Abstract
Unliganded thyroid hormone receptor (TR) actively represses transcription via the nuclear receptor corepressor (N-CoR)/histone deacetylase 3 (HDAC3) complex. Although transcriptional activation by liganded receptors involves chromatin remodeling, the role of ATP-dependent remodeling in receptor-mediated repression is unknown. Here we report that SNF2H, the mammalian ISWI chromatin remodeling ATPase, is critical for repression of a genomically integrated, TR-regulated reporter gene. N-CoR and HDAC3 are both required for recruitment of SNF2H to the repressed gene. SNF2H does not interact directly with the N-CoR/HDAC3 complex, but binds to unacetylated histone H4 tails, suggesting that deacetylase activity of the corepressor complex is critical to SNF2H function. Indeed, HDAC3 as well as SNF2H are required for nucleosomal organization on the TR target gene. Consistent with these findings, reduction of SNF2H induces expression of an endogenous TR-regulated gene, dio1, in liver cells. Thus, although not apparent from studies of transiently transfected reporter genes, gene repression by TR involves the targeting of chromatin remodeling factors to repressed genes by the HDAC activity of nuclear receptor corepressors.
Keywords: chromatin, HDAC, nuclear receptor, repression, SNF2H
Introduction
Thyroid hormone receptor (TR) functions as a critical transcription factor during development, growth, and metabolism (Zhang and Lazar, 2000). In the presence of its thyroid hormone ligand, TR interacts with specific coactivators that function to promote gene transcription. By contrast, unliganded TR binds to corepressor proteins that actively repress transcription (Wu and Koenig, 2000). This ligand induced exchange of cofactors leads to alterations in chromatin that signal changes in gene transcription (Glass and Rosenfeld, 2000).
Several histone modifying enzymes have been implicated in transcriptional activation by nuclear receptors (NRs) (Aranda and Pascual, 2001). In particular, specific histone acetyltransferase enzymes, such as SRC-1, associate with TR and stimulate local histone hyperacetylation in the presence of ligand (Jeyakumar et al, 1997; Spencer et al, 1997). Conversely, the conformation of unliganded TR favors interactions with the corepressors nuclear receptor corepressor protein (N-CoR) or silencing mediator for retinoid and thyroid receptor (SMRT) (Chen and Evans, 1995; Horlein et al, 1995; Hu and Lazar, 2000). While TR preferentially binds and utilizes N-CoR for repression (Cohen et al, 2000; Webb et al, 2000; Ishizuka and Lazar, 2003), both N-CoR and SMRT stably associate with and activate histone deacetylase 3 (HDAC3) through a SANT (SWI3, ADA2, N-CoR, TFIIB) motif-containing region termed the deacetylase activation domain (Webb et al, 2000; Guenther et al, 2001; Ishizuka and Lazar, 2005). The deacetylation of local histones by HDAC3 results in repression of gene transcription (Ishizuka and Lazar, 2003). A second SANT domain in the N-terminus region of N-CoR and SMRT mediates a direct interaction with unacetylated histone H4 N-terminus tails, suggesting a feedforward model for corepressor binding (Yu et al, 2003; Hartman et al, 2005).
In addition to enzymes that covalently modify histone proteins, NR-mediated gene activation has been shown to require ATP-dependent chromatin remodeling enzymes (Urnov and Wolffe, 2001; Chen et al, 2006). Remodelers utilize ATP hydrolysis to alter the structure of chromatin, through mechanisms such as nucleosomal dissociation, sliding, or relocation (Johnson et al, 2005). Several studies have identified the SWI/SNF ATPases as critical factors in ligand mediated activation by NRs, including retinoid receptor, glucocorticoid receptor, thyroid receptor, and androgen receptor (Yoshinaga et al, 1992; Muchardt and Yaniv, 1993; Fryer and Archer, 1998; Dilworth et al, 2000; Huang et al, 2003).
Another extensively studied group of chromatin remodeling enzymes is the imitation switch (ISWI) family of ATPases. These ATPases are distinguished from the SWI/SNF ATPases by the presence of SANT domains in the C-terminus region and are found in complexes that have been implicated in an array of biologic processes, including chromatin assembly, maintenance of higher order chromatin, as well as transcription activation and repression (Corona and Tamkun, 2004; Dirscherl and Krebs, 2004). In vitro studies have suggested a role for ISWI ATPases in NR regulation and the Drosophila ISWI containing complex, NURF, was found to be critical for activation of a set of ecdysone responsive genes, likely through its ligand dependent interaction with the ecdysone NR (Di Croce et al, 1999; Dilworth et al, 2000; Badenhorst et al, 2005).
Although the importance of ATP-dependent chromatin remodeling has been examined with respect to ligand-induced transcriptional activation, very little is known about the role of remodeling in NR repression. The Mi-2/NURD complex, which contains a chromodomain ATPase as well as histone and DNA modifying enzymes, functions predominantly in transcriptional repression and has been suggested to play a role in NR mediated repression (Mazumdar et al, 2001; Li et al, 2002; Johnson et al, 2004). In yeast, Drosophila, and mammalian systems, the ISWI ATPases and ISWI containing complexes have also been implicated in repression and, in fact, ISWI preferentially associates with silenced regions of the Drosophila chromosome (Deuring et al, 2000; Goldmark et al, 2000; Zhou et al, 2002). Furthermore, recent genetic studies suggest that NURF as well as ISWI itself are involved in both activation and repression (Badenhorst et al, 2005; Xi and Xie, 2005).
Two ISWI ATPases, termed SNF2H and SNF2L, have been identified in mammalian systems. SNF2H, the more extensively studied mammalian ISWI ATPase, exists in at least six known complexes and demonstrates significant functional differences when compared to the SWI/SNF ATPase BRG1 (Aalfs et al, 2001; Dirscherl and Krebs, 2004; Fan et al, 2005). The lethal phenotype of the SNF2H null mice demonstrates the critical role for this ATPase in mammalian development (Stopka and Skoultchi, 2003). Here, we utilize a stably integrated luciferase reporter gene under regulation by the Gal4-TR receptor to demonstrate a role for the ATPase SNF2H in mammalian NR repression. SNF2H recruitment to the TR repressed gene depends on N-CoR, but SNF2H does not form a stable interaction with the corepressor complex. Instead, SNF2H interacts with endogenous histone H4 and preferentially binds to unacetylated histone H4 tails. HDAC3 is required for recruitment of SNF2H, suggesting that histone deacetylation by HDAC3 favors the binding of SNF2H to histone tails within repressed genes. Furthermore, depletion of SNF2H as well as HDAC3 by RNA interference changes the MNase sensitivity of the repressed promoter. Consistent with this model, SNF2H functions in repression of the endogenous TR regulated gene type I iodothyronine deiodinase (dio1) and recruitment of SNF2H to the dio1 promoter is dependent on HDAC activity. Thus, histone deacetylation by the N-CoR complex represses transcription from chromosomal genes in part by recruitment of SNF2H.
Results
Repression of an integrated gene by a stably expressed TR requires N-CoR
We utilized a 293T cell line that has an integrated (Gal4 UAS x5)-sv40 promoter-luciferase reporter gene (UAS-Lucif) (Ishizuka and Lazar, 2003). The integrated region of the sv40 genome was limited to the sv40 promoter (5175 to 130, NC_001669) and did not contain the 72 bp tandem repeat enhancer sequence. From the UAS-Lucif 293T line, we established a subline (GalTR/UAS-Lucif), which co-expressed the TRβ-ligand binding domain fused to the Gal4 DNA binding domain (DBD) (GalTR). Addition of T3 was sufficient to activate the reporter gene in the GalTR/UAS-Lucif cells (Figure 1A). Chromatin immunoprecipitation (ChIP) following 24 h treatment with T3 demonstrated that activation corresponded to increased recruitment of RNA polymerase II and decreased association of the corepressor complex proteins, N-CoR, and HDAC3, with the integrated reporter gene (Figure 1B). The role of the association of N-CoR in the absence of T3 was explored by depleting N-CoR using small interfering RNA (siRNA) (Figure 1C). Loss of N-CoR markedly increased luciferase activity, but not in the receptor deficient cell line (UAS-Lucif) (Figure 1D), demonstrating the role of N-CoR in actively repressing the reporter gene in the presence of the TR.
Figure 1.
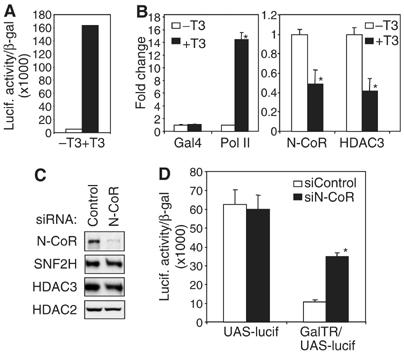
GalTR/UAS-Lucif model system requires N-CoR for inhibiting transcription in unliganded state. (A) T3-dependent luciferase activity of a 293T cell line with stably integrated (UASx5)-sv40 promoter-luciferase reporter gene and Gal4-TRβ fusion receptor (GalTR/UAS-Lucif). Cells were incubated with T3 (final concentration 10 nM) for 24 h (B) ChIP analysis at the integrated gene in GalTR/UAS-Lucif cells in the absence or presence of T3 (10 nM, 24 h). Samples were analyzed by real-time PCR, normalized to the 36B4 gene, and plotted as fold change relative to no ligand (−T3). *P<0.05 versus no ligand. (C) Immunoblot following transfection of cells with siRNA for N-CoR. (D) Transcription assay following treatment with siRNA for N-CoR in control UAS-Lucif cells (lack receptor) and GalTR/UAS-Lucif cells. Luciferase activity was normalized to β-gal expression and then plotted. *P<0.05 versus siRNA control.
SNF2H functions in repression of a stably integrated NR regulated gene
We next tested whether the chromatin remodeling ATPase SNF2H played a role in repression of the integrated luciferase gene in GalTR/UAS-Lucif cells. Using siRNA, we successfully reduced SNF2H protein levels in the cells (Figure 2A). Relative to cells treated with control siRNA, depletion of SNF2H increased luciferase activity in cells expressing Gal-TR (Figure 2B), suggesting that SNF2H was required to mediate repression of the integrated reporter when under the regulation of GalTR. Interestingly, SNF2H was not needed for repression by GalTR of a transiently transfected luciferase gene (Figure 2C) whereas the repressive function of N-CoR could be detected on this gene (Figure 2D). Transiently transfected genes do incorporate histones to form nucleosomes, and indeed we have previously shown that the repressive functions of N-CoR and HDAC3 can be observed in this system (Ishizuka and Lazar, 2003). However, the presently demonstrated context-dependence of SNF2H effects on repression underscores the notion that transiently transfected genes do not assemble into the same higher ordered chromatin structure that exists for chromosomal genes (Fryer and Archer, 1998; Cervoni and Szyf, 2001). For this reason, transiently transfected genes may not provide an adequate template on which to study the mechanism of chromatin remodeling.
Figure 2.
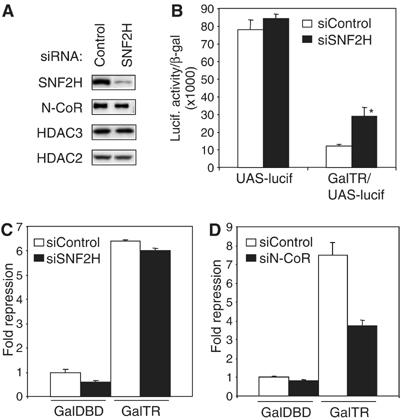
SNF2H mediates repression of a stably integrated TR regulated gene. (A) Immunoblot following transfection of cells with siRNA for SNF2H. (B) Transcription assay following treatment with siRNA for SNF2H in control UAS-Lucif cells (lack receptor) and GalTR/UAS-Lucif cells. Luciferase activity was normalized to β-gal expression and then plotted. *P<0.05 versus siRNA control. (C, D) Transcription assay in transient reporter system. The (UASx5)-sv40 promoter-luciferase reporter was transiently transfected along with Gal4-DBD or Gal4-TR into 293T cells treated with siRNA for (C) SNF2H or (D) N-CoR. Luciferase activity was normalized to β-gal expression and plotted as fold repression relative to Gal4-DBD.
SNF2H recruitment depends on N-CoR
We hypothesized that the repressive function of SNF2H in the context of the integrated TR-regulated reporter might work in cooperation with the N-CoR corepressor. To test this, we performed ChIP analysis in the GalTR/UAS-Lucif cells with or without siRNA depletion of N-CoR. As expected, decreased levels of endogenous N-CoR led to decreased recruitment of HDAC3 to the promoter region (Figure 3). Importantly, loss of N-CoR also decreased recruitment of SNF2H, demonstrating that recruitment of SNF2H is dependent on N-CoR.
Figure 3.
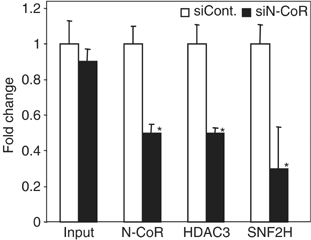
N-CoR is required for SNF2H recruitment. ChIP analysis at the integrated gene in GalTR/UAS-Lucif cells following treatment with control or N-CoR siRNA. Samples were analyzed by real-time PCR, normalized to the 36B4 gene, and plotted as fold change relative to control siRNA treatment. *P<0.05 versus siRNA control (n=3).
SNF2H does not stably associate with the N-CoR corepressor complex
N-CoR exists in high molecular weight, multiprotein complexes (Guenther et al, 2000; Li et al, 2000, 2005; Zhang et al, 2002; Goodson et al, 2005), and hence we hypothesized that the reliance on the N-CoR complex for SNF2H recruitment to the repressed reporter gene might be due to an interaction of SNF2H with the N-CoR complex. In order to examine whether SNF2H could stably interact with N-CoR, cells were transfected with Flag tagged N-CoR and subjected to immunoprecipitation, followed by immunoblot analysis. As expected, HDAC3 strongly associated with N-CoR; however, there was no interaction detected with SNF2H (Figure 4A). Moreover, when endogenous SNF2H was immunoprecipitated, no stable association was found between SNF2H and the corepressor proteins (Figure 4B). These findings suggested that the N-CoR complex does not recruit SNF2H to the promoter through a direct interaction.
Figure 4.
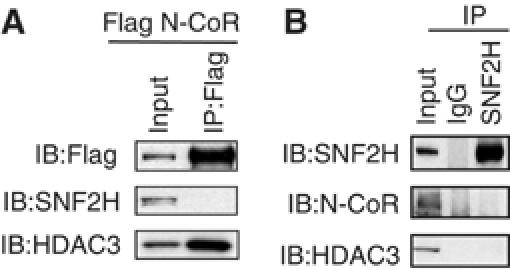
SNF2H does not bind N-CoR/HDAC3 complex. (A) 293T cells were transfected with pCMX-Flag-N-CoR and whole-cell lysates were immunoprecipitated with anti-Flag agarose beads. Bound proteins were separated by SDS–PAGE and immunoblotted with Flag, SNF2H or HDAC3 antibodies. Input is 1%. (B) Immunoprecipitation of 293T whole-cell lysates was performed with normal rabbit IgG or rabbit SNF2H antibody and followed by immunoblot analysis. Input is 3%.
SNF2H preferentially binds unacetylated histone H4 tails
Studies of Drosophila ISWI containing complexes have shown that histone H4 N-terminus tails are essential for ISWI remodeling activity and that lysine acetylation may interfere with ISWI function (Georgel et al, 1997; Clapier et al, 2001; Corona et al, 2002; Shogren-Knaak et al, 2006). Mammalian complexes have also suggested links between SNF2H activity and HDACs (Zhou et al, 2002, 2005; Santoro and Grummt, 2005). Consistent with this, endogenous SNF2H bound histone H4 in our mammalian system (Figure 5A). The C-terminus of ISWI contains a SANT domain that has been suggested to bind histone tails (Grune et al, 2003). As N-CoR and SMRT associate preferentially with unacetylated histone tails via a SANT domain (Yu et al, 2003), we hypothesized that SNF2H interactions with histone may also be regulated by acetylation state. Indeed, as previously reported for the corepressors, recombinant SNF2H bound with much higher affinity to unacetylated histone H4 tails in comparison to tetra-acetylated H4 tails (Figure 5B).
Figure 5.
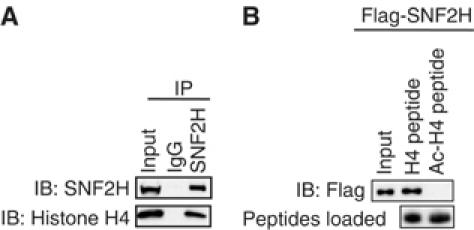
SNF2H preferentially binds unacetylated histone H4 tails. (A) Immunoprecipitation of 293T whole-cell lysates was performed with normal rabbit IgG or rabbit SNF2H antibody and followed by immunoblot analysis for SNF2H or histone H4. Input is 3%. (B) Recombinant Flag-SNF2H was incubated with bead-bound biotinylated unacetylated histone H4 peptide (H4 peptide) or tetra-acetylated H4 peptide (Ac-H4 peptide). Beads were washed and bound protein was separated by SDS–PAGE and immunoblotted. Input is 10%.
SNF2H recruitment depends on HDAC3
Since SNF2H associates with repressed genes but is not directly recruited by N-CoR, the observation that SNF2H binds to hypoacetylated histones suggested that its recruitment might be dependent upon deacetylation of H4 N-terminal tails by HDAC3. This was tested by depleting HDAC3 protein (Figure 6A). Consistent with the known function of HDAC3, H4 hyperacetylation was observed upon loss of HDAC3 from the cells (Figure 6B) (Hartman et al, 2005). Moreover, HDAC3 knockdown also decreased recruitment of SNF2H to the TR target gene (Figure 6B). Together with the observation that SNFH binds to unacetylated histone H4 tails, this finding suggests that recruitment of SNF2H to the repressed chromatinized gene depends upon HDAC3 providing a preferred template for SNF2H binding to nucleosomal histones.
Figure 6.
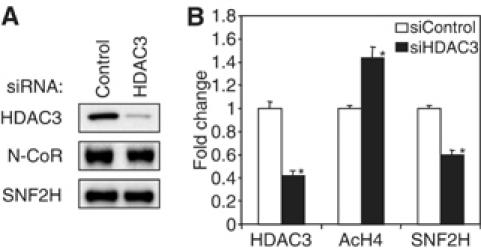
HDAC3 is required for SNF2H recruitment. (A) Immunoblot following transfection of cells with siRNA for HDAC3. (B) ChIP analysis at the integrated gene in GalTR/UAS-Lucif cells following treatment with control or HDAC3 siRNA. Samples were analyzed by real-time PCR, normalized to the 36B4 gene, and plotted as fold change relative to control siRNA treatment. *P<0.05 versus siRNA control (n=3).
SNF2H and HDAC3 regulate MNase sensitivity of the TR-repressed gene
We next determined the role of chromatin remodeling at the TR target gene using micrococcal nuclease (MNase), which preferentially digests the linker DNA between nucleosomes. Digestion was measured by PCR amplification of a 90 bp region (promoter) or 120 bp region (start site), which is less than the DNA length protected by a single nucleosome (Figure 7A). Reduction in SNF2H levels significantly decreased MNase protection in the region containing the transcriptional start site, and concomitantly increased protection in the promoter region (Figure 7B). Although this assay does not distinguish changes in nucleosomal position from alterations in nucleosomal occupancy, stability, or structure, these data clearly reveal that the specific chromatin architecture within the promoter of the TR-repressed gene is normally dependent upon SNF2H.
Figure 7.
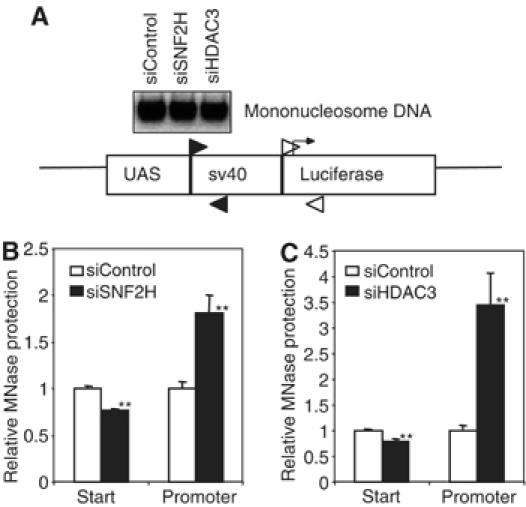
SNF2H and HDAC3 regulate chromatin architecture of a TR-repressed gene. (A) Nuclei from GalTR/UAS-Lucif cells were collected following treatment with control and (B) SNF2H siRNA or (C) HDAC3 siRNA and digested with MNase. After DNA purification, complete digestion to mononucleosome size was confirmed by agarose gel electrophoresis. Samples were analyzed by real-time PCR using primers that amplified a 90 bp region in the sv40 promoter (black arrows) or a 120 bp region containing the luciferase start site (white arrows). Results were normalized to the 36B4 gene and plotted as fold change relative to control siRNA treatment. **P<0.01 versus siRNA control (n=3).
Since HDAC3 is crucial for SNF2H recruitment, next we determined whether HDAC3 played a role in this remodeling activity. To do this, we conducted the MNase sensitivity assay following treatment with HDAC3 siRNA (Figure 7C). Similar to the change in MNase sensitivity observed with SNF2H knockdown, depletion of HDAC3 led to increased protection from MNase digestion at the promoter, consistent with our finding that SNF2H recruitment depends on HDAC3. As HDAC3 itself does not possess any known remodeling activity, our data suggest that the change in MNase sensitivity with HDAC3 knockdown resulted, in part, from increased histone tail acetylation and a corresponding decrease in stable SNF2H nucleosome binding and remodeling.
SNF2H functions in repression of endogenous TR target gene
The dio1 gene, which encodes type I iodothyronine deiodinase, a critical selenoenzyme involved in the metabolism of thyroid hormone, is one of the few well characterized direct targets of TR. (Berry et al, 1991; Zavacki et al, 2005). A classic thyroid hormone response element (TRE), consisting of two direct repeat half-sites separated by 4 bp (DR+4), has been identified within the distal hdio1 promoter and has been shown to bind TR and mediate T3 responsiveness (Toyoda et al, 1995; Jakobs et al, 1997). To compare our findings using the integrated reporter model with an endogenous target gene regulated by native TR, we examined the role of SNF2H in regulation of dio1 expression in human hepatocellular carcinoma HepG2 cells. SNF2H siRNA plasmid electroporation of HepG2 cells led to efficient reduction in SNF2H without altering protein levels of TRβ (Figure 8A). Notably, however, knockdown of SNF2H led to a significant increase in dio1 expression (Figure 8B), demonstrating a functional role for SNF2H in repression of an endogenous TR regulated gene.
Figure 8.
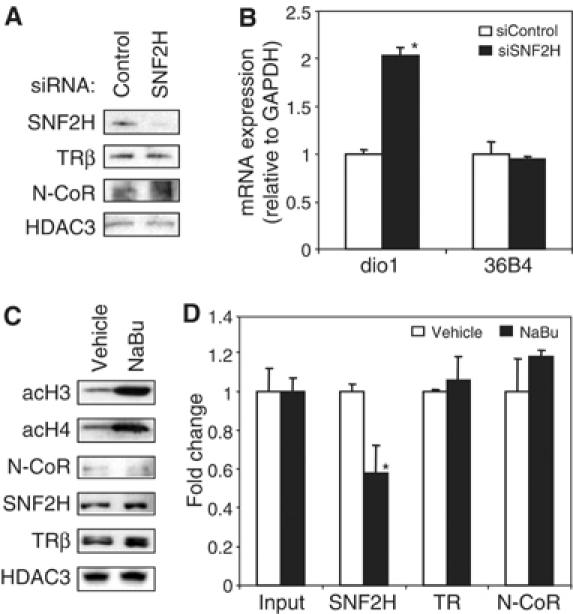
SNF2H represses endogenous dio1 gene and recruitment requires HDAC activity. (A) Immunoblot following electroporation of HepG2 cells with siRNA for SNF2H. (B) Reverse transcriptase analysis of dio1 and 36B4 mRNA expression in HepG2 cells following treatment with control or SNF2H siRNA. Samples were analyzed by real-time PCR, normalized to Gapdh, and plotted as fold change relative to control siRNA treatment. *P<0.05 versus siRNA control (n=3). (C) Immunoblot following incubation of HepG2 cells with vehicle (water) or sodium butyrate (5 mM, 5 h). (D) ChIP analysis at the hdio1 promoter in HepG2 cells following treatment with sodium butyrate (5 mM, 5 h). Samples were analyzed by real-time PCR, normalized to the 36B4 gene, and plotted as fold change relative to vehicle treatment. *P<0.05 versus vehicle (n=3).
SNF2H recruitment to endogenous TRE requires HDAC activity
In order to test whether HDAC activity is required for SNF2H recruitment to the dio1 promoter, HepG2 cells were treated with the HDAC inhibitor, sodium butyrate (5 mM, 5 h). As expected, inhibition of HDAC activity led to increased histone acetylation (Figure 8C). Interestingly, treatment of cells with sodium butyrate led to decreased recruitment of SNF2H to the TRE region of the dio1 promoter without altering recruitment of TRβ or N-CoR (Figure 8D). Thus, consistent with our findings that SNF2H recruitment to the integrated TR reporter gene depends on HDAC3 (Figure 6B), recruitment of SNF2H to the dio1 promoter requires HDAC enzymatic activity.
Discussion
We have demonstrated a novel role for the chromatin remodeling ATPase SNF2H in NR mediated repression of an integrated reporter gene as well as an endogenous TR regulated gene. N-CoR is necessary for recruitment of SNF2H, but the ATPase does not stably associate with the corepressor complex itself. Instead, SNF2H interacts with unacetylated histone H4 N-terminus tails and, interestingly, depletion of the HDAC3 enzyme or inhibition of HDAC activity decreases SNF2H recruitment. Consistent with these findings, depletion of SNF2H as well as HDAC3 leads to similar changes in MNase sensitivity at a repressed promoter. This work suggests a model in which receptor bound HDAC3 directs deacetylation of histone H4 tails, repressing transcription but also permitting stable binding of the chromatin remodeler SNF2H (Figure 9).
Figure 9.
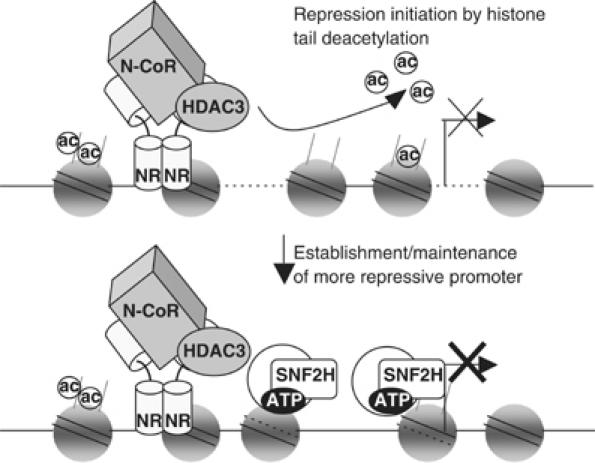
The N-CoR/HDAC3 corepressor couples histone deacetylation to SNF2H-dependent chromatin remodeling in NR repression. The N-CoR/HDAC3 complex associates with unliganded NR and deacetylates local histones, resulting in active repression of the gene. Deacetylation of histone tails also permits stable recruitment of the ATP-dependent chromatin remodeler SNF2H, which can further modify chromatin, possibly by sliding nucleosomes as depicted in the model.
The prevailing model for the mechanism of ISWI remodeling proposes that these ATPases physically alter chromatin structure by sliding nucleosomes along the DNA, in contrast to SWI/SNF ATPases such as BRG1, which are believed to loop the DNA away from the histone octamer (Hamiche et al, 1999; Langst et al, 1999; Fan et al, 2003; Fazzio and Tsukiyama, 2003; Kassabov et al, 2003; Stockdale et al, 2006). Therefore, it is probable that SNF2H recruitment to genes repressed by TR, and potentially other NRs, leads to local nucleosomal movement in a manner that enhances repression (Figure 9). However, in addition to or instead of changes in nucleosome position or occupancy, it is also possible that SNF2H represses by establishing a more stable higher order chromatin structure (Varga-Weisz and Becker, 2006). Furthermore, our model does not eliminate the possibility that, on other promoters, SNF2H could alter chromatin character such that it counters the repressive effects of histone deacetylation, consistent with its role in both transcriptional activation and repression (Corona and Tamkun, 2004). As the regulatory regions and nucleosome positions of more mammalian TR- regulated genes become characterized, it will be important to determine the promoter specificity of SNF2H as well as the mechanism of how it remodels these promoters.
Studies in Xenopus oocytes have revealed that the nucleosomal architecture of a thyroid hormone responsive promoter is integral to its regulation (Wong et al, 1995, 1997). The Mi2/NURD remodeling complex, which also possesses HDAC activity, has been suggested to play a nonspecific role in repression of TR-regulated genes via a constitutive association with chromatin (Li et al, 2002). Interestingly, it has been reported that the N-CoR/HDAC3 complex contains components of the SWI/SNF complex, including the ATPase BRG1 (Underhill et al, 2000). A direct interaction between this SWI/SNF ATPase and the N-CoR complex suggests that the complex itself may contain chromatin remodeling activity that could play a role in repression. However, intrinsic remodeling activity of the N-CoR/HDAC3 complex or a role for BRG1 in NR-mediated repression has yet to be shown. Here, instead, we show that a different remodeling ATPase, SNF2H, is indirectly recruited through N-CoR/HDAC3 activity and contributes to repression of an integrated and an endogenous T3-responsive gene.
Since SNF2H recruitment to the repressed gene does not occur through a direct interaction with the N-CoR/HDAC3 complex, it was somewhat surprising to find equivalent decreases in recruitment of both HDAC3 and SNF2H with depletion of N-CoR by RNAi. The findings that depletion of HDAC3 and inhibition of HDAC activity also decrease SNF2H recruitment suggest that SNF2H recruitment is actually dependent on a functional N-CoR/HDAC3 complex. This complex has other functions, and indeed depletion of N-CoR had a greater effect than depletion of SNF2H on transcription. Nevertheless, although we did not detect a decrease in N-CoR recruitment to the promoter on reduction in SNF2H levels (data not shown), it is possible that SNF2H could also be involved in stabilization of the receptor or N-CoR/HDAC3 complex at the promoter.
Following treatment with thyroid hormone, there is an increase in histone acetylation. However, in this context, we did not detect a decrease in SNF2H recruitment to the integrated reporter gene (data not shown). Since we have found that increased histone H4 acetylation decreases the interaction between SNF2H and H4 N-terminus tails, this recruitment of SNF2H likely occurs through an association with T3 recruited coactivator proteins. Indeed, although ISWI can stimulate remodeling on its own, the ATPase has been purified in a variety of complexes that can modulate its recruitment under different cellular conditions (Langst and Becker, 2001; Varga-Weisz, 2001; Dirscherl and Krebs, 2004).
Regulation of transcription involves coordination between covalent modification of nucleosomal histones and changes in chromatin remodeling (Eberharter et al, 2005; Wysocka et al, 2006). Our findings suggest that the N-CoR/HDAC3 corepressor complex couples histone deacetylation to SNF2H-dependent chromatin remodeling, and that both contribute to repression (Figure 9). The chromatin remodeling activity of ISWI is facilitated by histone interaction (Georgel et al, 1997; Clapier et al, 2001), but acetylation of H4-K16 has an inhibitory effect (Corona et al, 2002; Shogren-Knaak et al, 2006). More recently, deacetylation of H4-K16 has been suggested to act as a switch from a less condensed chromatin architecture to a more condensed higher order structure (Shogren-Knaak et al, 2006). Intriguingly, H4-K16 deacetylation at an NR-repressed promoter occurs after deacetylation of other H4 tail lysines (Hartman et al, 2005). This may serve to ensure that SNF2H mediated chromatin remodeling of NR-regulated promoters occurs after histone deacetylation is complete.
Materials and methods
Plasmids
pCMX-Gal4-DBD, pCMX-Gal4-TRβ1 and pCMX-Flag-mouse N-CoR constructs have been described previously (Hu et al, 2001; Ishizuka and Lazar, 2003). The Gal4-TR-hygro construct was created using restriction digestion followed by ligation into the pcDNA 3.1 Hygro vector (Invitrogen).
Mammalian cell culture and stable transfection
293T and HepG2 cells were maintained in DMEM (high glucose) (GIBCO BRL) supplemented with 10% fetal bovine serum (Sigma). Cells were grown at 37°C in 5% CO2. For stable transfection, ‘UAS-lucif' cells, containing the integrated (Gal4 UAS x5)-sv40 promoter (5175 to 130, NC_001669)-luciferase reporter gene (Ishizuka and Lazar, 2003) were grown in 10 cm dishes and transfected with Gal4-TR-hygro using Lipofectamine2000 (Invitrogen). Cells were cultured for 48 h prior to selection with hygromycin (Sigma, 200 μg/ml) for 3 weeks. For T3 or sodium butyrate (Sigma) experiments, cells were incubated with DMEM supplemented with 10% charcoal/dextran stripped serum for 18–24 h, followed by addition of either T3 (final concentration 10 nM) for 24 h or sodium butyrate (final concentration 5 mM) for 5 h.
Immunoprecipitation
Cells were washed in PBS and lysed in BC100 buffer (100 mM KCl, 20 mM HEPES, 0.2% NP40, 0.2 mM EDTA, 20% glycerol) containing protease inhibitor cocktail (Roche) on ice for 30 min or HERR buffer (50 mM KCl, 20 mM HEPES, 2 mM EDTA, 0.1% NP40, 10% glycerol, 0.5% nonfat dry milk) followed by sonication. Lysates were clarified by centrifugation at 12 000 g for 10 min at 4°C. Supernatants were incubated with anti-Flag agarose beads (Sigma) at 4°C overnight or anti-SNF2H (Abcam)/(Hakimi et al, 2002) at 4°C overnight followed by 1 h incubation with protein A agarose beads at 4°C. Immunoprecipitates were washed and subjected to immunoblot analysis.
In vitro interaction assays
For the histone tail binding assay, 1 μg of biotinylated histone H4 tail peptides, unacetylated or tetraacetylated (Upstate), was immobilized on streptavidin beads (Pierce). Recombinant Flag-SNF2H (0.15 μg) (Hakimi et al, 2002) was incubated with streptavidin bound histone H4 tails at 4°C for 2 h in HERR buffer. Bound proteins were eluted and subjected to immunoblot analysis.
Immunoblot analysis
Proteins were harvested in either passive lysis buffer (Promega) or modified RIPA followed by sonication and were separated by SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes with HMW transfer buffer. Blots were probed with the following primary antibodies in Tris-buffered saline containing 0.15% Tween-20 and 3% nonfat dry milk, followed by HRP-conjugated anti-Rabbit (Chemicon) or anti-Mouse (Santa Cruz) antibodies and ECL reagent (Amersham): rabbit N-CoR (Ishizuka and Lazar, 2003) rabbit HDAC2, mouse TRβ (Santa Cruz), rabbit SNF2H, rabbit HDAC3 (Abcam), mouse HDAC3 (BD Biosciences), rabbit histone H4, rabbit acetyl-histone H3, rabbit acetyl-histone H4 (Upstate). Alternatively, the streptavidin-HRP (Upstate) and Flag-HRP (Sigma) conjugates were used for direct detection.
RNA interference
Control, N-CoR and HDAC3 siRNA constructs were described previously (Ishizuka and Lazar, 2003). The SNF2H siRNA construct was created by inserting an annealed hairpin DNA oligonucleotide containing the SNF2H target sequence GAGGAGGATGAAGAGCTAT into the pSUPER vector (Oligoengine). 293T cells in 12-well plates were transfected with 1.5 μg of the siRNA construct and 0.1 μg of the β-galactosidasae (β-gal) expression vector using Lipofectamine2000 (Invitrogen). At 48 h after transfection, cells in each well were divided. After an additional 24 h, a repeat transfection was performed. At 48 h after the second transfection, cells were harvested for luciferase assays and immunoblot analysis. Light units were normalized to the co-transfected β-gal expression plasmid. For transient transfection of the luciferase reporter and Gal4 fusion receptor into 293T cells, the second transfection was performed with 0.8 μg of siRNA, 0.3 μg of UAS-sv40-Lucif, 0.4 μg of pCMX-Gal4-DBD or pCMX-Gal4-TR, and 0.1 μg of β-gal. siRNA treated 293T cells intended for ChIP were split into six-well plates 48 h after the first transfection, transfected a second time after an additional 24 h, and then harvested 48 h following the second transfection. For siRNA experiments in HepG2 cells, 3 μg of the siControl or siSNF2H construct were prepared with Nucleofector Solution V (Amaxa biosystems) and 2 × 106 cells were electroporated according to the manufacturer's protocol and transferred onto one well of a six-well plate. After 18 h, media was changed and 48 h later, cells were harvested for RNA and protein analysis.
ChIP assay
ChIP assays were performed according to the protocol of Upstate Biotechnology with minor modifications. Immunoprecipitation was performed with the following antibodies: anti-Pol II (Covance); anti-N-CoR (Ishizuka and Lazar, 2003); anti-Gal4, anti-HDAC3, anti-TRβ (Santa Cruz); anti-acetylated histone H4 (Upstate); and anti-SNF2H (Abcam)/(Hakimi et al, 2002). Samples were analyzed by real-time PCR using Sybr Green Mastermix (Applied Biosystems) and primers specific for: UASx5-sv40-luciferase reporter: (5′-TTCCGGTACTGTTGGTAAAATGG-3′ and 5′-ACCAGGGCGTATCTCTTCATAGC-3′); dio1 promoter: (5′-GAGGGAGAATTTAAGATGTGGAATCT-3′ and 5′-CCAATTCTTTGCCTGAGCTTTT-3′); and 36B4 control: (5′-ACGCTGCTGAACATGCTCAA-3′ and 5′-GATGCTGCCATTGTCGAACA-3′).
MNase protection assay
Nuclei were harvested by resuspending cells in a hypotonic lysis buffer (20 mM HEPES (pH 7.5), 0.25 M sucrose, 3 mM MgCl2, 0.2% NP-40, 3 mM 2-mercaptoethanol) with protease inhibitors (Roche) followed by 60 strokes of a Dounce B homogenizer. Nuclei were washed and resuspended in MNase digestion buffer (20 mM HEPES (pH 7.5), 100 mM NaCl, 1.5 mM CaCl2, 3 mM MgCl2). Aliquots (200 μl) of nuclei were digested with 4 U of MNase (Sigma) for 15 min at 37°C. The digestion was terminated with the addition of 200 μl stop buffer containing proteinase K and incubation for 2 h at 37°C. Samples were analyzed by real-time PCR using Sybr Green Mastermix (Applied Biosystems) and primers specific for the 5′ region of the sv40 promoter: (5′-CAACCATAGTCCCGCCCCTA-3′ and 5′-AAAATTAGTCAGCCATGGGG-3′) or the luciferase start site: (5′-TTCCGGTACTGTTGGTAAAATGG-3′ and 5′-ACCAGGGCGTATCTCTTCATAGC-3′), and 36B4 as a control.
RNA isolation and quantification
RNA was isolated from HepG2 cells using RNeasy Mini Kit (Qiagen) then subjected to reverse transcription (Clontech). mRNA transcripts for Gapdh and dio1 were quantified by real-time PCR analysis using TaqMan PCR Mastermix (Applied Biosystems) and TaqMan gene expression primer/probe assays that span an exon junction (Applied Biosystems). Primers and probe for h36B4: 5′-GAGGGCACCTGGAAAACAAC-3′ and 5′-CCTCCTTGGTGAACACAAAGC-3′, FAM-CTCTGGAGAAACTGCTGCCTCATATCCG-T AMRA.
Real-time PCR analysis
Samples were analyzed using a Prism 7900 thermal cycler and sequence detector (Perkin Elmer/ABI). Equivalent PCR efficiency was confirmed for each primer set from the slope of the standard curve of serial two-fold dilutions and dissociation curves always indicated the formation of a single PCR product. Data were analyzed with a threshold set in the linear range of amplification and then processed by the 2−ΔΔCt method (Livak and Schmittgen, 2001). Specifically, the cycle number at which the house keeping gene (36B4, GAPDH) was detectable (Ct) was subtracted from the Ct of the gene of interest (sv40, dio1), referred to as ΔCt. For each sample, duplicates were analyzed by real-time PCR and the Ct values were averaged prior to determining the ΔCt. The fold change of the gene of interest in the ‘treatment' sample (siN-CoR, siHDAC3, siSNF2H, sodium butyrate) relative to the ‘control' sample (siControl, vehicle) was calculated as 2−ΔΔCt, in which ΔΔCt=ΔCt (treatment)−ΔCt (control). This method results in the normalization of the control sample's fold change to 1. In order to present cumulative data from three separate experiments, the ΔCt values for control and treatment samples (n=3) were averaged to obtain the mean fold change relative to the control group by the 2−ΔΔCt method, which normalized the control mean fold change to 1. To obtain standard deviations, fold change for each individual experiment relative to the mean fold change was determined by the same 2−ΔΔCt method, subtracting the mean control ΔCt from each individual ΔCt to calculate ΔΔCt. Statistical analysis was performed using one-tailed, paired t-test of fold change for treatment relative to control.
Acknowledgments
We thank O Barak and R Shiekhattar for SNF2H reagents and helpful discussions, and D Steger for carefully reading the manuscript. This work was supported by NIH Grant DK43806 to MAL.
References
- Aalfs JD, Narlikar GJ, Kingston RE (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J Biol Chem 276: 34270–34278 [DOI] [PubMed] [Google Scholar]
- Aranda A, Pascual A (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81: 1269–1304 [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C (2005) The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19: 2540–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Larsen PR (1991) Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature 349: 438–440 [DOI] [PubMed] [Google Scholar]
- Cervoni N, Szyf M (2001) Demethylase activity is directed by histone acetylation. J Biol Chem 276: 40778–40787 [DOI] [PubMed] [Google Scholar]
- Chen J, Kinyamu HK, Archer TK (2006) Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol 20: 1–13 [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RN, Putney A, Wondisford FE, Hollenberg AN (2000) The nuclear corepressors recognize distinct nuclear receptor complexes. Mol Endocrinol 14: 900–914 [DOI] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun JW (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- Di Croce L, Koop R, Venditti P, Westphal HM, Nightingale KP, Corona DF, Becker PB, Beato M (1999) Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell 4: 45–54 [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P (2000) ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol Cell 6: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Dirscherl SS, Krebs JE (2004) Functional diversity of ISWI complexes. Biochem Cell Biol 82: 482–489 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferreira R, Becker P (2005) Dynamic chromatin: concerted nucleosome remodelling and acetylation. Biol Chem 386: 745–751 [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Fan HY, Trotter KW, Archer TK, Kingston RE (2005) Swapping function of two chromatin remodeling complexes. Mol Cell 17: 805–815 [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Tsukiyama T (2003) Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol Cell 12: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK (1998) Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393: 88–91 [DOI] [PubMed] [Google Scholar]
- Georgel PT, Tsukiyama T, Wu C (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J 16: 4717–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433 [DOI] [PubMed] [Google Scholar]
- Goodson M, Jonas BA, Privalsky MA (2005) Corepressors: custom tailoring and alterations while you wait. Nucl Receptor Signal 3, doi:10.1621/nrs.03003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW (2003) Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12: 449–460 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev 14: 1048–1057 [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R (2002) A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418: 994–998 [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97: 833–842 [DOI] [PubMed] [Google Scholar]
- Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA (2005) The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep 6: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377: 397–404 [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA (2000) Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab 11: 6–10 [DOI] [PubMed] [Google Scholar]
- Hu X, Li Y, Lazar MA (2001) Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol 21: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZQ, Li J, Sachs LM, Cole PA, Wong J (2003) A role for cofactor-cofactor and cofactor–histone interactions in targeting p300, SWI/SNF and mediator for transcription. EMBO J 22: 2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA (2003) The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol 23: 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA (2005) The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol Endocrinol 19: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Jakobs TC, Schmutzler C, Meissner J, Kohrle J (1997) The promoter of the human type I 5′-deiodinase gene--mapping of the transcription start site and identification of a DR+4 thyroid-hormone-responsive element. Eur J Biochem 247: 288–297 [DOI] [PubMed] [Google Scholar]
- Jeyakumar M, Tanen MR, Bagchi MK (1997) Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor. Mol Endocrinol 11: 755–767 [DOI] [PubMed] [Google Scholar]
- Johnson CN, Adkins NL, Georgel P (2005) Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol 83: 405–417 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Lovett JM, Hirsch M, Xia F, Chen JD (2004) NuRD complex component Mi-2beta binds to and represses RORgamma-mediated transcriptional activation. Biochem Biophys Res Commun 318: 714–718 [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B (2003) SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell 11: 391–403 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Li J, Lin Q, Wang W, Wade P, Wong J (2002) Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev 16: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J 19: 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qin J, Lazar MA (2005) Characterization of nuclear receptor corepressor complexes. NURSA Datasets 1, doi: 10.1621/datasets.01002. [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R (2001) Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol 3: 30–37 [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12: 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389: 194–198 [DOI] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T (2006) Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281: 16279–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI (2003) The ISWI ATPase SNF2H is required for early mouse development. Proc Natl Acad Sci USA 100: 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda N, Zavacki AM, Maia AL, Harney JW, Larsen PR (1995) A novel retinoid X receptor-independent thyroid hormone response element is present in the human type 1 deiodinase gene. Mol Cell Biol 15: 5100–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 275: 40463–40470 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Wolffe AP (2001) A necessary good: nuclear hormone receptors and their chromatin templates. Mol Endocrinol 15: 1–16 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P (2001) ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20: 3076–3085 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB (2006) Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 16: 151–156 [DOI] [PubMed] [Google Scholar]
- Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ (2000) The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol Endocrinol 14: 1976–1985 [DOI] [PubMed] [Google Scholar]
- Wong J, Li Q, Levi BZ, Shi YB, Wolffe AP (1997) Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J 16: 7130–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Shi YB, Wolffe AP (1995) A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev 9: 2696–2711 [DOI] [PubMed] [Google Scholar]
- Wu Y, Koenig RJ (2000) Gene regulation by thyroid hormone. Trends Endocrinol Metab 11: 207–211 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T (2005) Stem cell self-renewal controlled by chromatin remodeling factors. Science 310: 1487–1489 [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR (1992) Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258: 1598–1604 [DOI] [PubMed] [Google Scholar]
- Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA (2003) A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22: 3403–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavacki AM, Ying H, Christoffolete MA, Aerts G, So E, Harney JW, Cheng SY, Larsen PR, Bianco AC (2005) Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology 146: 1568–1575 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG (2002) The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA (2000) The mechanism of action of thyroid hormones. Annu Rev Physiol 62: 439–466 [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM (2005) Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J 24: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21: 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]


