Abstract
The addition of the propynyl group at the 5 position of pyrimidine nucleotides is highly stabilising. We have determined the thermodynamic stability of the DNA·RNA hybrid r(GAAGAGAAGC)·d(GCpUpUpCpUp CpUpUpC) where p is the propynyl group at the 5 position and compared it with that of the unmodified duplex and the effects of methyl substitutions. The incorporation of the propyne group at the 5 position gives rise to a very large stabilisation of the hybrid duplex compared with the analogous 5-Me modification. The duplexes have been characterised by gel electrophoresis and NMR spectroscopy, which indicate that methyl substitutions have a smaller influence on local and global conformation than the propynyl groups. The increased NMR spectral dispersion of the propyne-modified duplex allowed a larger number of experimental restraints to be measured. Restrained molecular dynamics in a fully solvated system showed that the propyne modification leads to substantial conformational rearrangements stabilising a more A-like structure. The propynyl groups occupy a large part of the major groove and make favourable van der Waals interactions with their nearest neighbours and the atoms of the rings. This enhanced overlap may account at least in part for the increased thermodynamic stability. Furthermore, the simulations show a spine of hydration in the major groove as well as in the minor groove involving the RNA hydroxyl groups.
INTRODUCTION
There is considerable interest in the potential of oligonucleotides as antisense agents both for disease control and for regulating gene expression as a research tool (1–3). However, to be effective therapeutically, they must be easy to synthesise, relatively inexpensive, stable, readily taken up by cells and form a stable hybrid with the target RNA. The stability of DNA·RNA hybrids has been recently addressed, and it has been shown that it is highly sensitive to the composition of the target strand. In general, mixed-sequence DNA·RNA hybrids are of lower stability than the DNA duplex counterpart (4,5). However, purine-rich RNA targets, such as found in HIV RNA primer sites that are RNAse H resistant (6) form much more stable hybrids than do pyrimidine rich ones (5,7,8), which has been related to the significantly different conformations adopted by these different kinds of duplex helices (8,9). It is desirable to understand the origin of this difference, and to find modifications that increase the overall stability of hybrid duplexes.
Numerous nucleotide modifications have been examined for increased thermodynamic and/or biochemical stability, including phosphorothioates (10), 2′-O-alkyl (11), 2′-F-arabinoside substitutions (12), cyclohexenyl sugar replacement (13) and base alkyl substitutions such as a C5-methyl on pyrimidines (5,7). The latter have quite modest effects on stability of DNA·RNA hybrid duplexes. However, the larger, unsaturated propyne group is much more stabilising (14), and it has been shown to be a useful modification for antisense oligonucleotides, as DNA molecules as short as 7–8 nt can be effective antisense agents, and also by enhancing mismatch penalties (15–18). It has also been shown that this modification may stabilise DNA triplexes (19,20).
We are interested in DNA·RNA hybrids because of their biological importance as intermediates in transcription and DNA replication, their potential in antisense therapy, and because we wish to understand the relationship between sequence, composition, stability and structure. We have recently described the solution properties of several DNA·RNA hybrid duplexes (8,9,21), and we have demonstrated a clear difference in average conformation between duplexes of the kind rR·dY and dR·rY (where R and Y are purine- and pyrimidine-rich strands, respectively). We have also shown that the C2′-OH groups in the RNA strand of these hybrid molecules give well resolved resonances in the NMR spectrum, due to slow exchange with solvent protons, and in addition show nuclear Overhauser enhancements (NOEs) to protons of the solute (22). In this article, we report a high resolution solution structure of the propyne-modified duplex d(GCpUpUpCpUpCpUpUPC)·r(GAAGAGAAGC), which is an analogue of one of the duplexes previously studied (8,9). We have used a combination of NMR and molecular modelling under fully solvated conditions. The molecular modelling also allows calculation of properties such as charge distribution and hydration that cannot be readily obtained by experiment. For comparison, we report spectroscopic and thermodynamic data on the C5-Me analogues, which are shown to be much less stabilising than 5-propyne. The results shed light on the origin of the stability of different DNA·RNA hybrid duplexes.
MATERIALS AND METHODS
The decamers dR10 [d(GAAGAGAAGC)], rR10 [r(GAAG AGAAGC)], rY10 [r(GCUUCUCUUC)] and dY10 [d(GCU UCUCUUC)] were synthesised and purified as previously described (8). Of the modified oligomers, rY(T)10 [r(GCTT CTCTTC)] contains C5-Me at every uracil base, dY(U)10 [d(GCUUCUCUUC)] is demethylated at every thymidine base and dYP10 [d(GCpUpUpCpUpCpUpUpC)] contains a pyrimidine base modified at the 5 position with the propyne group (–C≡C–CH3). rY(T)10 and dY(U)10 and dYP10 were synthesised using commercially available phosphoramidites (Applied Biosystems) and purified by reverse phase HPLC.
Absorption coefficients were determined by digestion with nuclease P1 (8). The appropriate duplexes were prepared by mixing equal quantities of oligonucleotides, annealing from 70°C. The annealed duplexes were then dialysed against 0.1 M KCl, 10 mM Na-phosphate, 0.1 mM EDTA pH 7, using 2 kDa molecular weight cut-off membranes (Spectrapor).
UV melting curves were obtained as previously described (8,23). The thermodynamic parameters were determined from the concentration-dependence of the melting temperature, Tm, according to:
1 / Tm = (ΔH / R)ln(Ct)1
where ΔH is the van’t Hoff enthalpy, R is the gas constant and Ct is the concentration of strands. The standard state is taken to be 1 µM strands at pH 7.
1H NMR spectra were recorded at 11.75 and 14.1 T on Varian UnityPlus and Unity spectrometers, respectively. Two-dimensional NMR experiments were recorded in the phase-sensitive mode using the methods of States et al. (24). For experiments in 1H2O, the intense solvent signal was suppressed using the gradient echo Watergate method (25). For deriving distances, NOESY spectra were recorded in D2O at 30°C at four mixing times: 50, 100, 150 and 250 ms. The acquisition times in t1 and t2 were 0.05 and 0.341 s, respectively, and the recycle time was 2.34 s. Peak volumes were measured in two ways: using a footprint of variable size to include the peak, and integration within that area, and second by peak fitting in the two orthogonal direction and estimating the volume as the product of the areas at the peak maxima, normalised to the maximum peak height.
Molecular modelling
Accurate nucleotide conformations were determined from scalar coupling and NOE data using a multi-spin optimisation procedure as previously described (8,9,21). Because the deoxyriboses exist as a mixture of (at least) two conformations, we have analysed the scalar coupling NOE data according to an ensemble model. This approach gives better agreement with experimental data than the assumption of a single conformation (9,26). Briefly, each nucleotide was treated as an equilibrium mixture of ‘N’ and ‘S’ sugar conformation, with different glycosyl torsion angles for each sugar state. The time courses for intranucleotide NOEs (base-sugar and intra sugar NOEs) plus sums of coupling constants were simultaneously fitted for the phase angle and glycosyl torsion angle of the dominant sugar pucker, the mole fraction of the ‘S’ state, for a variety of conformations of the ‘N’ state. Given the conformations of the nucleotides, appropriate torsion angles and distances could be determined for subsequent restrained MD calculations. Internucleotide distances were initially calculated by first extrapolating the NOE time courses to zero mixing time, and then using the 2-spin approximation for converting these values to distances. As the N/S conformational averaging was not fully determined for all residues, the derived distances were first given generous upper and lower bounds, i.e. 1.8 Å lower bound and target distance +20% (for r < 3 Å) or +40% for r > 3 Å for the upper bound. After preliminary refinement rounds, the bounds were narrowed to ±20%.
Two restraint sets were prepared for the restrained molecular dynamics (rMD) runs. The first (set 1) comprised distances derived from the NOEs directly, with torsion angles for the dominant ‘S’ state. The second (set 2) retained the NOEs, but used torsion angles for the minor ‘N’ state. Each restraint set was used with starting structures of both minimised A and minimised B hybrid duplexes. 210 experimental distances including those corresponding to H-bonds and 70 torsion angles including the glycosyl torsion angles and gamma or a total of 280 restraints (14/residue). To avoid bias no restraints were applied to the α/β/ε/ζ torsions. Although in principle there are 1024 distinct conformations when considering just the N/S equilibrium in the 10 deoxynucleotides, previous experiments have suggested that the individual nucleotides behave as though they are essentially independent of their neighbours, and therefore each nucleotide can be regarded as a mixture of two conformational types (26).
Simulations of the fully solvated duplex were run at 300 K using Amber 7.0 incorporating the particle mesh Ewald method for the electrostatics. The Amber force-field well represents the properties in DNA·RNA hybrids (27). Eight molecular dynamics trajectories were calculated: the unrestrained molecular dynamics run starting with the 5-propynyl DNA·RNA A- and B-form duplexes; the NMR-derived restrained run starting with the 5-propynyl DNA·RNA A- and B-form duplexes with two sets of NMR-derived restraints; the unrestrained unmodified DNA·RNA starting with A- and B-form duplexes. The A- and B-form starting models were generated using nucgen and modified to include the 5-propyne moiety. The partial charges and parameterisation for 5-propynyl-U and -C was calculated using the 6-311G* basis set within GAMESS. Models were hydrated in a 10 Å box with TIP3P waters using standard AMBER (28) rules; Na+ cations were added for charge neutrality. Parameter and starting coordinate files were adjusted to include 2802 waters for each model with RDPARM. Simulations were performed in the isothermal isobaric ensemble (300 K, 1 atm) with the AMBER 7.0 program (28) and parameters from parm98.dat, using periodic boundary conditions and the PME algorithm. Molecular dynamics (MD) simulations used the mpi version of Sander (1.5 fs time step), with SHAKE to freeze all bonds involving hydrogen. Initial equilibrium for 150 ps with gradual warming and removal of positional restraints was performed. The total and potential energies were checked for variation to ensure equilibrium had been reached. The production runs for unrestrained and NMR derived restrained systems using starting structures in the A- and B-forms were 2 ns in length and average structures for each complex (taken from 33 snapshots accumulated in the last 50 ps) were obtained and subsequently fully minimised to give the final structures.
Several calculations were carried out to assess the influence of the force-field, the experimental restraints and the starting structures. Thus, free MD runs (2 ns) were carried out for the propynyl-modified duplex starting from standard A- and B-forms as well as for the unmodified duplex, i.e. with standard bases. Independent MD simulations were also carried out on the free DNA strands to assess the influence of the propyne groups on the conformational ensemble.
Final structures were then used to calculate helical parameters using Curves5.1 (29), and energy contribution with the Amber force-field. The hydration density was calculated using the program Watden (30). This program divides the volume into a 1 Å grid. The number of water molecules falling within a grid element was determined over the last 1 ns of the MD production runs for the most well refined structures.
RESULTS
Thermodynamic stability
The thermodynamic stability of the DNA·RNA hybrids was determined by UV melting (see Materials and Methods). The melting temperature, Tm, was obtained from the maximum of the first derivative of the absorbance with respect to the absolute temperature. ΔH and ΔS were obtained from the concentration-dependence of Tm by linear regression to equation 1. ΔG could then be calculated at any desired temperature. The thermodynamic parameters are gathered in Table 1.
Table 1. Thermodynamic stability of DNA·RNA hybrids: effect of pyrimidine (C5) modifications.
| Sequence | ΔG(298) (kJ mol–1) | ΔH (kJ mol–1) | ΔS (kJ mol–1 K–1) | Tm (1 µM) (K) | ΔTm (K) | Modification | |
|---|---|---|---|---|---|---|---|
| R strand | Y strand | ||||||
| dR10·dY10a | –42.9 | –290 | –0.829 | 307 | +2 | – | +Me |
| dR10·dY(U)10 | –41.3 | –283 | –0.811 | 305 | 0 | – | – |
| dR10·rY10a | –31.5 | –238 | –0.693 | 294 | –11 | – | +OH |
| dR10·rY(T)10 | –39.9 | –275 | –0.789 | 304 | –1 | – | +OH, +Me |
| rR10·dY(U)10 | –65.3 | –397 | –1.113 | 323 | +18 | +OH | – |
| rR10·dY10a | –56.1 | –343 | –0.963 | 318 | +13 | +OH | +Me |
| rR10·dPY10 | –107.5 | –441 | –1.119 | 357 | +52 | +OH | +Propynyl |
| rR10·rY10a | –66.8 | –394 | –1.098 | 325 | +20 | +OH | +OH |
| rR10·rY(T)10 | –87.7 | –420 | –1.115 | 341 | +36 | +OH | +OH, +Me |
aData from Gyi et al. (8).
As Table 1 shows, adding methyl groups to the RNA strands is strongly stabilising; in rR10·rY10 the Tm value increased 16° and in dR10·rY10, it increased 10°. In contrast, removing the methyl groups from the DNA strands had a significantly smaller effect on stability. For example, in dR10·dY10, the Tm decreased only 2° on removing the methyl groups and in rR10·dY10 removing the methyl groups actually increased Tm by 5°. The results with the RNA strands are similar to literature values (7). However, our results with DNA are significantly different. This may be due to the difference in solution conditions. The previous study (7) included 10 mM magnesium, whereas we added no magnesium to the buffers.
The propyne-modified decamer melts at 357 K at 1 µM strands, which can be compared with the Tm values of 318 K for rR10·dY10 and 323 K for rR10·dY(U)10 (Table 1). Hence, the replacement of the 5-proton by the propyne function increases the Tm by 34 K, or 4.25 K per modification. This is a very large stabilisation, and is comparable to previous measurements on mixed sequence shorter oligonucleotides (16,17). The enthalpy difference on melting was –441 kJ mol–1 for the propyne derivative, compared with –397 kJ mol–1 for rR10·dY(U)10, i.e. 44 kJ mol–1. This difference in enthalpy largely accounts for the greater stability of the propyne-modified duplex compared with the 5-methyl compound (42.2 kJ mol–1). In contrast, the methyl group had a much smaller effect on stability (and was slightly destabilising). The question arises as to how this difference in stability is related to conformational properties of the duplexes.
Electrophoretic mobility
The electrophoretic mobility of short oligonucleotides depends on several factors including size, shape, charge, ionic strength and gel concentration (31). For a series of oligomers of the same number of base pairs the formal charge is the same, so that in the same gel, relative mobility is defined by differences in shape and ionic interaction with the mobile phase (8,32). In general, the effects of methyl groups on mobility are relatively small compared with the effect of C2′-OH. Thus, the mobility of dR10·rY(T)10 is 87% that of dR10·dY10, and rR10·rY10 has a mobility of 88.5% of rR10·dY(U)10. In contrast, the mobility of dR10·rY(T)10 is 95% that of dR10·rY10, and rR10·dY10 is 97% that of rR10·dY(U)10. The bulkier propyne modification further decreases the electrophoretic mobility [compare rR10·dPY(10) with rR10·dY(U)10, rR10·dY10 and dR10·rY(T)10]. As electrophoretic mobility is primarily a measure of a global conformational property, this indicates that the methyl group, situated in the major groove, has only a small influence on global conformation, whereas the presence of a C2′-OH in the minor groove has a substantially larger effect on mobility, correlating with the large differences in conformation between B-DNA duplexes and A-RNA duplexes.
We have previously argued (32) that the electrophoretic mobility does not correlate with the expected hydrodynamic frictional coefficient as calculated using the bead model (33), which predicts that the DNA duplexes should have the largest Stokes’ radius. This indicates that other factors determine the electrophoretic drag in the gel, such as hydration and/or ionic atmospheres, which themselves depend on conformation (8,32). In fact, it appears that the axial charge density, and therefore the net effective charge on the duplexes, largely accounts for the electrophoretic mobility of short oligonucleotides (31,32); the shorter the duplex, the higher the axial charge density and therefore the larger the fraction of condensed ions. Thus, A-form RNA migrates more slowly than B-form DNA, and DNA·RNA hybrids, which have intermediate structures, migrate with an intermediate mobility. To a first approximation, then, decreasing electrophoretic mobility should correlate with the helical pitch, and by implication the propyne-modified duplex may have a shorter pitch than the unmodified duplex.
NMR spectroscopy
NMR spectroscopy provides a wealth of detailed information about conformation. Chemical shift perturbations are very sensitive to conformational effects arising from small modifications. The NMR spectra of the unmodified duplexes have been previously assigned (8). We have now assigned the spectra of the duplexes with modifications from addition or removal of methyl groups, and with the propyne-modified DNA strand.
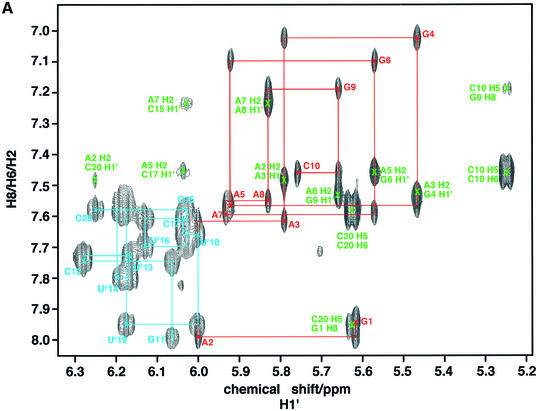
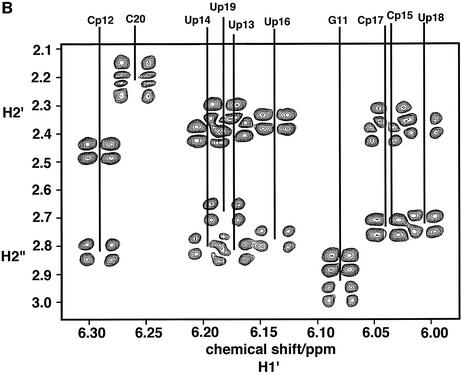
Compared with the parent unmodified duplexes, the propyne-modified hybrid duplex shows much better resolved NMR spectra (8). Figure 1A shows the base to anomeric proton region of a NOESY spectrum of the propyne-modified DNA·RNA hybrid (rR10·dPY10). The assignment of the base and H1′ resonances was therefore straightforward. The H2′ and H2” of the DNA strand were readily assigned both by NOESY and by COSY using the assigned H1′ coupling with H2′ and H2” (Fig. 1B). DNA H3′ were assigned using the sequential NOE pathway H8(i)–H3′(i)–H8(i + 1), and verified by scalar couplings with the assigned H2′. H4′ resonances were assigned by scalar coupling with assigned H3′ and dipolar interactions with H1′ of the same residue. It is clear from the fine structure present in the DQF-COSY spectrum and the presence of cross peaks between H1′ and H2′, H2” that the DNA strand is neither pure C3′-endo nor pure C2′-endo (and see below).
Figure 1.
NMR spectra of the propyne-modified DNA·RNA hybrid duplex. Spectra were recorded at 303 K and 600 MHz as described in the Materials and Methods. (A) NOESY spectrum recorded with a mixing time of 250 ms. The base-H1′ region is shown, with connectivities of the DNA and RNA strands. Red, RNA purine strand; cyan, modified DNA pyrimidine strand; green, Ade H2 intra and cross-strand NOEs. (B) Double quantum filtered COSY spectrum of the H1′ to H2′/H2” region showing the coupling patterns for the deoxyriboses. The acquisition time in t2 was 0.68 s.
The H2′ of the riboses were assigned from the strong H1′–H2′(i) NOE, and the intense sequential H2′(i)–H8(i + 1) NOE. H3′ and H4′ resonances were assigned using the NOEs between H1′ and H4′, H3′ and the base protons of residue i and i + 1, and scalar connectivity between H2′ and H3′. The H2′–H3′ scalar coupling was confirmed using a two-quantum COSY experiment as previously described for an RNA duplex (23). Exchangeable protons were assigned from NOESY spectra in H2O. The imino protons were readily assigned from the sequential NOEs, and differentiating AU from GC base pairs by noting the intense NOE to AC2H in the former, and a strong NOE to CN4H in the latter. The methyl-modified duplexes were assigned in a similar manner. The assignments are tabulated in the Supplementary Material.
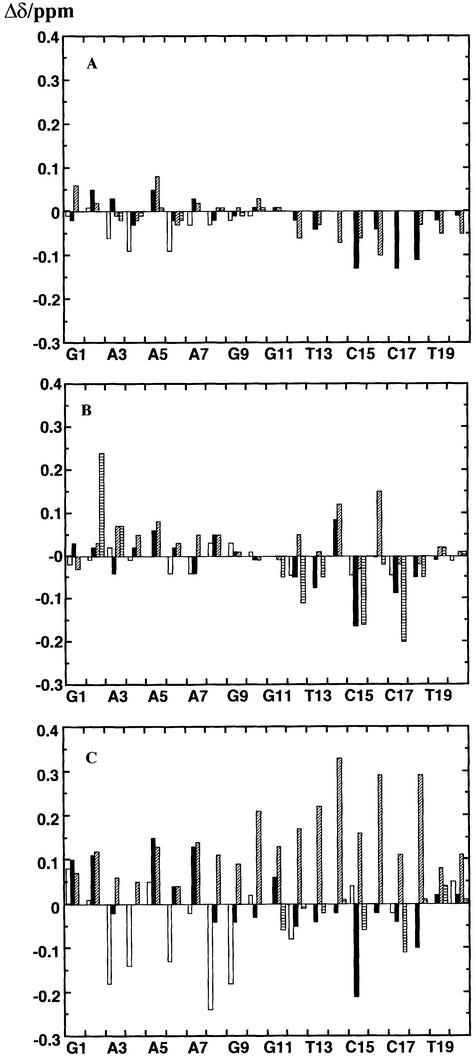
Chemical shift differences
Chemical shifts are sensitive indicators of conformational differences in DNA and RNA duplexes and hybrids (8). For the reference state, we have used the unmodified hybrid duplexes dR10·rY10 and rR10·dY10 previously studied (8,9). The changes in chemical shift due to adding or removing methyl groups are shown in Figure 2. Adding methyl groups to dR10·rY10 [i.e. dR10·rY(T)10] showed only small changes in the chemical shifts of the deoxypurine strand, all <0.1 p.p.m. and most <0.05 p.p.m. (Fig. 2A). The differences in shift for the rY10 strands are somewhat larger (the base protons have been ignored, as the addition of the methyl group will have a direct influence on the magnetic properties of the neighbouring H6), especially for the H1′ resonances in the minor groove. However, the comparatively small changes observed indicate that the conformation of dR10·rY(T)10 is similar to that of dR10·rY10. This is not surprising, as we have shown that most differences in the hybrids can be attributed to the DNA strand (8), which is unchanged in this molecule. There are perhaps small adjustments in the conformation of the RNA strand due to methylation of the bases.
Figure 2.
Chemical shift differences between modified and unmodified duplexes. Chemical shift differences were obtained for non-exchangeable protons at 30°C using the unmodified duplexes as the reference as described in the text. Open bars, H8/H6; closed bars, H1′; shaded bars, H2′; horizontal striped bars, H3”. (A) dR10·rMeY10–dR10·rY10. (B) rR10·dY10– rR10·dY(U)10. (C) rR10·dpY10–rR10·dY(U)10.
Significantly larger changes in chemical shifts were observed for protons of the deoxypyrimidine strand in rR10·dY(U)10, particularly for the H3′ and H4′ in the minor groove (Fig. 2B), but generally small changes in the purine strand. This suggests that there are some alterations in the conformation of the pyrimidine strand. It is notable that the effects of methyl groups on the proton chemical shifts are much smaller than the effect of introducing a C2′–OH, which certainly causes a major rearrangement of the global conformation (8,9), and therefore the influence of the ring currents of the bases. Hence, the effect of methyl groups on conformation appears to be relatively small.
Figure 2C shows the changes in chemical shifts due to substitution of the 5-propyne group in the deoxypyrimidine strand. Here the shift differences are much larger than the corresponding effect of a methyl group. That the changes are observed throughout the molecule and in both grooves is consistent not only with changes in the anisotropic shielding contributions from the propyne groups themselves (in the major groove), but also with significant changes in conformation of the molecule as a whole. This indicates that the large electron-rich propyne modification in the DNA strand has a greater influence on conformation in the hybrids than the smaller methyl group, and correlates with the much greater thermodynamic stability (see above).
Sugar conformations
The unmodified hybrid duplexes on average showed a greater degree of conformational averaging in the DNA strands than in the pure DNA duplex, especially in the pyrimidine strand (8,9). Coupling patterns in dR10·dY(U)10 are similar to those observed in rR10·dY10 (data not shown), indicating that the influence of the methyl group on the N/S preference of the DNA sugars is minimal.
The strand containing the propyne group, in contrast, showed an increased tendency to repucker toward a higher fraction of the ‘N’ conformer. The H1′ of the deoxy sugars in the propyne-modified duplex appear as triplets in the base-H1′ region of the NOESY spectrum (Fig. 1A), indicating significant coupling to both H2′ and H2”, and therefore not pure ‘N’ state. The H1′–H2′/H2” cross peaks are all present for the DNA strands (Fig. 1B). The cross-peak patterns, however, are not typical of a predominantly C3′-endo state, but rather a mixture of ‘S’ and ‘N’ pucker states, with a mean fraction of the ‘S’ state of 0.6–0.7 apart from the terminal nucleotides where the fraction is closer to 0.5.
As shown in Figure 1A, the base-H1′ cross-peaks for the RNA strand appear as singlets, indicating a small H1′–H2′ coupling. No COSY cross-peaks were observed between the H1′ and H2′ in the RNA residues of rR10·dPY10, indicating that the major conformation is N, which is not perturbed by addition or removal of methyl groups from either strand. In addition, intense H3′–H4′ cross-peaks were observed both in the DQF-COSY and in the proton INADEQUATE experiment, which is also consistent with a C3′-endo conformation for the riboses. This was confirmed by the relative intensities of the NOEs between base and ribose protons.
The scalar coupling data for all of the modified duplexes indicate that the deoxyriboses adopt neither pure ‘N’ nor pure ‘S’ states. This is in agreement with recent NMR data for a different purine·pyrimidine hybrid sequence (34), and contrasts with X-ray crystallographic data that all sugars are essentially C3′-endo (35).
Solution conformation of the propyne-modified DNA·RNA hybrid duplex
The spectral dispersion of rR10·dY(U)10 is too low to provide sufficient NOE restraints for a full structure calculation. However, the available evidence from CD (data not shown) and NMR indicates that the lack of methyl groups have a minor influence on conformation. The propyne modification, in contrast, increases the spectral dispersion presumably in part because of the shielding anisotropy of the triple bond, and alterations in conformation via interactions between neighbouring propyne groups (see above).
NOEs between base protons and H1′ and H2′ showed that all glycosyl torsion angles lie in the anti domain, with the torsion in the RNA strands ∼–160° typical of the local A conformation. In the DNA strands, the base-H2′ NOE is weaker than found in the DNA duplex, indicating a glycosyl torsion angle more negative than the ‘B’ values of –100 to –110°. The NOESY mixing time dependence of the base-sugar NOEs was analysed using NUCFIT (9,36) which showed that for non-terminal residues, the glycosyl torsion angles are mostly in the range –120 to –140°, i.e. similar to the values observed previously in the unmodified hybrid duplex (9).
In addition to the expected NOEs for an overall A-like duplex, there are NOEs to the propyne methyl groups. As these are highly characteristic, we have carefully quantified them.
Molecular modelling
We have built hybrid duplexes with and without the propyne group in A- and B-forms as described in the Materials and Methods. The molecules were fully solvated and energy minimised, equilibrated, followed by a 2 ns molecular dynamics trajectory at 300 K. These calculations show the influence of the force-field on the final structures, as well as the effects of the propyne group on the free dynamics runs.
The last 50 ps of the trajectories were then averaged and the resulting average structures were energy minimised to produce a ‘typical’ structure. These structures can be characterised by a few critical parameters, including the nucleotide conformations (sugar puckers, glycosyl torsion angle and gamma) and the helical parameters twist, rise, inclination and x-displacement as shown in Table 2. The energy minimised initial structures (prior to molecular dynamics) remain fairly close to the canonical A- and B-forms, which both give very large violations of distances and torsion angles. This is expected as minimisation will only find local minima and molecular dynamics is required to pass over energy barriers. The MD simulations of the propynyl-modified duplex in the absence of restraints behave differently over a period of 2 ns. The sugar puckers of the DNA strand in the A structure convert from the ‘N’ domain to the ‘S’ domain with a corresponding change in the glycosyl torsion angle. This change in nucleotide conformation is accompanied by a flattening of the base pair inclination and an increase in the rise. In contrast, the sugars of the RNA strand in the B structure did not convert to the ‘N’ domain during the trajectory though the helical twist decrease from ∼36 to ∼30° and the x-displacement increased from ∼–0.7 to ∼–2.6 Å. Thus, the rise and inclination are more B-like, whereas the twist and displacement are more A-like. However, both of these structures are intermediate between the energy minimised A- and B-forms and in this system much longer trajectories may be required for sugar conformation convergence. These structures also show significant violations of the experimental restraints (Table 2). The ‘B’ structure gave large torsion violations because the RNA sugars did not repucker. The ‘A’ structure gives much smaller violations because the DNA strand did repucker, giving the characteristic hybrid structure of the nucleotides. The ‘A’ structure also gave very much smaller violations than the energy-minimised structures.
Table 2. Curves nucleotide and helicoid parameters.
| MD run | P(D) (°) | χ(D) (°) | P(R) (°) | χ(R) (°) | γ (°) | Dx (Å) | η (°) | θ (°) | h (Å) | Rd (Å) | Rt (°) | Etot(Erst) (kcal mol–1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A(EM)_1 | 8 | –155 | 8 | –153 | 46 | –5.4 | 21 | 32.6 | 2.6 | 3.4 | 122.3 | |
| B(EM)_1 | 190 | –97 | 192 | –98 | 36 | –0.7 | –6 | 36.1 | 3.4 | 5.55 | 265.5 | |
| MD(A)f | 120 | –130 | 10 | –160 | 58 | –4.7 | 4.4 | 30 | 3.1 | 2.72 | 31.4 | |
| MD(B)f | 135 | –130 | 135 | –115 | 47 | –2.6 | –1.3 | 30 | 3.4 | 6.9 | 207.9 | |
| rMD(A)_1 | 130 | –125 | 9 | –160 | 57 | –4.1 | 6.4 | 32.4 | 3.12 | 0.13 | 0 | –29084(11.8) |
| rMD(A)_2 | 95 | –150 | 10 | –164 | 60 | –5.4 | 12.2 | 30.0 | 2.84 | 0.17 | 21 | –29028(26.8) |
| rMD(B)_1 | 125 | –125 | 9 | –160 | 57 | –4.0 | 5.5 | 31.3 | 3.41 | 0.14 | 0 | –29059(13.5) |
| rMD(B)_2 | 97 | –151 | 20 | –159 | 60 | –4.39 | 5.65 | 29.8 | 3.10 | 0.2 | 23 | –28999(30.2) |
| rR·dY | 130 | –130 | 9 | –160 | 50 | –3.9 | 5 | 31–34 | 3.0 |
Parameters were obtained using Curves5.1 on the average structures from the last 50 ps of the production run.
MD(A) and MD(B) are free MD runs starting from A- and B-form structures, respectively. rMD(B)_1 and rMD(B)_2 are restrained MD runs starting from the B-form structure with restraint sets corresponding to C2′-endo and C3′-endo sugars in the DNA strand, respectively (see Materials and Methods). rMD(A)_1 and rMD(A)_2 are the corresponding runs starting from the A-form. A(EM) and B(EM) are energy minimised initial A- and B-form hybrids. Rd and Rt are the total distance and torsion angle restraint violations, respectively. Etot(Erst) are the total and restraint energies.
In the presence of the experimental restraints, the final structures are different from the free dynamics runs, indicating that the restraints are driving the structures. Starting from either A- or B-forms, and using the restraints appropriate for the dominant (‘S’) sugars, no torsion violations were observed, and low distance violations compared with the free dynamics or energy minimisation calculations (Table 2). The two rMD structures have for the most part converged and are very similar both at the nucleotide level, and in terms of the x-displacement (∼–4 Å), low inclination and helical twist. The A-form using both sets of NMR restraints is energetically more stable relative to the B-form by 26–29 kcal when comparing average total energies over the last nanosecond of the trajectories. The A-form using the NMR restraint set 1 is 57 kcal more stable than using NMR restraint set 2. The low inclination results in the rise being substantially larger than in the canonical A-form, though the converged structures differ significantly in the final value. This indicates that the helical rise is not as well determined by the data as are the other parameters. In contrast, if the nucleotide conformations are restrained to an ‘A’-like state, the distance violations increase, and the torsion violations are severe, even though a compromise structure is produced by the efforts of the force-field and the distance restraints. This indicates that at the local nucleotide level, the DNA strand is more B-like than A-like.
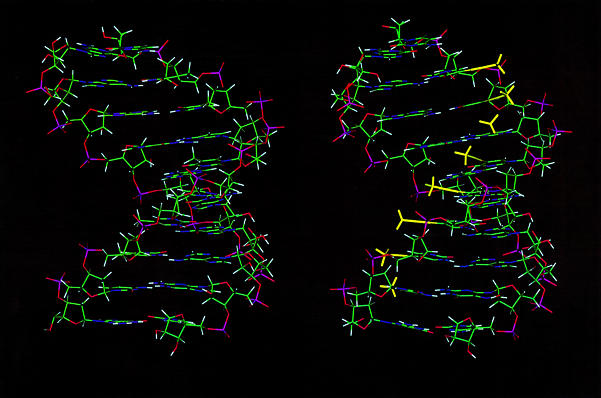
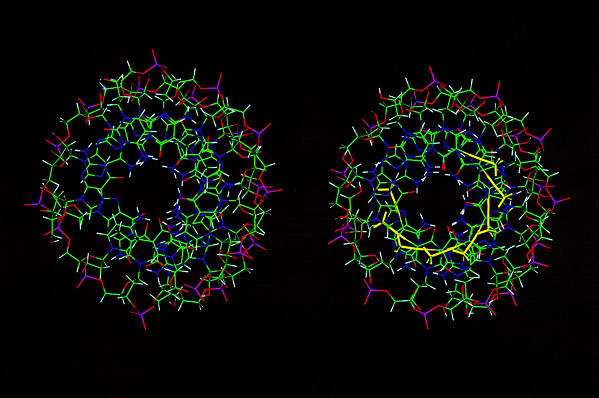
Figure 3 shows the averaged structures from the A and B runs, and for comparison, the NMR-derived structure of the unmodified version of the duplex (9). The structures appear similar (RMSD of the bases to the original structures = 1.7 Å). The propyne structure is slightly shorter and more A-like. The major groove is partly filled by the bulky propyne groups (yellow). These propyne groups clearly form a stack that runs on the inside edge of the major groove. The central hole in the duplex, which reflects the displacement of the base pair from the helix axis is large as expected for a globally A-like structure. However, the degree of base-stacking is greater in the presence of the propyne groups than in the rR·dY duplex.
Figure 3.
Structures of the propyne-modified hybrid duplex compared with dY·rR. Averaged structures are displayed looking into the major groove and down the helix axis. Left, rR·dY [from Gyi et al. (9)]. Right, rR·dpY. The propyne groups are shown in yellow.
It is notable that the snapshot structures do not account completely for the NMR data. Most importantly, the sugar puckers do not give rise to coupling constants that agree with observations. For the DNA strand, the observed coupling patterns can be interpreted as a mixture of N and S states, or slightly less successfully with an intermediate O4′-endo conformation. However, this O4′-endo conformation itself introduces errors into the distance estimates, especially the H1′–H4′ distance, which is <2.7 Å for O4′-endo, but according to the NOE data is >3 Å.
However, if the structures derived from the two restraint sets are used to derive averaged distances and coupling constants, better agreement can be obtained. For example, if the rMD(A)_1 and rMD(A)_2 structures are used to calculate distances as 〈r–6〉 averages and sums of coupling constants as linear averages, then a mixture of 50–70% of the state 1 and 30–50% of state 2 retains agreement with the distances, but gives much better agreement with the coupling constant data. This is because an intermediate structure gives rise to O4′-endo-like sugar puckers, which do not agree with the coupling constants, nor the critical H1′–H4′ distance. In the limit of no cooperativity between N and S type nucleotide conformations, and for moderately low restraint densities available, the problem can be simplified to solving the structure as just two molecular species (i.e. all N and all S for the DNA strands), but the net result should be viewed as an equilibrium at the nucleotide level. The all S and all N states are in fact low population. Given the good agreement with the data it seems that a more sophisticated fitting procedure (26) is not needed. However, to represent the ensemble, the two sets of structures can be mixed at the nucleotide level and energy minimised to produce any of the possible mixed states.
Propyne stacking
The derived structures provide a rationalisation for the pronounced thermal stability of the propyne-modified structure. The Lennard–Jones component of the overall energy of the 5-propyne groups interacting with neighbouring 5-propyne groups indicates that there is a net stabilisation. In contrast, the electrostatic component of the 5-propyne–5-propyne is slightly destabilising. Moreover, as the base charge distributions are different for U and C, parsing the electrostatic and Lennard–Jones contributions from each 5-propyne base shows that there is net stabilisation from base-stacking, i.e. that outweighs the unfavourable propyne–propyne electrostatic interaction. Furthermore, the 5-propyne bases also show increased Lennard–Jones and electrostatic stabilisation over the C and T bases in the same calculations for the dR10·dY10 duplex.
Hydration
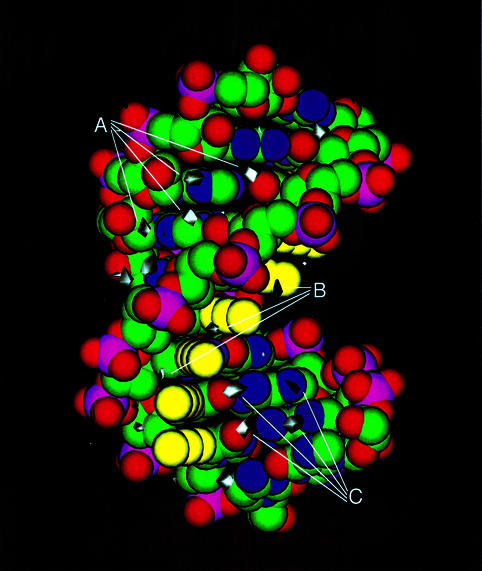
Analysis of the solvated NOE-derived restrained molecular dynamics trajectories show several distinct hydration patterns. The spine of hydration in the minor groove is reproduced well (Fig. 4), although it is more associated with the bases than being bifurcated. Additional hydration in the minor groove is associated with the RNA strand. The major groove also shows significant hydration associated with the purine RNA strand located mainly near the N6 of adenine and O6 of guanine. There is a hydration pattern associated with the 2′-OH and the phosphate backbone of the RNA strand on the minor groove side, which may correspond to the experimental decrease in proton exchange by NMR (22). The 5-propyne group on the DNA strand creates a small groove between itself and the phosphate backbone. This is stabilised by bridging water molecules between the 5-propyne group and the i – 1 phosphate group creating a spine of hydration. The 5-propyne groups have favourable small Lennard–Jones energies and larger electrostatic interactions with the solvent water as well as an increase in overall hydration stability when compared to the same calculations for the dR10·dY10 duplex.
Figure 4.
Hydration rR·dpY. Hydration density was calculated as described in the Materials and Methods. The white contours for water are drawn at 6-fold greater than average water density. (A) Minor groove hydration. (B) Hydration in the 5-propyne:pyrimidine phosphate backbone groove. (C) Major groove hydration.
We have previously presented evidence that both the major and minor grooves of this duplex are hydrated. Direct interactions between water protons and H8 and H6 of bases in the major groove were detected, as well as interactions with the methyl groups of the propyne functions. In addition, significant interactions between water and the AdeC2H and H1′ in the minor groove were detected (22). Thus, the simulations are in substantial agreement with the available experimental data.
DISCUSSION
The stabilisation of the propyne-modified duplex compared with rR·dY(U) is very large, and seems to be primarily enthalpic in origin. Thus, the difference free energy is ∼42.5 kJ mol–1 at 298 K, and the enthalpy difference is ∼44 kJ mol–1. These differences represent the energy differences associated with adding the propyne groups to the DNA strand in the duplex states versus the single strand state. This is more favourable in the duplex state than in the strand state. The electrophoretic mobility implies global conformation differences as in B/A/hybrid-forms. The propyne derivative should be somewhat squatter (more A-like) than the unmodified duplex. The spectroscopic data indicate that the propyne group causes substantial changes in conformation of the hybrid, both at a local and at a more global level, as a consequence of the interactions this group can make in the major groove. We have also previously reported that the propyne-modified duplex gives rise to increased water stabilisation in the minor groove (22), presumably as a consequence of the change in conformation, which is supported by the explicit solvent molecular dynamics calculations. There are numerous possibilities to account for the stabilisation that is mainly enthalpy driven. These have to account for the observation that there are differences in the structure of the propyne and methyl containing duplexes, as the propyne groups form a stack along the major groove. This appears to include a long-range cooperativivity (16,17). Although the specific electrostatic interaction among these groups is unfavourable, this is more than offset by the favourable base electrostatic, Lennard–Jones and solvation energies. Furthermore, we might expect that the polarisability of the propyne functions might lead to additional stabilisation that is not accounted for in the Amber force-field. The readjustments to the structure that allow optimal propyne stacking and thereby altered base-pair stacking can in large measure account for the overall increase in favourable potential energy in the duplex compared with the isolated strand state. In contrast with the propyne groups, methylation of the DNA strand has only small effects on thermodynamic stability and conformation, which may be localised to the modified strand. The possibilities for methyl–methyl interactions along the major groove are much more limited than for the propyne stacks, as the dipolar interactions will not be as extensive as for the longer, electron-rich propyne groups. Methylation at the 5 position of pyrimidines generally does not produce large changes in structure (37) as we have observed in these DNA·RNA hybrids.
These observations are important in the design of optimal antisense agents, where maximum thermodynamic stability is needed (16,17). Clearly it is important to know the structural consequences of any modification. This work indeed shows that substantial sized groups can have significant effects on conformation, and thereby couple to large changes in thermodynamic stability. The propyne group appears to stabilise a more A-like structure than do the methyl groups, which is associated with increased thermodynamic stability. Moreover, the additional interactions involving the propynes must also contribute favourably to the stability, as the modified DNA·RNA hybrid is now more stable than the RNA duplex.
COORDINATES
The coordinates have been deposited at Brookhaven Data Bank, accession no. 1007.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
NMR spectra were recorded at the MRC Biomedical NMR Centre, Mill Hill, London UK. This work was supported by the Medical Research Council of the UK.
REFERENCES
- 1.Dagle J.M. and Weeks,D.L. (2001) Oligonucleotide-based strategies to reduce gene expression. Differentiation, 69, 75–82. [DOI] [PubMed] [Google Scholar]
- 2.Read M.L. and Bremner,K.H. (2002) Antisense strategies and non-viral gene therapy for cancer. Expert Opin. Ther. Pat., 12, 379–391. [Google Scholar]
- 3.Zinker B.A., Rondinone,C.M., Trevillyan,J.M, Gum,R.J., Clampit,J.E., Waring,J.F., Xie,N., Wilcox,D., Jacobson,P., Frost,L. et al. (2002) PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose and improves insulin sensitivity in diabetic mice. Proc. Natl Acad. Sci. USA, 99, 11357–11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall K.B. and McLaughlin,L.W. (1991) Thermodynamic and structural properties of pentamer DNA.RNA, RNA.RNA and DNA.DNA duplexes of identical sequence. Biochemistry, 30, 10606–10613. [DOI] [PubMed] [Google Scholar]
- 5.Lesnick E.A. and Freier,S.M. (1995) Relative thermodynamic stability of DNA, RNA and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry, 34, 10807–10815. [DOI] [PubMed] [Google Scholar]
- 6.Hughes S.H. and Arnold,E. (2001) Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J., 20, 1440–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S. and Kool,E.T. (1995) Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2′-hydroxyl effects. Biochemistry, 32, 4125–4132. [DOI] [PubMed] [Google Scholar]
- 8.Gyi J.I., Conn,G.L., Lane,A.N. and Brown,T. (1996) DNA.RNA hybrids containing purine rich and pyrimidine rich strands with DNA and RNA duplexes. Biochemistry, 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- 9.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- 10.Kanaori K., Tamura,Y., Wada,T., Nishi,M., Kanehara,H., Morii,T., Tajima,K. and Makino,K. (1999) Structure and stability of the consecutive stereoregulated chiral phosphorothioate DNA duplex. Biochemistry, 38, 16058–16066. [DOI] [PubMed] [Google Scholar]
- 11.Venkateswarlu D., Lind,K.E., Mohan,V., Manoharan,M. and Ferguson,D.M. (1999) Structural properties of DNA:RNA duplexes containing 2′-O-methyl and 2′-S-methyl substitutions: a molecular dynamics investigation. Nucleic Acids Res., 27, 2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trempe J.F., Wilds,C.J., Denisov,A.Y., Pon,R.T., Damha,M.J. and Gehring,K. (2001) NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J. Am. Chem. Soc., 123, 4896–4903. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Verbeure,B., Luyten,I., Lescrinier,E., Froeyen,M., Hendrix,C., Rosemeyer,H., Seela,F., Van Aerschot,A. and Herdewijn,P. (2000) Cyclohexene nucleic acids (CeNA): serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. J. Am. Chem. Soc., 122, 8595–8602. [Google Scholar]
- 14.Conn G.L. (1996) Structural and thermodynamic studies of RNA. PhD Thesis, University of Edinburgh, UK.
- 15.Wagner R.W., Mateuci,M.D., Grant,D., Huang,T. and Froehler,B.C. (1996) Potent and selective inhibition of gene expression by an antisense heptanucleotide. Nat. Biotechnol., 14, 840–844. [DOI] [PubMed] [Google Scholar]
- 16.Barnes T.W. and Turner,D.H. (2001) C5-(1-propynyl)-2′-deoxy-pyrimidines enhance mismatch penalties of DNA:RNA duplex formation. Biochemistry, 40, 12738–12745. [DOI] [PubMed] [Google Scholar]
- 17.Barnes T.W. and Turner,D.H. (2001) Long-range cooperativity in molecular recognition of RNA by oligodeoxynucleotides, with multiple C5-(1-propynyl) pyrimidines. J. Am. Chem. Soc., 123, 4107–4118. [DOI] [PubMed] [Google Scholar]
- 18.Flanagan W.M., Wolf,J.J., Olson,P., Grant,D., Lin,K.Y., Wagner,R.W. and Matteucci,M.D. (1999) A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 96, 3513–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacroix L., Lacoste,J., Reddoch,J.F., Mergny,J.L., Levy,D.D., Seidman,M.M., Matteucci,M.D. and Glazer,P.M. (1999) Triplex formation by oligonucleotides containing 5-(1-propynyl)-2′-deoxyuridine: Decreased magnesium dependence and improved intracellular gene targeting. Biochemistry, 38, 1893–1901. [DOI] [PubMed] [Google Scholar]
- 20.Mills M., Arimondo,P.B., Lacroix,L., Garestier,T.S., Klump,H. and Mergny,J.L. (2002) Chemical modification of the third strand: differential effects on purine and pyrimidine triple helix formation. Biochemistry, 41, 357–366. [DOI] [PubMed] [Google Scholar]
- 21.Lane A.N., Ebel,S. and Brown,T. (1993) NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur. J. Biochem., 215, 297–306. [DOI] [PubMed] [Google Scholar]
- 22.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Identification and hydration of C2′-OH in RNA and RNA·DNA hybrids by NMR. Nucleic Acids Res., 26, 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conte M.R., Conn,G.L., Brown,T. and Lane,A.N. (1996) Hydration of the RNA duplex r(CGCAAAUUUGCG)2 determined by NMR. Nucleic Acids Res., 24, 3693–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.States D.J., Haberkorn,R.A. and Ruben,D.J. (1982) A two-dimensional nuclear Overhauser experiment with pure absorption phase in 4 quadrant. J. Magn. Reson., 48, 286–292. [Google Scholar]
- 25.Piotto M., Saudek,V. and Sklenar,V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. Struct., 2, 661–665. [DOI] [PubMed] [Google Scholar]
- 26.Lane A.N. (1996) The influence of conformational averaging on 1H-1H NOEs and structure determination in DNA. Magn. Res. Chem., 34, S3–S10. [Google Scholar]
- 27.Cheatham T.E. and Kollman,P.A. (1997) Molecular dynamics simulations highlight the structural differences among DNA:DNA, RNA:RNA and DNA:RNA hybrid duplexes. J. Am. Chem. Soc., 119, 4805–4825. [Google Scholar]
- 28.Cornell W.D., Cieplak,P., Bayly,C.I., Gould,I.R., Merz,K.M., Ferguson,D.M., Spellmeyer,D., Fox,T., Caldwell,W.J. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 29.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 30.Shields G.C., Laughton,C.A. and Orozco,M. (1997) Molecular dynamics simulations of the d(T.A.T) triple helix. J. Am. Chem. Soc., 119, 7463–7469. [Google Scholar]
- 31.Gast F.U. and Hagerman,P.J. (1991) Electrophoretic and hydrodynamic properties of duplex ribonucleic-acid molecules transcribed in vitro—evidence that A-tracts do not generate curvature in RNA. Biochemistry, 30, 4268–4277. [DOI] [PubMed] [Google Scholar]
- 32.Bonifacio G., Brown,T., Conn,G.L. and Lane,A.N. (1997) Comparison of the electrophoretic and hydrodynamic properties of DNA and RNA oligonucleotides. Biophys. J., 73, 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia de la Torre J., Navarro,S. and Lopez Martinez,M.C. (1994) Hydrodynamic properties of a double helical model for DNA. Biophys. J., 66, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hantz E., Larue,V., Ladam,P., Le Moyec,L., Gouyette,C. and Dinh,T.H. (2001) Solution conformation of an RNA–DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand. Int. J. Biol. Macromol., 28, 273–284. [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y. and Sundaralingam,M. (2000) Crystal structure of a DNA center dot RNA hybrid duplex with a polypurine RNA r(gaagaagag) and a complementary polypyrimidine DNA d(CTCTTCTTC). Nucleic Acids Res., 28, 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane A.N. (1990) Determination of solution structures of nucleic acids by NMR. Biochim. Biophys. Acta, 1049, 189–204. [DOI] [PubMed] [Google Scholar]
- 37.Derreumaux S., Chaoui,M., Tevanian,G. and Fermandjian,S. (2001) Impact of CpG methylation on structure, dynamics and solvation of cAMP DNA responsive element. Nucleic Acids Res., 29, 22314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.