Abstract
Human and mouse genomes share similar long-range sequence organization, and have most of their genes being homologous. As alternative splicing is a frequent and important aspect of gene regulation, it is of interest to assess the level of conservation of alternative splicing. We examined mouse transcript data sets (EST and mRNA) for the presence of transcripts that both make spliced-alignment with the draft mouse genome sequence and demonstrate conservation of human transcript-confirmed alternative and constitutive splice junctions. This revealed 15% of alternative and 67% of constitutive splice junctions as conserved; however, these numbers are patently dependent on the extent of transcript coverage. Transcript coverage of conserved splice patterns is found to correlate well between human and mouse. A model, which extrapolates from observed levels of conservation at increasing levels of transcript support, estimates overall conservation of 61% of alternative and 74% of constitutive splice junctions, albeit with broad confidence intervals. Observed numbers of conserved alternative splicing events agreed with those expected on the basis of the model. Thus, it is apparent that many, and probably most, alternative splicing events are conserved between human and mouse. This, combined with the preservation of alternative frame stop codons in conserved frame breaking events, indicates a high level of commonality in patterns of gene expression between these two species.
INTRODUCTION
The availability of human and mouse genomes has fuelled interest in understanding the fashion and extent to which these genomes have common genes and organization. These two genomes do share similar long-range sequence organization (1–3), with >90% of the mouse and human genomes having been partitioned into corresponding regions of conserved synteny (4). It has been shown that the number of exons is the same in as many as 95% of genes from a data set of 117 human–mouse orthologous gene pairs (5), and in as many as 86% of genes from a larger data set of 1506 human–mouse orthologous genes (4). Furthermore, most of the genes from these two species are homologous to one another (6,7), with genome-wide analysis indicating that the proportion of mouse genes without any homolog currently detectable in the human genome (and vice versa) is <1% (4). As alternative splicing is often an important component in the expression of eukaryotic genes, and acts to generate a large set of transcript and protein isoforms (8–19), it is essential to understand the extent to which alternative splicing is conserved.
We address this question by examining human splice junctions, from our AltExtron data set of human transcript-confirmed constitutively and alternatively spliced introns and exons (16), for conservation in mouse. First we examine mouse transcript sequences for the presence of transcripts that match a human sequence tag comprising the flanking exons of a human splice junction, and second we ascertain that the matching mouse sequence tag makes a gapped alignment with the draft mouse genome sequence (thus confirming conservation of both the transcript isoform and the intron position). A transcript coverage model, which extrapolates from the observed levels of conservation, indicates that 74% of constitutive human splice junctions and 61% of alternative human splice junctions are conserved in mouse (with 95% confidence intervals of 71–78% and 47–86%, respectively). These estimates indicate high levels of conservation for alternative splicing.
MATERIALS AND METHODS
Data sets
Within the AltExtron data set (16) there are 16 269 transcript-confirmed introns (from 2793 genes) of which 2050 are unannotated introns (from 1045 genes). Of these 2050 transcript-confirmed introns, 1492 (with at least 35 bases of each flanking exon defined) overlapped with 1440 transcript-confirmed annotated introns and were thus taken as confirmed alternative forms (confirmed annotated forms considered as constitutive forms). The splice junctions corresponding to these 2932 (= 1492 + 1440) introns (from 786 genes) were examined for occurrence in mouse transcript data (EST and full-length mRNA) obtained from the EMBL nucleotide sequence database (October 2001) (20).
Identification of conserved splice junctions
Identification of conserved human splice junctions is carried out in two main steps; first we search for mouse transcripts that are homologous to human transcript fragments spanning splice junctions in the human gene, and second we ensure that the mouse gene also contains an intron in this region.
For the first step, a sequence tag of length up to 140 nt was constructed for each confirmed human splice junction by concatenating the flanking exon regions (using up to 70 nt from each exon). Mouse transcript sequences were searched for matches to these human sequence tags using FASTA (21). Tag-transcript matches were considered as two component matches—one to each exon—and it was required that each component showed nucleotide identity of ≥77% over a length spanning ≥35 bases (thus having full matches of at least 70 bases). Since alignment gaps that occur close to the splice junction can be a consequence of false alignment with a transcript actually demonstrating a small extension or truncation of an exon, alignments with more than one gap occurring close to the splice junction were rejected (a region encompassing the nucleotides –7 to +7 relative to the splice junction was considered as ‘close’; this being the region that we have found to be potentially subject to erroneous alignment gaps due to the similarity that often exists in the sequence at the 3′ end of a donor exon and the 5′ end of an acceptor exon, and the way that this can act to confound alignment tools if the alignment is not perfect in this region), while cases with one gap of length one base were corrected to reflect the nucleotide usage in the human sequence (this usually increased amino acid similarity). For each of the tag-transcript matches, the amino acid identity and similarity were evaluated. Physico-chemical properties of amino acid residues were used to assign similar residues: aliphatic, I, L, V; aromatic, F, Y, W, H; positive, H, K, R; negative, D, E; and tiny, A, C, T, S, G. The matches that satisfied the following criteria were retained: (i) matches with nucleotide identity ≥85% were accepted outright; (ii) matches with nucleotide identity of 77–85% were further scrutinized for amino acid identity ≥70% or similarity ≥80% (with lower allowed values of 65 and 70%, respectively).
When distinct human sequence tags share a high level of nucleotide identity, it is possible that both may align with a common mouse transcript, and thus indicate the presence of false-positive alignments. Hence, we performed checks on the tag-transcript matches to identify false positives of the following types: (i) where sequence tags corresponding to splice junction (intron) isoforms match to the same transcript; (ii) where sequence tags corresponding to splice junctions from distinct genes match to the same transcript. Such false positives may occur if the sequence tags share nucleotide identity. There were five cases of the former and in each of these the match statistics (in terms of length and percent of nucleotide identity) clearly distinguished between the true and false alignments. There were 37 cases of the latter, and in all such cases the common transcripts were removed from consideration.
The above analysis led to identification of 1198 human splice junctions (from 565 genes), for which the corresponding sequence tag had at least one acceptable match with a mouse transcript sequence. In the second phase, these 1198 splice junctions were considered further to examine whether they really exist as splice junctions in mouse. For each of these human splice junctions the corresponding mouse sequence tag was retrieved from the matching mouse transcripts and was aligned with the draft mouse genome. In the case where the splice junction is conserved in mouse, we expect the mouse sequence tag to align as two distinct fragments (corresponding to the 5′ and 3′ exonic regions) separated by a gap (the intron). In the case where the intron position is not conserved (through either intron loss in mouse, gain in human or because the alignment is with a mouse pseudogene), we expect to observe a contiguous alignment of the mouse sequence tag to the mouse genome. It is noted that we do not identify the exact exon boundaries in mouse, and have thus not screened for the (presumably) rare cases where splice junctions have arisen independently at close, but not exactly the same, positions in each of mouse and human.
We used Sequence Search and Alignment by Hashing Algorithm (SSAHA) (22) in order to align the mouse transcript sequence tags with the draft mouse genome. SSAHA is a tool for very fast matching and alignment of nucleotide sequences to identify exact or ‘almost exact’ matches. This tool is available as a web server (http://www.ensembl.org/Mus_musculus/ssahaview), and is a part of the mouse EnsEMBL resources (23). We used draft mouse genome sequence release MGSC v3 (January 2003), which contains 2740 contigs with a sequence quality of 7-fold coverage and with an estimated coverage of 96% of the euchromatic DNA.
The SSAHA matches were scrutinized with the following criteria: (i) each of the 5′ and 3′ exon regions of the sequence tag match to the same chromosome region with a gap on the genome sequence between the matches; (ii) the orientation of the matches (forward–forward; reverse–forward) remains the same for both the 5′ and 3′ exon-region regions of a sequence tag and for all the tags from the same human gene; (iii) all the splice junctions from a human gene map to the same chromosome region in the same positional order as that in the human gene. It was generally the case that the length of the gap was consistent with the length of the human intron (see the web data) in accordance with the observations of others (4)—namely that there is strong correlation between the lengths of orthologous introns from human and mouse.
Of the 1198 human splice junctions checked, the SSAHA matches showed gapped alignments in 1134 cases. For a further 25 splice junctions, use of standard WU-BLAST (as implemented in the mouse EnsEMBL BLAST server at http://www.ensembl.org/Mus_musculus/blastview) showed gapped alignments satisfying the conditions listed above (though in 11 of these 25 cases, the average identity was <96%). The remaining 39 human splice junctions (35 constitutive and four alternative) for which the mouse sequence tags failed to show gapped alignments (with the draft mouse genome sequence) were carefully examined. In one case, the region on the mouse genome to which the splice junction showed gapped alignment was different from the one with which the other splice junctions of the gene mapped; in 25 cases, either the 5′ or the 3′ (but not both) exonic regions of the splice junction mapped to the mouse sequence; in six cases, the mouse sequence matched with the mouse genome sequence without a gap; and in the remaining seven cases, no significant matches could be identified. Possible reasons for these anomalies are given below.
(i) Cases where only the 5p or the 3p exonic region matches with the draft mouse sequence (25 entries—21 constitutive and four alternative splice junctions). Considering that both the 5′ and 3′ exonic regions are observed in a mouse transcript sequence, failure to observe the match for one of these two regions can not be a case of exon-loss, nor can it be a case of exon variation between human and mouse. The draft mouse genome has 96% coverage, and it is probable that the missing exons map to gaps, or low quality regions, in the draft.
(ii) Cases where the sequence tag matches as a long contiguous stretch on the mouse genome (six cases—all constitutive splice junctions). Absence of a gap between the 5′ exon match and the 3′ exon match probably indicates that either the human intron does not exist in mouse or the alignment is to a mouse pseudogene. The pseudogene possibility arises only if ungapped mouse alignment is observed with every splice junction from the human gene. In three of six instances of splice junctions showing ungapped mouse alignments other splice junctions from the gene were observed as conserved, and thus these three instances can be considered as definite ‘intron loss/gain’ between human and mouse. In the remaining three cases we did not observe any related splice junction, and it seems probable that these alignments are to mouse pseudogenes. It is to be noted that all six of these cases are constitutive splice junctions.
(iii) Cases where the sequence tag does not show a significant match with the mouse genome (seven entries—all constitutive splice junctions). Considering that these mouse exon sequence tags are observed in mouse transcript sequences, the absence of significant matches with the draft mouse genome sequence is probably due to sequence quality issues in the draft mouse genome sequence [as for (i) above].
Transcript coverage model
The observed conserved splice junctions represent only that fraction of conserved splice junctions that available mouse transcript data allows. In order to account for this we constructed a simple statistical model based on the idealization that human and mouse genes have identical expression profiles and that this is reflected in the respective transcript libraries. An important further idealization is also necessary, and that is to assume that both the probability of a gene having alternative isoforms, and the probability of such an isoform being conserved, is independent of the level of transcript coverage. The probability of observing a given human splice junction in mouse can thus be considered as the product of the chance that the splice junction is conserved in mouse (let this chance have a value ‘A’), and the chance that the splice junction is observed within available mouse transcript data given that it does exist in mouse genes. By assumption, we consider the expected number of transcripts demonstrating the corresponding splice junction in mouse to be a constant portion of the number seen in human. Let this portion have a value ‘θ’ and be interpreted as the ratio of the sizes of the transcript libraries in the ideal case of these libraries being very large random samples of transcript space. Thus, the probability of observing a mouse homolog to a human splice junction demonstrated by Nhum transcripts can be written as:
Prob(splice junction exists in mouse) · Prob(observe splice junction given it exists)
⇒A · [1 – Prob(don’t observe splice junction even though it exists)]
⇒A · [1 – Prob(observe 0 | expected value of θ*Nhum)]
The probability of observing no mouse transcripts when θ*Nhum are expected is considered simply as being described by a Poisson distribution with parameter θ*Nhum. Thus, the desired probability is:
⇒A · [1 – exp( – θ*Nhum)]
where A is the overall fraction of splice junctions conserved and Nhum is the number of transcripts demonstrating the splice junction in human.
RESULTS AND DISCUSSION
The alignment data
We have previously constructed a data set (AltExtron) of transcript-confirmed constitutively and alternatively spliced introns and exons from human (16). This was done through careful spliced alignment of gene and transcript sequences (EST and mRNA). In this current study we examine the extent and nature of conservation of these human alternative splice events in mouse genes.
A human splice junction has been considered as conserved in mouse if a sequence tag from human (constructed by concatenating flanking exon regions) has acceptable matches with one or more mouse transcript sequences (EST and mRNA), as determined by careful analysis of the match data, and that the matching mouse sequence tag makes a gapped alignment with the draft mouse genome sequence (see Materials and Methods). We find that, of 2932 human splice junctions considered, the sequence tags corresponding to a total of 1198 splice junctions show acceptable matches with a total of 15 323 mouse transcripts.
It is important to note that false positives can arise if a transcript is wrongly associated with a paralogous gene, or aligned incorrectly with the homologous gene. The way in which this issue of false positives has been examined is 4-fold, and these are discussed at different points through the remainder of this paper. In summary, (i) first the match statistics have been carefully processed—this is all that can be done initially (an alternative approach is to utilize synteny data, although this does not necessarily help). The remaining three pieces of evidence demonstrate that the level of false positives is very low, and these points are: (ii) the mouse transcripts confirming the constitutive and alternative splice junctions from a human gene mapped to the same region on the mouse genome sequence; (iii) the high quality of matches (in the common regions) between mouse transcripts that confirm the constitutive and alternative splice junctions of an alternative splice event; and finally, (iv) the consistency that is found between the observed and expected numbers of observed conserved alternative splicing events (this consistency ties the quality of the subset back to that of the entire set of matches).
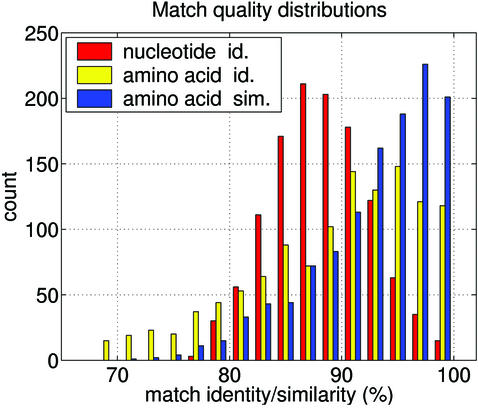
The distribution of the above 1198 matched human sequence tags against the average percentage values of nucleotide identity, amino acid identity, and amino acid similarity are shown in Figure 1. While the nucleotide identity distribution peaks at 86–88%, the distributions for amino acid identity and similarity not only peak at higher values but also have a greater range. For 83% of the conserved splice junctions there existed at least one mouse transcript match showing a minimum of 85% nucleotide identity. In 80% of instances there existed at least one mouse transcript match showing a minimum of 85% amino acid identity. In 92% of instances there existed at least a single mouse transcript showing a minimum of 85% amino acid similarity. Note that the multiple transcript sequences matching a sequence tag can differ in start and end positions and thus may have different statistics; however, each of the matching transcript sequences pass the minimal match criteria as set out in the Materials and Methods.
Figure 1.
Distribution of matched human sequence tags [as constructed by concatenating exon regions that flank an intron (see Materials and Methods)] as per the average nucleotide percent identity (red), amino acid identity (yellow), and amino acid similarity (blue). For each of the tags the given match statistics represent the average over all transcript matches.
The statistics quoted above are very similar to what has been reported from curated data sets of orthologous human and mouse genes (5,24); these reports observe a mean nucleotide identity (in coding regions) of 85% with a range of 61–98% (with the non-coding regions showing lesser values), and a mean amino acid identity of 86% with a range of 41–100%. The statistics observed in this work (Fig. 1) compare favorably with these reported figures, and this is consistent with the identified matches containing a low level of false positives.
While the above analysis indicates that the matching mouse sequence tag is expressed in mouse, it is necessary to ascertain that the splice junction actually exists in mouse genes. The matching mouse sequence tags were examined to see whether they make gapped alignments with the draft mouse genome sequence. This analysis (see Materials and Methods) revealed that of the 1198 human splice junctions under consideration there were 1159 cases where the splice junctions were unambiguously seen to be conserved, six cases where the splice junctions were seen not to be conserved, and 33 ambiguous cases, most of which are expected to be conserved cases (see Materials and Methods). The intron-loss cases constitute only a tiny fraction of the data set, and we have retained them in our further analysis.
Conservation of splice junctions
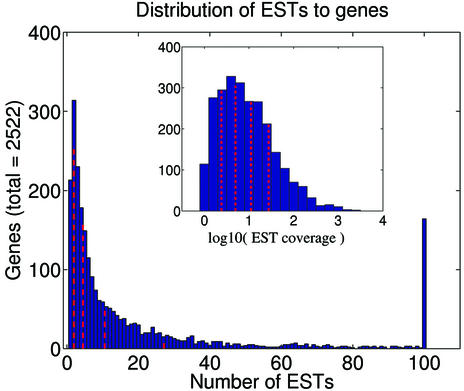
The results presented so far indicate that, of 1440 constitutive and 1492 alternative human splice junctions considered (from AltExtron), a total of 968 constitutive and 230 alternative splice junctions show conservation in mouse. This represents observed conservation of 67% of the constitutive splice junctions and 15% of the alternative splice junctions. The observed level of conservation of human splice junctions in mouse is patently effected by the level of transcript coverage (Table 1), with constitutive splice junctions having better transcript support in both human and mouse than do alternative splice junctions. Although the (EST) transcript libraries contain a very large number of individual sequences, they fail to provide good coverage of the gene sets. The distribution of ESTs to genes is highly skewed (Fig. 2), with a small number of genes being represented by large numbers of transcripts, while a large number of genes are represented by only one or a few transcripts (or none at all).
Table 1. Levels of human transcript coverage versus extent of conservation in mouse.
| Human transcript coveragea,b | Fraction of considered human splice junctions that are conserved in mousec | ||
|---|---|---|---|
| All splice junctions (constitutive and alternative) | Constitutive splice junctions | Alternative splice junctions | |
| 1 | 0.173 | 0.492 | 0.082 |
| 2 | 0.366 | 0.594 | 0.165 |
| 3 | 0.526 | 0.664 | 0.337 |
| 4 | 0.574 | 0.710 | 0.327 |
| 5–9 | 0.629 | 0.723 | 0.326 |
| ≥10 | 0.749 | 0.779 | 0.530 |
aOnly 19% of constitutive splice junctions as opposed to 64% of alternative splice junctions have transcript coverage of 1. As high as 51% of constitutive splice junctions as opposed to 10% of alternative splice junctions have a high transcript coverage of ≥5.
bThe extents of conservation of splice junctions, irrespective of the human transcript coverage, is 41% for all 2932 (constitutive and alternative) splice junctions, 67% for the 1440 constitutive splice junctions, and 15% for the 1492 alternative splice junctions.
cThe extent of conservation increases with the increasing human transcript coverage, irrespective of the type of splice junction (constitutive or alternative).
Figure 2.
Distribution of ESTs to genes in the AltExtron data set of transcript confirmed human introns and exons. The insert shows the distribution with the coverage transformed by log to the base 10. The spike at 100 ESTs represents the sum of all genes with 100 or more aligned ESTs.
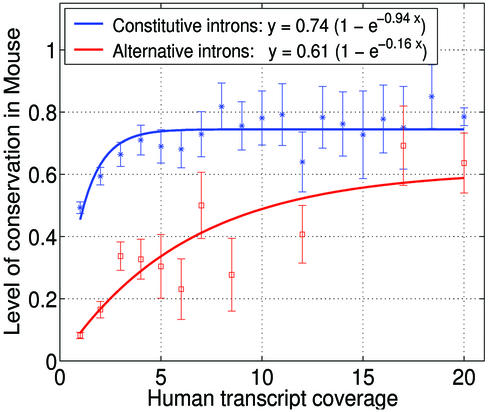
In order to address the question of the extent to which alternative splicing events are conserved between human and mouse, we plotted the level of conservation as a function of transcript coverage, for each of the constitutive and alternative groups (Fig. 3). The observed data points consist of the considered human splice junctions grouped by transcript coverage, with an associated overall level of conservation observed for each group. There is substantial variation in the uncertainty associated with these observations, due to the differing number of splice junctions associated with each of these data points. This data is fitted against a model that describes expected levels of observed splice junction conservation as a function of transcript coverage (see Materials and Methods). This model requires that the level of transcript coverage for homologous human and mouse genes is correlated, and, although not perfect, this was found to be the case (see Table 2).
Figure 3.
The observed level of splice junction conservation as a function of transcript coverage for the constitutive and alternative splice junctions. The final data point of each curve groups all splice junctions with 20 or more human transcripts. Further grouping of data was performed for some of the alternative curve data points, and one of the constitutive curve data points, to allow calculation of sample variance. The grouping was done manually and ensures that each point represents at least 10 human splice junctions. The number of observations at each plotted point is given below as (transcript coverage in human, number of human splice junctions with this coverage value). For the alternative curve: (1, 958), (2, 230), (3, 98), (4, 55), (5, 23), (6, 26), (7, 22), (8–9, 18), (10–14, 12), (15–19, 17) and (20+, 14). For the constitutive curve: (1, 274), (2, 202), (3, 134), (4, 100), (5, 84), (6, 69), (7, 48), (8, 44), (9, 41), (10, 32), (11, 24), (12, 25), (13, 23), (14, 21), (15, 11), (16, 18), (17, 12), (18–19, 20), (20+, 251).
Table 2. Correlation between the human and mouse transcript coverage for the conserved splice junctions.
| Human transcript coveragea | Fraction of human conserved splice junctions with a mouse transcript coverageb,c | |
|---|---|---|
| ≥5d | <2e | |
| All (constitutive and alternative) splice junctions | ||
| 1 | 0.33 | 0.46 |
| 2 | 0.47 | 0.34 |
| 3 | 0.51 | 0.27 |
| 4 | 0.64 | 0.21 |
| ≥5 | 0.84 | 0.10 |
| Constitutive splice junctions | ||
| 1 | 0.42 | 0.38 |
| 2 | 0.53 | 0.28 |
| 3 | 0.58 | 0.21 |
| 4 | 0.66 | 0.19 |
| ≥5 | 0.85 | 0.09 |
| Alternative splice junctions | ||
| 1 | 0.17 | 0.61 |
| 2 | 0.26 | 0.50 |
| 3 | 0.30 | 0.42 |
| 4 | 0.56 | 0.33 |
| ≥5 | 0.73 | 0.16 |
aHuman transcript coverage being the number of human transcript sequences that confirm a splice junction in human.
bMouse transcript coverage being the number of supporting mouse transcript sequences that confirm a splice junction in mouse.
cThe data indicates a strong positive correlation between the transcript coverage levels in human and mouse; as the human transcript coverage increases from a value of 1 to ≥5, the fraction of conserved splice junctions with mouse coverage value of ≥5 transcripts increases steadily while the fraction at ≤2 decreases steadily. The data indicates correlation coefficients of 0.64 for the constitutive splice junctions and 0.60 for the alternative splice junctions (with the distributions made normal).
dA value of ≥5 transcripts has been chosen to indicate high coverage.
eA value of ≤2 transcripts has been chosen to indicate low coverage.
The model is fitted to the data using maximum likelihood (thus taking account of the different uncertainties associated with the data points). This involves calculating the probability of each observation (for a given model curve) for each data point, and using the (negative of the) sum of the logs of the probabilities as the metric for curve fitting. The fitted curves (Fig. 3) provide estimates that 74% of constitutive human splice junctions, and 61% of alternative human splice junctions, are conserved in mouse—with 95% confidence intervals estimated at 71–78% and 47–86%, respectively. (The confidence intervals were calculated using a non-parametric bootstrap resampling approach with 1000 curves, meaning that 1000 resampled data sets were generated from the actual data (with replacement) and that each of these was fitted as above. The resultant distribution of parameter values was examined to identify the 95% confidence interval.) We point out that the lower bound (47%) of the 95% confidence interval for the estimated extent of conservation of alternative introns is much higher than the value of 15% actually observed.
Studies of gene conservation between human and mouse—such as that of Mural et al. (6), that of Dehal et al. (7), and that of Mouse Genome Sequencing Consortium (4) suggest that only a few percent of human genes do not have mouse homologs. Analysis of data sets of human–mouse orthologous gene pairs has shown that the number of exons can be the same in 86–95% of instances (4,5). These observations suggest that the level of constitutive splice junction conservation may be >85%—this being substantially greater than the figure of 74% derived here. Two reasons why such a discrepancy might arise are: (i) While the transcript coverage model considers the transcript libraries for each organism as a single pool of transcripts, in actual fact they are made up of many transcript collections that are specific to given tissue types, developmental stages and physiological conditions. Alternative splicing events that are specific to some of these states may not be observed as conserved unless the relevant transcript libraries exist for both human and mouse. (ii) The transcript coverage model contains an implicit assumption that both the probability of a gene having alternative forms, and the probability of these forms being conserved, is independent of the level of transcript support, and hence also of gene expression level. It is sometimes suggested that genes in low G+C regions are largely tissue specific, and that such genes have a higher level of alternative splicing (25). If this is indeed the case, then it may be expected that the asymptote at 61% identified for alternative splice junctions (see Fig. 3) would be less than the true level of conservation.
Also, it must be made clear that the observed transcript coverage is only tenuously related to actual (average) transcript abundance in cells. For example, some EST libraries are normalized, while others are not, and the inclusion of mRNA sequences further weakens the link. It should be understood that the use of transcript coverage as a parameter in the analysis here is a useful average property that allowed us to proceed with the problem of extracting the asymptotes in Figure 3 in a reasoned and insightful way. Given the broad confidence interval for the alternative splice junctions, the complexity of the data and other confounding issues (as discussed below), the extrapolated levels of conservation are not to be taken as precise measurements but as indicating a high level of conserved alternative splicing between human and mouse.
Finally, some comments on the fitted values of θ are in order (see Materials and Methods). The fitted values of θ are 0.96 and 0.16 for the constitutive and alternative splice junction data, respectively. The human and mouse transcript data sets used in this work were of similar size, and this is reflected in the fitted value of θ being close to 1 for the constitutive splice junctions [human transcript data set, extracted from GenBank release 117 (April 2000) (26), contained 1 990 202 EST and mRNA sequences; and mouse transcript data set, extracted from EMBL release 68 (October 2001) (20), contained 2 098 943 EST and mRNA sequences]. In the case of the alternative splice junctions, the lower fitted value of θ is to be interpreted as either indicating that, on average, conserved splice junctions have six times greater coverage in human than in mouse, or that one or more of the assumptions underlying the model is being substantially violated. Examination of the raw transcript coverage data clearly indicated that, on average (but with substantial variation), the observed conserved alternative splice junctions are supported by similar numbers of transcripts in human and mouse.
Recall that θ is interpreted as the ratio of the sizes of the transcript libraries in the ideal case of these libraries being very large random samples of transcript space. However, alternative splicing is largely specific to physiological conditions (e.g. tissue type, developmental stage or disease states) (27–30), and many of these states are not well sampled by the transcript libraries. For low abundance transcripts in particular, this sparse sampling can act to reduce the chance of observing conservation (because the state of an observed human form may not even have been sampled in the mouse transcript data). This is exactly the effect that we observe (note that the effect of lowering θ is to reduce the steepness of the curve at low transcript coverage) (see Fig. 3), and hence we consider this to be the most parsimonious explanation for the observed low value of θ in the case of alternative splice junctions.
Conserved alternative splicing events
The identified conserved splice junctions were further examined to determine which of our previously reported human alternative events (from the AltExtron data set) can be seen as conserved in mouse. For a human alternative event to be observed as conserved it is required that the constituent splice junctions of both constitutive and alternative splice patterns be observed in mouse data, with a further requirement that constituent splice junctions of a splice pattern are observed in a common transcript sequence. We found: (i) 39 intron isoform events, where an overlapping pair of introns demonstrates truncation or extension of one (or both) of the flanking exons; (ii) one intron retention event, where an intron is retained; (iii) 47 cassette exon events, where an exon is included in some transcripts and excluded in others; (iv) five alternating exon events (and one of the two isoforms of a sixth alternating exon event), where each of the two isoforms contains one of two mutually exclusive exons. These above 92 conserved events occur in 76 genes.
It is possible that the presence of duplicated genes (in either or both of mouse and human) could lead to detection of false-positive conserved events. For example, if a mouse gene orthologous to the human gene under consideration has undergone duplication, then a false positive could arise with the constitutive splice junctions(s) being demonstrated through matches to one mouse gene and with the alternative splice junction(s) being demonstrated by matches to the duplicate gene. Such a situation is feasible given that the matches between human sequence tags and mouse transcripts display a certain degree of divergence (18.3% of conserved splice junctions are supported solely by matches with nucleotide identity of <85%). It is to be noted that we have already shown that all the splice junctions (constitutive and alternative) from each human gene (with one exception) mapped to the same region in the draft mouse genome sequence (see Materials and Methods). We carried out an additional test, as below, to confirm that these observed conserved events do not contain false positives due to gene duplication events.
The check relies on the fact that, while the sequence identity between human and mouse transcript sequences averages ∼85%, the sequence identity between mouse transcripts from the same gene is expected to be high (≥98%). If the mouse transcripts representing the constitutive and alternative forms correspond to the same gene, it will usually be the case that these mouse transcripts overlap substantially [over the exon region(s) not affected by alternative splicing]. We found that, except in one case, there existed one or more such pairs of transcript sequences with one or more such matching overlap regions (with mean and median lengths of 610 and 459 bases). These results demonstrate that the mouse transcript sequences that confirm the constitutive splice junctions and those that confirm the corresponding alternative splice junctions are derived from the same gene.
The transcript coverage model can also be applied (using the fitted parameter values) to derive expected levels of observation for the conserved alternative events. We consider that the probability of observing an event is the product of the probabilities of observing the two isoforms, with the probability of observing an isoform derived from that of the constituent splice junctions. Summing the probabilities of observation over each group of considered human events gives the expected number of observed events. In the case of intron isoform events, each isoform involves a single splice junction and hence the probability of observing each isoform is simply that of observing the corresponding splice junction. In the case of cassette and alternating exon events, at least one of the isoforms involves multiple splice junctions (introns), and we consider the probability of observing such isoforms as that of the ‘least likely splice junction’. This analysis, carried out on the different categories of human events, gave the following expected numbers: for intron isoform events expect 39.9 (observed 39), for simple cassette exon events expect 42.0 (observed 47), and for alternating exon events expect 14.6 (observed 11). These numbers are in excellent agreement, further demonstrating both a lack of false-positive alignments as well as the utility of the model in describing the overall transcript coverage dynamics.
Conserved alternative stop codons and alternative frame of translation
Use of alternative stop codons (31,32) and alternative frames of translation (33,34) are two important factors that modulate the expression of genes into protein isoforms. Modrek et al. (14) have reported that, in a data set of transcript-confirmed alternative splice events, frame-shift events can extend the protein C-terminus sequence in 6% of instances, and that use of an alternative stop codon to replace the protein C-terminus sequence occurs in 20% of instances. Further, Stamm et al. (35) found 22% of alternatively spliced exons compiled from the literature contained a stop codon or introduced a frame-shift resulting in a premature stop codon. While some premature stop codons are used as markers for the nonsense-mediated decay of the mRNA (36), it is quite clear from the reports cited above that some also modify protein function.
We examined the observed conserved alternative events occurring within annotated CDS and found that 70 events preserved the frame of translation, while 19 events changed the frame of translation for the downstream exons. Each of these 19 frame-shift events leads to use of an alternative stop codon, as do a further three of the 70 frame-preserving events (see Table 3). The lengths of regions that can be potentially translated in an alternative frame are: five cases of zero length, four cases of <10 codons, three cases of 10–14 codons, two cases of 19–21 codons, two cases of 34–35 codons, and three cases of >50 codons (including one case at 100 codons); it may be that some of these cases (particularly the longer ones) represent genuine use of an alternative frame of translation. It was further seen that the corresponding mouse transcript sequences preserve these features, though there may be variations (as seen in six cases; Table 3) in the positions of the alternative stop codons and the extent of regions that are (possibly) translated in an alternative frame. These observations suggest that the use of alternative frames of translation is important in the evolution of proteins. Consistent with this suggestion is our previously reported observation that 36% of human exons are translatable in multiple frames (16).
Table 3. Characterization of conserved events that introduce alternative stop codonsa.
| Event type [extent of change due to the event (in nt)] | Position of alternative stop codon from the position of changed codon in the transcript (in codons)b | Extent of region possibly translated in alternative frame (in codons)b | |
|---|---|---|---|
| I. Frame-breaking events | |||
| I.1. Intron isoform events | |||
| 1. | Exon truncation (590) | 100 (54) | 100 (54) |
| 2. | Exon truncation (14) | 71 (mouse EST is short and cannot detect the stop codon) | 71 (>77) |
| 3. | Exon truncation (5) | 51 (58) | 51 (58) |
| 4. | Exon extension (11) | 39 (39) | 35 (35) |
| 5. | Exon extension (65) | 184 (184) | 34 (34) |
| 6. | Exon truncation (28) | 21 (21) | 21 (21) |
| 7. | Exon truncation (37) | 19 (53) | 19 (53) |
| 8. | Exon truncation (79) | 2 (2) | 2 (2) |
| 9. | Exon extension (59) | 12 (6) | 0 (0) |
| 10. | Exon extension (182) | 1 (1) | 0 (0) |
| I.2. Cassette exon events | |||
| 1. | Skipped exon (74) | 14 (14) | 14 (14) |
| 2. | Skipped exon (64) | 12 (12) | 12 (12) |
| 3. | Cryptic exon (73) | 35 (35) | 10 (10) |
| 4. | Cryptic exon (92) | 36 (36) | 6 (6) |
| 5. | Skipped exon (130) | 5 (5; additional at –12) | 5 (0) |
| 6. | Skipped exon (146) | 4 (4) | 4 (4) |
| 7. | Cryptic exonc (115) | 5 (5) | (0) (0) |
| 8. | Cryptic exonc (110) | 32 (32) | (0) (0) |
| 9. | Cryptic exonc (122) | 20 (38) | (0) (0) |
| II. Frame-preserving events | |||
| II.1. Intron isoform events | |||
| 1. | Exon extension (258) | 3 (3) | Not applicable |
| 2. | Exon extension (195) | 4 (4) | Not applicable |
| II.2. Intron retention event | |||
| 1. | Intron retention (78) | 9 (11) | Not applicable |
aOf the 93 observed conserved events, 89 occurred in CDS regions. In 70 of these 89 events, the inserted/deleted nucleotides were in multiples of three (and hence they are frame-preserving events).
bValue given in parenthesis are as seen in mouse sequences; this is shown in bold font when it differs from human.
cIn these three events, the cryptic exon is itself not translatable and hence there is no alternative frame of translation in these cases.
CONCLUSION
The work presented here demonstrates that the extent of conservation of alternative splicing between human and mouse is high, with upwards of one half of human alternative splice junctions being conserved in mouse. It may be that patterns of alternative splicing are conserved at similar levels to genes and gene structures. Further work will act to improve the accuracy of this estimate, although it should be understood that this will primarily depend on the availability of transcript data sets of much greater size and depth. Even at the current level of accuracy this result has an important consequence, namely it indicates that comparative analysis of human and mouse gene sequences will be useful in the identification of alternative splicing signals.
We did not carry out the reciprocal comparison between mouse and human, as we did not have an independent set of confirmed mouse splice junctions to work with. More broadly what is required is a wider analysis that assesses the extent of conservation of alternative splicing between mammals in general. Pending the availability of such measurements it remains that a fraction (between 14 and 53% according to the data and analysis described here) of human alternative splice junctions are not conserved in mouse. It is an intriguing possibility that the actual differences in patterns of alternative splicing, when individually determined, will provide much insight into the processes of genomic evolution (and speciation among mammals).
We have also observed that levels of transcript coverage are reasonably correlated between human and mouse, and this is indicative of overall gene expression patterns being similarly correlated, although further work is required to deal with the methodological issues involved in quantifying such a link. Also, the use of alternative stop codons and alternative frames of translation is preserved. These results suggest a high degree of commonality in gene expression patterns among closely related species.
AVAILABILITY OF THE DATA SETS
The paper presents a high quality data set of transcript-confirmed human constitutive and alternative splice junctions that are conserved in mouse, and presents a high quality data set of conserved alternative events and conserved modulations (such as use of an alternative frame of translation and an alternative stop codon) in the protein sequences. These data sets are of particular use for studying, through comparative sequence analysis, the signals that modulate splicing, and also as training data for software that aims to predict alternative splice events.
The generated data on the conserved splice junctions and alternative events is presented with our AltExtron web data (http://www.ebi.ac.uk/asd/altextron/data/index.html and at http://www.bit.uq.edu.au/altExtron/ as flat files, and at http://www.ebi.ac.uk/asd/altextron/access.html as query web pages with links to other data resources).
The work reported here is part of the activities of the ASD consortium (see http://www.ebi.ac.uk/asd/asd-ec/index.html). The consortium is committed to providing the community with genome-wide data sets of alternative splice events for human and other model species.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rolf Apweiler, Peter Stoehr and Kevin Burrage for ongoing support and encouragement, and Angus Ng and Geoff McLachlan for discussion on the transcript coverage model. This work is supported by a European Commission grant (QLK3-CT-2002-02062) to T.A.T.
REFERENCES
- 1.Nadeau J.H. and Taylor,B.A. (1984) Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl Acad. Sci. USA, 81, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Gregory S.G., Sekhon,M., Schein,J., Zhao,S., Osoegawa,K., Scott,C.E., Evans,R.S., Burridge,P.W., Cox,T.V., Fox,C.A. et al. (2002) A physical map of the mouse genome. Nature, 418, 743–750. [DOI] [PubMed] [Google Scholar]
- 4.Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 5.Batzoglou S., Pachter,L., Mesirov,J.P., Berger,B. and Lander,E.S. (2000) Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res., 10, 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mural R.J., Adams,M.D., Myers,E.W., Smith,H.O., Miklos,G.L.G., Wides,R., Halpern,A., Li,P.W., Sutton,G.G., Nadeau,J. et al. (2002) A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science, 296, 1661–1671. [DOI] [PubMed] [Google Scholar]
- 7.Dehal P., Predki,P., Olsen,A.S., Kobayashi,A., Folta,P., Lucas,S., Land,M., Terry,A., Zhou,C.L.E., Rash,S. et al. (2001) Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science, 293, 104–111. [DOI] [PubMed] [Google Scholar]
- 8.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 9.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbrück,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 11.Black D.L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell, 103, 367–370. [DOI] [PubMed] [Google Scholar]
- 12.Croft L., Schandorff,S., Clark,F., Burrage,K., Archtander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the prevalence of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 13.Smith C.W.J. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 14.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark F. and Thanaraj,T.A. (2002) Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet., 11, 451–464. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T. and Tasic,B. (2002) Alternating pre-mRNA splicing and proteome expansion in metazoans. Nature, 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 18.Caceres J.F. and Kornblihtt,A.R. (2002) Alternative splicing regulation: multiple control mechanisms and involvement in human diseases. Trends Genet., 18, 186–193. [DOI] [PubMed] [Google Scholar]
- 19.Roberts G.C. and Smith,C.W.J. (2002) Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol., 6, 375–383. [DOI] [PubMed] [Google Scholar]
- 20.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V. et al. (2002) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning Z., Cox,A.J. and Mullikin,J.C. (2001) SSAHA: a fast search method for large DNA databases. Genome Res., 11, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard T., Barker,D., Birney,E. and Cameron,G. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makalowski W. and Boguski,M.S. (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and mammalian sequences. Proc. Natl Acad. Sci. USA, 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernardi G. (2001) Misunderstandings about isochores. Part 1. Gene, 276, 3–13. [DOI] [PubMed] [Google Scholar]
- 26.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Q., Modrek,B. and Lee,C. (2002) Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res., 30, 3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamm S., Zhu,J., Nakai,K., Stoilov,P., Stoss,O. and Zhang,M.Q. (2000) An alternative-exon database and its statistical analysis. DNA Cell Biol., 19, 739–756. [DOI] [PubMed] [Google Scholar]
- 29.Cartegni L., Chew,S.L. and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 30.Stoilov P., Meshorer,E., Gencheva,M., Glick,D., Soreq,H. and Stamm,S. (2002) Defects in pre-mRNA processing as causes and predisposition to diseases. DNA Cell Biol., 21, 803–818. [DOI] [PubMed] [Google Scholar]
- 31.Lu X. and Rubin,C.S. (1990) Cloning, characterization and expression of the gene for the catalytic subunit of camp-dependent protein kinase in Caenorhabditis elegans. Identification of highly conserved and unique isoforms generated by alternative splicing. J. Biol. Chem., 265, 6896–6907. [PubMed] [Google Scholar]
- 32.Morrison M., Harris,K.S. and Roth,M.B. (1997) smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 94, 9782–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quelle D.E., Zindy,F., Ashmun,R.A. and Sherr,C.J. (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell, 83, 993–1000. [DOI] [PubMed] [Google Scholar]
- 34.Pianese L., Tammaro,A., Turano,M., Biase,I.D., Monticelli,A. and Cocozza,,S. (2002) Identification of a novel transcript of X25, the human gene involved in Friedreich ataxia. Neurosci. Lett., 320, 137–140. [DOI] [PubMed] [Google Scholar]
- 35.Stamm S., Zhu,J., Nakai,K., Stoilov,P., Stoss,O. and Zhang,M.Q. (2000) An alternative exon database and its statistical analysis. DNA Cell Biol., 19, 739–756. [DOI] [PubMed] [Google Scholar]
- 36.Maquat L.E. (2002) Nonsense-mediated mRNA decay. Curr. Biol., 12, 196–197. [DOI] [PubMed] [Google Scholar]