indicated, having uncongressed chromosomes at T=24 min. (D) Mitotic phenotype following treatment with siRNAs as indicated. Cells were scored as either having undergone normal anaphase (green), having missegregated chromosomes in anaphase (blue) or becoming arrested prior to anaphase onset (as scored at T=120 min). (E) Quantification of Nnf1R protein levels, as described in Figure 2A, on KTs on prometaphase in siLaminA-, siNnf1R-1- or siNnf1R-3-treated cells.
We have previously noted similar variation in phenotype following siRNA-mediated depletion of Nuf2R or Ndc80Hec1. In that case, phenotypic variation arose from differences in the degree of protein depletion: partial depletion of Nuf2R or Ndc80Hec1 engaged the spindle checkpoint, whereas complete depletion inactivated the checkpoint, probably by preventing KT recruitment of Mad2 (Meraldi et al, 2004). To determine if this might also be true of Nnf1R, we measured the levels of Nnf1R on individual KTs in 3D images of immunostained cells transfected with siNnf1R-1 or siNnf1R-3 oligos. We have previously demonstrated that immunostaining of individual KTs is a better measure of protein depletion than Western blotting (Meraldi and Sorger, 2005). siNnf1R-1 transfection was observed to deplete 80% of the anti-Nnf1R fluorescence signal on ca. 95% of KTs, whereas siNnf1R-3 transfection led to greater than 99% depletion (Figure 3E) on essentially all KTs. We conclude that transfecting cells with siNnf1R-1 caused partial Nnf1R depletion, whereas siNnf1R-3 caused full depletion. To establish that mitotic arrest under conditions of partial depletion involved activation of the spindle assembly checkpoint, rather than mechanical failure of the spindle or some other process, we showed that co-transfecting cells with siRNA against Mad2 and siNnf1R-1 abrogated the mitotic arrest observed with siNnf1R-1 alone (Figure 4A). To further demonstrate that complete Nnf1R depletion (following siNnf1R-3 transfection) inactivated the checkpoint, we performed two functional tests. First, cells were treated with the MT poison nocodazole for 16 h and the fraction of arrested cells measured. Whereas treatment of control cells with nocodazole led to a five-fold increase in mitotic index reflecting activation of the spindle checkpoint (50–60 versus 8–12% for untreated cells), treatment of siNnf1R-3-transfected cells with nocodazole did not increase mitotic index, suggesting an absence of cell-cycle arrest despite MT depolymerization (Figure 4B). As a second test of checkpoint function, cells were co-transfected with siRNA against the MT associated protein ch-TOG1 and with either siNnf1R-3 or control siRNA. We have previously shown ch-TOG1 depletion to disrupt KT–MT attachment and provoke a robust Mad- and Bub-dependent checkpoint response. Under conditions of ch-TOG1 depletion, >95% of cells remain arrested for >6 h, as judged by live-cell imaging (Gergely et al, 2003; Meraldi et al, 2004). However, cells co-transfected with siNnf1R-3 and sich-TOG1 proceeded through mitosis with similar kinetics to unperturbed cells or cells singly transfected with siNnf1R-3, demonstrating checkpoint inactivation (Figure 4C, dotted red line). The combination of chromosome misalignment and checkpoint failure caused severe chromosome segregation errors in siNnf1R-3/sich-TOG1 co-transfected cells. From these data we conclude that partial Nnf1R depletion (with siNnf1R-1) provokes the spindle checkpoint, whereas complete Nnf1R depletion (with siNnf1R-3) inactivates the checkpoint. In this respect, Nnf1R strongly resembles Nuf2R and Ndc80Hec1 (see Discussion).
Figure 4.

Spindle checkpoint signaling fails following depletion of Nnf1R. (A, C) Cumulative frequency plots of anaphase onset times in siRNA-treated cells, as indicated, with NEB=T0, as determined from live-cell movies. (B) Percentage mitotic arrest in siLaminA-, siNnf1R-1-, siNnf1R-3 or siMad2-transfected cells treated with nocodazole for 16 h. (D) siLaminA-, siNnf1R-1- or siNnf1R-3-treated cells stained with CENP-A (KT marker), Mad2, Mad1, Bub1 or BubR1 antisera following treatment with nocodazole and MG132. Scale bar=10 μm. (E) Quantification of immunofluorescence signals as described in Figure 2A.
In the case of KT structural proteins examined to date, loss of spindle checkpoint function is associated with failure to localize at least one Mad or Bub checkpoint protein to KTs. A simple way to ascertain which checkpoint proteins require Nnf1R for KT binding is to transfect cells with siNnf1R-3 or control siRNA, depolymerize MTs to activate the checkpoint and then stain KTs with antisera against Mad and Bub proteins. However, Nnf1R depletion causes cells to pass rapidly into anaphase and enter a different cell-cycle state than control cells in which MTs have been depolymerized. To avoid this problem, control and siNnf1R-3-transfected cells were synchronized in the S phase, released into medium containing the proteasome inhibitor MG132, which blocks cells uniformly at the metaphase to anaphase transition (Rock et al, 1994), and then treated with nocodazole to cause KT accumulation of checkpoint proteins. Under these conditions, siNnf1R-3 transfection was observed to prevent Mad1, Mad2, Bub1 and BubR1 from associating with KTs (Figure 4D and E). In contrast, partial Nnf1R depletion with siNnf1R-1 was similar to nocodazole treatment alone in arresting cells with high levels of all four checkpoint proteins present on KTs (Figure 4D and E). From these data, we conclude that Nnf1R is necessary for the recruitment of Mad1, Mad2, Bub1 and BubR1 to KTs, explaining the requirement for Nnf1R in the checkpoint response.
Role of Nnf1R in chromosome congression
Next, we sought to understand the role played by Nnf1R in KT–MT attachment and chromosome congression. To quantify congression defects, synchronized cells were arrested at the metaphase–anaphase transition with MG132 and the percentage of cells with uncongressed chromosomes determined. Whereas <10% of control or partially Nnf1R-depleted cells failed to align their chromosomes on a metaphase plate, over 60% of cells completely depleted of Nnf1R contained misaligned chromosomes (Figure 5A–C). Chromosome misalignment can arise from several mechanistically distinct defects in KT–MT association including monopolar, syntelic or merotelic attachment. No direct biochemical assays exist for these defects, but they can be at least partly distinguished by morphology and inter-KT distances (a readout of the net force exerted on chromatid pairs). Many studies have relied on Mad2 staining as a marker for detached KTs (e.g. Hoffman et al, 2001), but this is not possible in the case of Nnf1R-depleted cells, because Mad2 requires Nnf1R for KT association. Inter-KT distances averaged 2.0±0.3 μm in control cells and 0.6±0.1 μm in nocodazole-treated cells, as expected for bi-oriented and detached chromatid pairs respectively (Figure 5D; Meraldi et al, 2004). In siNnf1R-1- and siNnf1R-3-transfected cells, the subset of chromatid pairs that managed to congress to the spindle equator had inter-KT distances intermediate between that of bi-oriented and detached controls (d=1.4±0.2 μm; Figure 5D and E), implying MT attachment by Nnf1R-depleted KTs, but without the generation of normal pulling forces. Unaligned chromatid pairs had inter-KT distances (d=1.0±0.2 μm) only slightly greater than detached pairs while remaining closely associated with the spindle. This implies MT binding without bi-orientation by Nnf1R-depleted sister KTs (detached KTs are normally ejected from the spindle; Figure 5E and data not shown). Thus, Nnf1R depletion is associated with two defects in chromosome segregation: (1) accumulation of chromatid pairs that are MT bound but not bi-orientated or congressed; (2) congression of a subset of bi-oriented chromatids, but without the generation of normal pulling forces. Thus, unlike the NDC80 complex, Nnf1R is not required for chromosome attachment per se, but rather for the alignment of KTs at the metaphase plate and for the correct generation of inter-KT forces.
Figure 5.

Nnf1R depletion leads to defects in KT-based force generation. (A) Chromosomes in metaphase cells were counted as unaligned if they were located outside of the central 30% of the mitotic spindle (dotted lines in schematic) or if their KTs were aligned perpendicular to the spindle axis. (B) Fraction of cells, treated with siLaminA, siNnf1R-1 or siNnf1R-3, having uncongressed chromosomes following 1 h treatment with MG132. (C) Representative immunofluorescence images of cells treated as described in (C) and stained with DAPI (blue), β-tubulin (green) and CENP-E (red) antisera. Scale bar=10 μm. (D, E) Inter-KT distances of chromosomes from cells arrested at metaphase with MG132 and treated with nocodazole or, siLaminA or siNnf1R-3. Inter-KT distances were measured using the outer KT marker CENP-E. They are shown as whisker plots and were derived by measuring at least 40 KT pairs in a total of eight cells. The central bar indicates the mean, the red box the 50th central percentile and the error bars the extremes.
Mcm21R is required for chromosome congression and spindle checkpoint function
Next, we compared the functions of Mcm21R to those of Nnf1R. When cells were transfected with either of two RNA oligos directed against Mcm21R and examined by 3D deconvolution imaging using anti-Mcm21R antibodies, KT staining was reduced at least 20-fold in >95% of cells (Figure 6C). Mcm21R depletion in H2B-GFP-expressing HeLa cells led to severe chromosome congression defects (96% of the cells had uncongressed chromosomes at T=24 min versus 10% in control cells; Figure 6A, B and D). Following anaphase onset, two separated DNA masses were visible in 45% of cells as were errors in chromosome disjunction (Figure 6A), while in the remaining cells, anaphase was highly asymmetric with nearly all chromosomes pulled toward one daughter (Figure 6B). We interpret this unusual state of segregation to represent anaphase because cells with a single DNA mass had three characteristics of cells that had exited mitosis: (1) low levels of CyclinB1, as revealed by immunofluorescence (data not shown), (2) onset of poleward chromosome movement and (3) chromosome decondensation at about the same time as in normal cells. The failure of Mcm21R-depleted cells to delay anaphase onset implies failure of checkpoint control (Figure 6A, E and F), an interpretation confirmed by data showing that Mcm21R depletion with either of two siRNA oligos bypassed the mitotic arrest imposed by either nocodazole treatment or ch-TOG1 depletion (Figure 7A and B). Moreover, when synchronized cells depleted of Mcm21R were arrested in metaphase with MG132 and treated with nocodazole, Mad2 was seen to be absent from KTs even though Mad1, Bub1 and BubR1 were present at high levels (Figure 7C and D). We therefore conclude that Mcm21R depletion severely disrupts chromosome congression and that it also prevents KT binding by Mad2, thereby inactivating the checkpoint.
Figure 6.
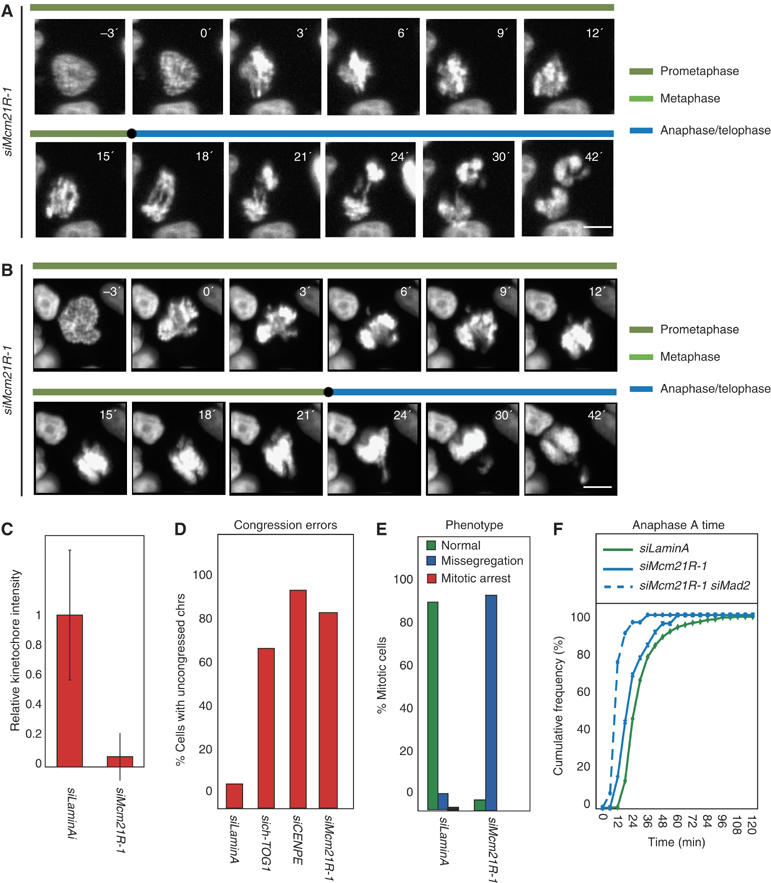
Mcm21R depletion causes chromosome misalignment, missegregation and bipolar spindle assembly failure. (A, B) Successive frames every 3 min from live-cell movies of H2B-GFP-expressing HeLa cells transfected with siMcm21R-1. Scale bar=10 μm. (C) Quantification of Mcm21R protein levels, as described in Figure 2A, on KTs of prometaphase cells in siLaminA- or siMcm21R-1 treated cells. (D) Cumulative frequency plots of anaphase onset times in siRNA-treated cells, as indicated, with NBD=T0, as determined from live-cell movies. Note that siMcm21R-1 does not affect mitotic timing as opposed to siMad2 (solid blue lines). (E) Fraction of cells, treated with siRNA as indicated having uncongressed chromosomes at T=24 min. (F) Mitotic phenotype following treatment with siRNA as indicated. Cells were scored as either having undergone normal anaphase (green), having missegregated chromosomes in anaphase (blue) or becoming arrested prior to anaphase onset (as scored at T=120 min).
Figure 7.

Spindle checkpoint failure in Mcm21R-depleted cells. (A) Percentage mitotic arrest in siLaminA-, siMcm21R-1- or siMad2-transfected cells treated with nocodazole for 16 h. (B) Cumulative frequency plots of anaphase onset times in siRNA-treated cells, as indicated, with NBD=T0, as determined from live-cell movies. (C) Quantification, as described in Figure 2A, of Bub1, BubR1, Mad1 and Mad2 proteins levels on KTs following siLaminA or siMcm21R-1 treatment. (D) Representative immunofluorescence images of cells from (C) stained with CREST, Mad2, Mad1, Bub1 or BubR1 antisera following treatment with nocodazole and MG132. Scale bar=10 μm.
Monopolar spindles in Mcm21R-depleted cells as a consequence of KT defects
A particularly striking feature of mitosis in Mcm21R-depleted cells was the asymmetric anaphase movement of sister chromatids in roughly half of all cells (Figure 6B), which suggested a potential defect in spindle formation. The locations of centrosomes relative to spindle fibers and DNA were therefore determined in control and Mcm21R-depleted cells by staining with anti-γ-tubulin and anti-β-tubulin antibodies, DAPI and CREST antisera. Mcm21R depletion caused a significant increase in the fraction of cells with unseparated centrosomes and monopolar spindles to which chromosomes were attached (60% monopolar versus 3% in control cells; Figure 8A and B). To study the origins of this phenotype in live cells, spindle formation and chromosome dynamics were followed by time-lapse microscopy in cells expressing H2B-GFP to mark chromosomes and α-tubulin-mRED to mark spindles. In >90% of control transfected cells (n=50 cells) bipolar spindles assembled within 12 min of NBD and chromosomes subsequently congressed to the spindle then segregated correctly at anaphase (Figure 9A and C, Supplementary movie 1). In Mcm21R-depleted cells, however, >50% of cells (n=36) failed to assemble a spindle and separate the poles in a timely manner. The great majority of these cells (45% of all cells) initiated anaphase in the presence of a monopolar spindle, while a subset (∼8% of all cells) slowly formed a spindle (Figure 9B, Supplementary movie 2). Thus, we conclude that Mcm21R depletion leads to a severe delay in pole separation and formation of a bipolar spindle.
Figure 8.
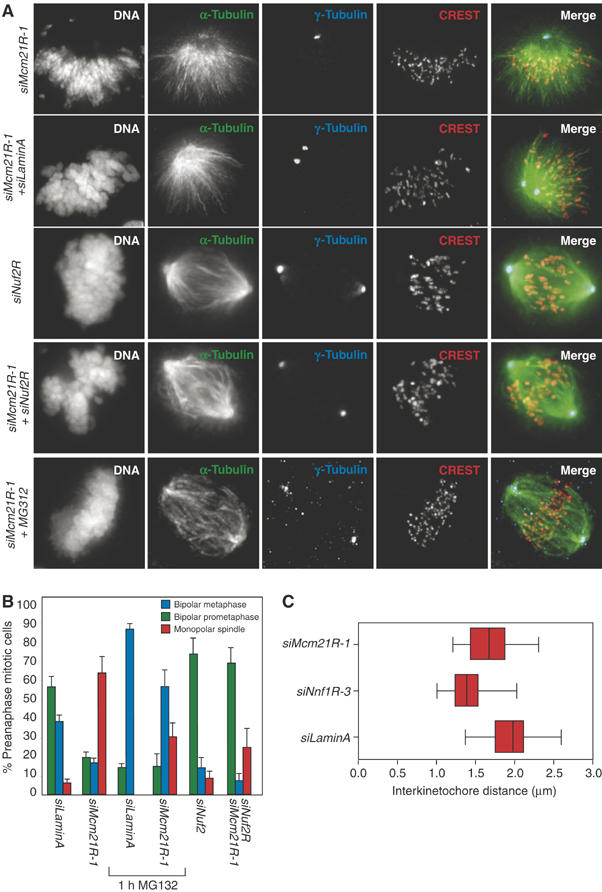
Monopolar spindles in Mcm21R-depleted cells are suppressed by loss of MT–KT attachment. (A) Typical siMcm21R-1, siLaminA/siNuf2R, siLaminA or siMcm21R-1/siNuf2R metaphase cells stained with antisera against γ-tubulin or CREST (red), β-tubulin (green) and DAPI. siMcm21R-1/siLaminA cells were also stained for immunofluorescence following treatment with MG132 for 1 h (see sixth row). Scale bar indicates 10 μm. (B) Fraction of cells with either normal metaphase spindles (blue bars), bipolar prometaphase spindles (green bars) or monopolar spindles (red bars) determined following treatment with siRNAs as indicated in the presence and absence of MG132. (C) Inter-KT distances were calculated as described in Figure 4A following treatment with siLaminA, siNnf1R-3 or siMcm21R-1 after incubation with MG132 for 1 h.
Figure 9.
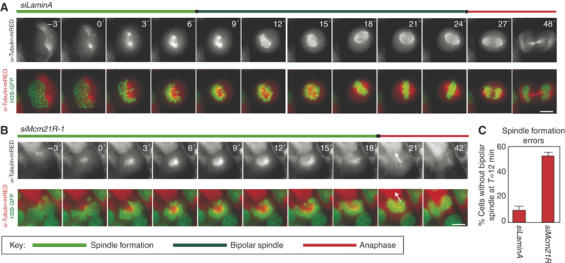
Dynamics of spindle assembly in Mcm21R-depleted cells. Successive frames every 3 min from live-cell movies of H2B-GFP/α-tubulin-mRED-expressing HeLa cells transfected with siLaminA (A) or siMcm21R-1 (B). Shown are the composite images for H2B-GFP (green) and α-tubulin-mRED (red) or the single channel for α-tubulin-mRED (lower panels). Arrows indicate anaphase movement in siMcm21R-1-treated cells. Note chromosome decondensation in both cells 18 min after anaphase onset. Scale bar=10 μm. (C) Quantification of bipolar spindle formation errors at T=12 min after NBD.
This is an unexpected phenotype since it has previously been shown that bipolar spindles can form in the absence of KTs (Heald et al, 1996; Bucciarelli et al, 2003). Indeed, complete disruption of KT–MT attachment by Ndc80Hec1 (DeLuca et al, 2003) or Nuf2R depletion (Figure 8A) does not appreciably alter spindle structure. We reasoned that monopolar spindles in Mcm21R-depleted cells might arise because defective MT–KT attachments were acting in a dominant manner that interfered with spindle assembly. If this hypothesis is correct, then detaching chromosomes from MTs in Mcm21R-depleted cells, by RNAi of a component of the NDC80 complex (Nuf2R), should block formation of dominant-interfering attachments and rescue spindle assembly. When this experiment was attempted, we observed that >80% of HeLa cells co-transfected with siRNA against both Nuf2R and Mcm21R contained bipolar spindles, demonstrating substantial rescue of spindle assembly (Figure 8A and B). Chromosomes in Nuf2R/Mcm21R double-depleted cells were detached from the spindle as previously described for Nuf2R depletion alone (Meraldi et al, 2004). Rescue of spindle assembly was not a consequence of competition between the RNA oligos as levels of both Mcm21R and Nuf2R protein were reduced to similar levels in single and double siRNA transfections (Supplementary Figure 1).
A second prediction of the hypothesis that MT–KT attachments in Mcm21R-depleted cells interfere with spindle assembly is that it should be possible to rescue assembly by extending the period in mitosis in which normal error-correcting processes function. Consistent with this idea, we observed that the fraction of cells with bipolar spindles rose dramatically when mitosis was extended to >1 h by the application of MG132 (Figure 8A and B). Those sister chromatid pairs that managed to congress in MG132-treated cells had inter-KT distances nearly as great as control cells (1.8±0.2 μm separation in Mcm21R-depleted cells versus 2.0±0.3 μm in control cells; Figure 8C) demonstrating bi-orientation and the imposition of pulling forces. From these data, we conclude that Mcm21R depletion causes a defect in KT–MT attachment that acts in a dominant manner to interfere with an otherwise intact spindle assembly pathway and that the overall effect is to delay rather than block the establishment of bipolarity. Because Mcm21R-depleted cells lack a functional spindle checkpoint, they do not wait for repair to take place unless an exogenous cell-cycle arrest is imposed (by MG132 in our case).
Independent recruitment of Nnf1R, Mcm21R and Nuf2R onto KTs
Having established that Nnf1R, Mcm21R and Nuf2R perform distinct functions in KT–MT binding, we wondered whether, like their S. cerevisiae counterparts, the proteins were recruited to KTs independent of each other. The levels of Nnf1R, Mcm21R or Nuf2R on KTs were determined by quantitative 3D-deconvolution imaging in cells treated with control, siNnf1R-3, siMcm21R-1 or siNuf2R siRNA oligos. We observed that Nnf1R levels were not affected by Nuf2R or Mcm21R depletion, but that Mad2 was lost from KTs—a positive control for the efficiency of the depletion (Figure 10, see also Figures 5D, E and 7C, D). Similarly, Nuf2R was recruited to KTs in the absence of Mcm21R or Nnf1R, and Mcm21R was KT bound in the absence of Nuf2R and Nnf1R (Figure 10). We therefore conclude that Nnf1R, Mcm21R and Nuf2R bind to KTs independent of each other, just like their orthologs in S. cerevisiae.
Figure 10.

Independent recruitment of Mcm21R, Nnf1R and Nuf2R onto KTs. Representative images of cells treated with siLaminA (A), siNnf1R-3 (B), siNuf2R (C) or siMcm21R-1 (D) and stained with Nnf1R, Mcm21R or Nuf2R antisera. Scale bar=10 μm. (E–G) Quantification of KT signals for Nnf1R, Mcm21R and Nuf2R as described in Figure 2A–D.
Discussion
In this study, we use a combination of RNAi, live-cell imaging and quantitative immunofluorescence to compare the functions of three human proteins orthologs to subunits of the S. cerevisiae MIND, COMA and NDC80 complexes. Live- and fixed-cell analysis of HeLa cells show that Nuf2R, Mcm21R and Nnf1R are recruited to KTs independent of each other and that they have distinct profiles of KT association during the cell cycle. Whereas Nuf2R is found on KTs only from prophase through anaphase A, Mcm21R is KT bound throughout the cell cycle and Nnf1R is bound during all but a brief period in telophase. RNAi of Nuf2R, Mcm21R or Nnf1R individually does not result in the loss of the other two proteins from KTs, demonstrating independent assembly onto CEN DNA. Moreover, unlike many KT-associated MAPs (kMAPs) and motors (Kaplan et al, 2001; Tirnauer et al, 2002), neither Nuf2R, Mcm21R nor Nnf1R requires MTs for KT association. The three proteins also have different functions in mitosis. Depletion of Nuf2R causes KTs to dissociate completely from MTs, but Mcm21R and Nnf1R are not essential for MT binding per se. We do not yet know how these phenotypic differences arise, but preliminary data suggest that Nuf2R, Mcm21R and Nnf1R are involved in the recruitment of distinct sets of kMAPs and motors to KTs (VM Draviam and PK Sorger, unpublished observations). Considered together, our data fall short of proving that Mcm21R, Nnf1R and Nuf2R are components of different multiprotein complexes, but they are highly suggestive. Moreover recent biochemical studies in Xenopus egg extracts show that subunits of MIND and NDC80 do not associate directly (Emanuele et al, 2005) and our preliminary data in human mitotic extracts show that Nnf1R, Ndc80Hec1 and Mcm21R fractionate as distinct species (S McClelland and AD McAinsh, unpublished observations). We therefore speculate that Mcm21R, Nnf1R and Nuf2R belong to distinct functional units of the KT with similarities to budding yeast COMA, MIND and NDC80 complexes.
Regulation of mitotic spindle formation by KTs
The observation that Mcm21R depletion interferes with bipolar spindle assembly is unanticipated. Spindle assembly can proceed efficiently in the absence of KTs, even though the primary function of the spindle is to bi-orient KT pairs (Heald et al, 1997; Bucciarelli et al, 2003). Spindle assembly is thought to involve four partly redundant processes (for a review, see Rieder et al, 2001; Gadde and Heald, 2004; Maiato et al, 2004): (1) positioning of duplicated centrosomes on opposite ends of the mitotic spindle; (2) nucleation of MTs by chromatin-bound factors in the vicinity of mitotic chromosomes; (3) attachment of MTs originating from centrosomes to KTs and (4) focusing of MTs nucleated by KTs and that extend toward the spindle poles. Which spindle assembly processes is disrupted by Mcm21R depletion? Our data show that KTs are involved, since the monopolar spindle phenotype in Mcm21R-depleted cells is suppressed by abolishing MT–KT attachment in double RNAi experiments. MT attachment by Mcm21R-depleted KTs appears to act in a dominant manner to block an otherwise functional spindle assembly process. A defect in spindle assembly has previously been observed following inhibition of the kMAP Clasp1, a protein that promotes nucleation of MTs from KTs. In the absence of Clasp1, bipolar spindles form transiently, but they then collapse (Maiato et al, 2002, 2003, 2005). This implies that KT-nucleated MTs are necessary for spindle stability, at least when KTs are present to exert a contractile force on the spindle. However, live-cell imaging of spindle dynamics in Mcm21R-depleted cells reveals that the formation of bipolar spindles is inefficient, but that once they form, they are stable and able to exert pulling forces on sister chromatids. Thus, the defects in spindle assembly caused by Clasp1 and Mcm21R depletion are distinct. We speculate that Mcm21R-depleted KTs bind MTs too avidly and too early in mitosis so that MTs arising from paired centrosomes are constrained to point in the same direction. Imposition of such a geometric constraint would interfere with centrosome separation and spindle assembly. Future live-cell experiments will be required to test this speculation.
The KT as an integrated platform for spindle checkpoint sensing and signaling
One critical function of KTs is to recruit and regulate spindle checkpoint proteins. Several studies in recent years have sought to identify KT structural proteins required for KT binding by Mad and Bub checkpoint proteins. Attention is usually drawn to situations in which KT association is impaired on the not unreasonable assumption that proteins required for binding Mad and Bub proteins to KTs might play a direct role in the poorly understood processes that generate a checkpoint signal. To date, the NDC80 complexes, Zw10 and CENP-I, have been implicated in KT binding by checkpoint proteins (Martin-Lluesma et al, 2002; Liu et al, 2003; Meraldi et al, 2004; Buffin et al, 2005; Kops et al, 2005), leading to the idea that a linear pathway links KT structural subunits and Mad–Bub proteins. This idea has been most clearly articulated in the case of the KT-associated CENP-E kinesin, which interacts directly with BubR1 (Mao et al, 2003).
However, we have established that in addition to members of the NDC80 complex, Nnf1R and Mcm21R are required for checkpoint signaling (Figure 11A). It seems highly unlikely that all of these KT structural proteins, most of which appear to be components of distinct multiprotein complexes, interact directly with Mad and Bub proteins. Instead, we propose that large segments of the inner and central domains of the KT are required for checkpoint signaling, presumably because many KT proteins act cooperatively to assemble the necessary quaternary structure. Nonetheless, a number of the interdependencies among checkpoint proteins and structural proteins are quite specific and appear rather surprising. For example, Mcm21R depletion prevents Mad2 binding to KTs without affecting Mad1. It has been well established that Mad1 binds tightly to Mad2 in vitro, and it has been shown in vivo that Mad1 is the Mad2 receptor on KTs (De Antoni et al, 2005). Why Mad1 is unable to bind Mad2 on Mcm21R-depleted KTs (even when kMTs are depolymerized) is not at all obvious. Perhaps, an as-yet unknown Mcm21R-sensitive modification or conformational change is involved.
Figure 11.

Speculative model for the interface between spindle checkpoint and core KT factors. (A) Model of vertebrate KT organization based on electron micrographs and adapted from (Emanuele et al, 2005). (B) Assembly model for core KT components and spindle checkpoint components. Black arrows represent dependencies; a red ‘X' indicated a lack of dependency (Martin-Lluesma et al, 2002; Liu et al, 2003; Meraldi et al, 2004; Kops et al, 2005). (C) Model for the effect of partial and complete KT protein depletion on the spindle checkpoint.
One complication in probing connections between KT structural proteins and Mad/Bub checkpoint proteins by RNAi is the extraordinary variation in phenotype caused by seemingly small differences in KT protein abundance. We have previously shown that 20-fold depletion of Nuf2R or Ndc80 activates the checkpoint, whereas 50- to 100-fold protein depletion inactivates it. We now show that this is also true of Nnf1R. In each case, the amount of protein required for correct MT binding is much greater than for correct checkpoint function. Very few molecules of Nuf2, Ndc80 or Nnf1R are sufficient to sustain a checkpoint signal, suggesting a high degree of amplification (Figure 11B). The extreme variability in phenotype with the extent of protein depletion helps to explain apparent contradictions between phenotype reported for different MIND proteins: Mis12 has been reported to cause chromosome missegregation without provoking the checkpoint (Goshima et al, 2003), whereas depletion of the associated Dsn1R protein causes defects in congression and provokes mitotic arrest (Obuse et al, 2004). Presumably, the full-depletion phenotype for MIND depletion is checkpoint inactivation.
Evolutionary conservation of core KT functions
Do MIND, COMA and NDC80 complexes have similar functions in yeast and human cells? In both S. cerevisiae and human cells, mutations in any one of the four subunits of the NDC80 complex block assembly of functional MT attachment sites and they inactivate the spindle checkpoint. NDC80 complexes also function as linkers to recruit checkpoint proteins, MAPs and motors onto KTs. In yeast, Nnf1p and Mtw1p mutants cause a defect in force generation, but not MT–KT detachment (De Wulf et al, 2003). Chromosome alignment, but not MT attachment, requires Nnf1 (KBP-2) and Nsl1 (KBP-1) orthologs in C. elegans (Cheeseman et al, 2004). We show here that in Nnf1R-depleted human cells, KTs bind spindle MTs but fail to congress to the metaphase plate, and similar phenotypes have been described for Mis12 and Dsn1R depletion (Goshima et al, 2003; Obuse et al, 2004). Human MIND proteins are required for assembly of checkpoint proteins and the Rod-Zw10-Zwilch complex onto KTs (Goshima et al, 2003; Cheeseman et al, 2004; Obuse et al, 2004; Wang et al, 2004; Emanuele et al, 2005), implying a role for MIND as a linker complex. In the case of Mcm21R, inactivation of the S. pombe ortholog Mal2 causes a very high rate of chromosome missegregation and interferes with spindle checkpoint function (Fleig et al, 1996). Formation of monopolar spindles has not been observed, but significant differences between spindle assembly in organisms with open and closed mitosis are a likely explanation. Based on these as-yet preliminary findings, we conclude that yeast and human KTs are likely to share a set of common protein complexes, corresponding to the ‘middle' of the KT, which play roughly similar roles. Linker proteins serve to build a platform for KT binding by a multiplicity of MAPs, motors and checkpoint proteins, thereby leading to the formation of fully functional MT attachment sites. With the isolation of many human linker proteins, several key steps in KT assembly are now open to molecular dissection.
Materials and methods
Antibody production
Nnf1R (accession number: NM_007221) and Mcm21R (accession number: BC002870) cDNAs were obtained from ATCC or PCR amplified from HeLa cell cDNA library (Invitrogen). Rabbit antibodies were made against full-length GST-Nnf1R and His6-Mcm21R expressed in Escherichia coli (Covance). GST-Nnf1R was purified as inclusion bodies and His6-Mcm21R was purified on Nickel Sepharose Beads (Qiagen).
Cell culture, siRNA transfection, MG132 and nocodazole treatments
HeLa and HeLa H2B-GFP cells were grown as described (Meraldi et al, 2004). To obtain double-stable H2B-GFP/α-tubulin-mRED cells line, H2B-GFP cells were transfected with α-tubulin-mRED-IRES-Puro vector (kind gift of D Gerlich) using Fugene 6 (Roche) and positive cells selected with puromycin. siRNA oligonucleotides (Qiagen or Invitrogen) are described in Supplementary data and were transfected as described (Elbashir et al, 2001). For MG132 experiments, HeLa cells were synchronized and treated with MG132 as described (Meraldi et al, 2004). To test spindle checkpoint activity, cells were treated with 100 ng/ml nocodazole for 16 h and the fraction of rounded-up cells determined by phase-contrast microscopy.
Immunofluorescence microscopy and immunoblotting
Cells were prepared as described previously (Meraldi et al 2004). Primary antibodies are described in Supplementary data. Cross-absorbed secondary antibodies were used (Molecular probes). Images were acquired as described (Martinez-Exposito et al, 1999). KT signals were quantified as described in Supplementary data. Whole-cell extract preparation and immunoblotting were carried out as described previously (Meraldi et al, 2004), using primary antibodies as described in Supplementary data and anti-mouse and anti-sheep HRP-conjugated secondary antibodies (Amersham Pharmacia).
Live-cell time-lapse imaging and analysis
Cells were imaged in ΔT 0.15 mm dishes (Bioptechs) or LabTech II (LabTech) chambers in CO2-independent medium (GibcoBRL) at 37°C. For H2B-GFP HeLa cell movies, pictures were acquired every 3 min for 6 h using a 20 × NA 0.75 objective on a Olympus Applied Precision Deltavision microscope equipped with a Mercury 100 W lamp, GFP-long-pass filter set (Chroma) and Coolsnap HQ camera. For the double H2B-GFP/α-tubulin-mRED HeLa cells, pictures were acquired on the same microscope with a 40 × oil NA 1.3 objective, and FITC and TexasRed filter sets (Chroma).
Supplementary Material
Supplementary data
Supplementary movie 1
Supplementary movie 2
Supplementary Figure 1
Acknowledgments
We thank D Gerlich, S Taylor and W Earnshaw for gifts of reagents, the Light Microscopy Centre of the ETH Zurich for technical support, Sarah McClelland for help with antibody characterization and the members of the McAinsh, Meraldi and Sorger labs for helpful discussions. ADM was supported by a Jane Coffin Childs Fund for Medical Research fellowship and PM by an EMBO long-term fellowship. This work was supported by NIH Grants CA84179 and GM51464 (PKS), Swiss National Science Foundation Grant 3100A0-107912/1 (PM) and Marie Curie Cancer Care (ADM).
References
- Bucciarelli E, Giansanti MG, Bonaccorsi S, Gatti M (2003) Spindle assembly and cytokinesis in the absence of chromosomes during Drosophila male meiosis. J Cell Biol 160: 993–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 [DOI] [PubMed] [Google Scholar]
- Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15: 589–598 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR III, Oegema K, Desai A (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280: 29088–29095 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A (2005) The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225 [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK (2003) Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 17: 2902–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED (2003) Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol 13: 2103–2109 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT (2005) Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 16: 4882–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U, Sen-Gupta M, Hegemann JH (1996) Fission yeast mal2+ is required for chromosome segregation. Mol Cell Biol 16: 6169–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T (2004) Assembly of kinetochores in vertebrate cells. Exp Cell Res 296: 21–27 [DOI] [PubMed] [Google Scholar]
- Gadde S, Heald R (2004) Mechanisms and molecules of the mitotic spindle. Curr Biol 14: R797–R805 [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW (2003) The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev 17: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M (2003) Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol 160: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M (2000) Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100: 619–633 [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK (2001) Molecular analysis of kinetochore–microtubule attachment in budding yeast. Cell 106: 195–206 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A (1997) Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol 138: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T (2003) Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci 116: 3347–3362 [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E (2001a) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 20: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, Schramm C, Schiebel E (2001b) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 20: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS (2001) A role for the adenomatous polyposis coli protein in chromosome segregation. Nat Cell Biol 3: 429–432 [DOI] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR III, Tagaya M, Cleveland DW (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Hittle JC, Jablonski SA, Campbell MS, Yoda K, Yen TJ (2003) Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat Cell Biol 5: 341–345 [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL (2005) Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol 7: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Earnshaw WC, Sunkel CE (2003) How do kinetochores CLASP dynamic microtubules? Cell Cycle 2: 511–514 [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE (2002) MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol 157: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW (2003) Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114: 87–98 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Martinez-Exposito MJ, Kaplan KB, Copeland J, Sorger PK (1999) Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc Natl Acad Sci USA 96: 8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19: 519–539 [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT (2003) The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Sorger PK (2005) A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J 24: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV (2003) Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004) A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6: 1135–1141 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Faruki S, Khodjakov A (2001) The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol 11: 413–419 [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78: 761–771 [DOI] [PubMed] [Google Scholar]
- Scharfenberger M, Ortiz J, Grau N, Janke C, Schiebel E, Lechner J (2003) Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J 22: 6584–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ (2002) EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell 13: 4308–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu X, Ding X, Dou Z, Yang Z, Shaw AW, Teng M, Cleveland DW, Goldberg ML, Niu L, Yao X (2004) Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J Biol Chem 279: 54590–54598 [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC (2005) Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA 102: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary movie 1
Supplementary movie 2
Supplementary Figure 1


