Abstract
Carotenoids are widely distributed pigments in nature and their biosynthetic pathway has been extensively studied in various organisms. The recent access to the overwhelming amount genomic data of cyanobacteria has given birth to a novel approach called comparative genomics. The putative enzymes involved in the carotenoid biosynthesis among the cyanobacteria were determined by similarity-based tools. The reconstruction of biosynthetic pathway was based on the related enzymes. It is interesting to find that nearly all the cyanobacteria share quite similar pathway to synthesize β-carotene except for Gloeobacter violaceus PCC 7421. The enzymes, crtE-B-P-Qb-L, involved in the upstream pathway are more conserved than the subsequent ones (crtW-R). In addition, many carotenoid synthesis enzymes exhibit diversity in structure and function. Such examples in the families of ζ –carotene desaturase, lycopene cylases and carotene ketolases were described in this article. When we mapped these crt genes to the cyanobacterial genomes, the crt genes showed great structural variation among species. All of them are dispersed on the whole chromosome in contrast to the linear adjacent distribution of the crt gene cluster in other eubacteria. Moreover, in unicellular cyanobacteria, each step of the carotenogenic pathway is usually catalyzed by one gene product, whereas multiple ketolase genes are found in filamentous cyanobacteria. Such increased numbers of crt genes and their correlation to the ecological adaptation were carefully discussed.
Keywords: Cyanobacteria, Comparative genomics, Carotenoid pathway
1. Introduction
Carotenoids have important functions in photosynthesis, nutrition, and protection against photooxidative damage 1. They are produced by all photosynthetic organisms–plants, algae and bacteria as well as many species of nonphotosynthetic eubacteria. Cyanobacteria are a group of eubacteria that can be traced back 3.5 billion years, based on the fossil and molecular evidence 2,3. Carotenoids in cyanobacteria have two main functions: they serve as light –harvesting pigments in photosynthesis and they protect against photooxidative damage 4.Thus, over hundreds of millions ago, cyanobacteria had photosynthetic activity.
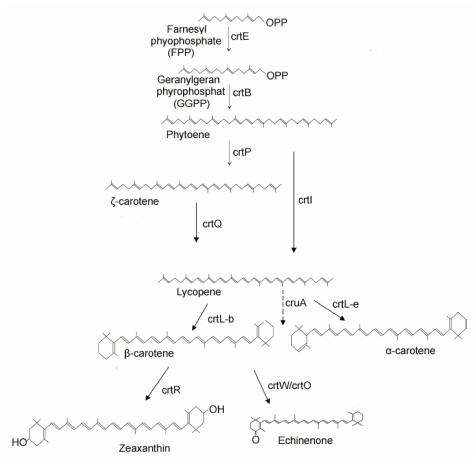
Extensive studies have been done on the biosynthetic pathway for carotenoids (Fig.1) 5-7. Farnesyl pyrophosphate (FPP) combining with C5-isoprenoid units is extended to C20 molecules, geranylgeranyl pyrophosphate (GGPP) by geranylgeranyl pyrophosphate synthase (crtE). The common C40 carbon results from the condensation of two C20 molecules by phytoene synthase (crtB). The sequential desaturation steps and cyclization of the ends of the molecule to generate carotenes are catalyzed by phytoene desaturase (crtP/crtI), ζ-carotene desaturase (crtQ) and lycopene cyclase. Finally, the carotenes are further modified by the enzymes such as β-carotene ketolase (crtW or crtO) and β-carotene hydroxylase (crtR) to generate a various C40 carotenoids. The general aspects of chemical structures, functions, molecular genetics of carotenoids and molecular evolution of enzymes involved in carotenoid biosynthesis have been reported 8-12. More recently the cyanobacterial genomes results in the focus on the enzymes involved in carotenoid biosynthesis from cyanobacteria. CrtQs from Anabaena sp. PCC 7120 (crtI-like sequence) 13, Synechocystis sp. PCC 6803 (plant crtQ-like) 14, and crtI-type phytoene desaturase from Gloeobacter violaceus PCC 74215 have been functionally identified. Although only one monoketolase CrtO from Synechocystis has been functionally characterized 15, two distinct β-carotene ketolases-crtO and crtW from Anabaena sp. PCC 7120 16, and two carotenoid ketolase genes (crtW) from Nostoc punctiforme PCC 73102 17 have been characterized. In generally, the biosynthetic pathways of the carotenoids are similar, but the carotenoids among these species are different in composition and diverse in category.
Fig 1.
Putative biosynthetic pathway of carotenoids in cyanobacteria. The names of enzymes are according to the crtE, geranylgeranyl pyrophosphate synthase; crtB, phytoene synthase; crtP, phytoene desaturase; crtQ, zeta-carotene desaturase; crtL, lycopene beta cyclase, ; lycopene epsilon cyclase; cruA, the most like candidate for lycopene cyclase by comparison to CT0456 in the species lacking crtL. crtO/crtW, beta-carotene ketolase; crtR, beta-carotene hydroxylase.
Now whole-genome information is being generated for a number of cyanobacteria.16 cyanobacterial genomes have been fully sequenced and 2 in the draft format and more than 20 are in the process of being sequenced (http://img.jgi.doe.gov/pub/main.cgi?Page=restrictedMicrobes&domain=Bacteria; http://www.ncbi.nlm.nih.gov/).The complete genome sequences of cyanobacteria allowed us to obtain a comprehensive data set of genes encoding enzymes in the carotenoid biosynthetic pathway. Moreover, even if experimental studies have become possible to reconstruct the pathway on the basis of a prediction of the genes and its function from the complete genome sequence data. Genome-wide screening of crt genes based on the genome-sequencing project provided us a new and comprehensive insight into the cyanobacterial carotenoid biosynthetic pathway. In this article, emphasis is centered on the comparative analysis of cyanobacteria and shedding light on the diversity of the carotenoid biosynthesis pathway based on the information of genomes.
2. Materials and methods
Data sources
The genomes of 18 cyanobacteria included Synechocystis, Synechococcus, Prochlorococcus, Anabaena, Nostoc, Trichodesmium, Gloeobacter and Crocosphaera were downloaded from IMG database. Each genome was fed into the program formatdb 18 to create an organism-species database. A set of crt genes was obtained from IMG v.1.1 (http://img.jgi.doe.gov/v1.1/main.cgi) and GenBank database. This dataset, including well-characterized and putative enzymes encoded by cyanobacterial crt genes, was used to construct a query protein set. Each protein in this query dataset was used to search the potential novel sequences in all cyanobacterial species with whole genome sequences available, by using the BLASTP and TBLASTN programs, with e-value<1-10. The best hits were identified as homologs in the species. Results of sequence similarity searches were parsed and the orthologues were extracted for each species. Positions of crt genes were manually inspected for each species. Similarity searches of the above databases also led to identification copies of crt genes in these species.
Multiple sequence alignment and phylogenetic analysis
Multiple protein sequence alignment was performed using ClustalX program with the implanted BioEdit 19, 20 for each of caroteniod biosynthetic pathway genes. Motifs of these enzymes across the domains were determined by NCBI BLAST search or SMART (http://smart.embl-heidelberg.de/) 21. Phylogenetic trees were reconstructed using neighbor-joining method 22, as implemented in the program MEGA 2.1 23. Bootstrap support was estimated using 1000 replicates for distance analyses.
Tertiary structure prediction
To well understand the evolution of certain enzyme, protein structure was analyzed using homology modeling. The protein sequences of lycopene cyclase from Prochlorococcus MIT 9312 and Arabidopsis thaliana were submitted to the protein model server: RCSB protein data bank Web server (http://www.rcsb.org/pdb/Welcome.do) with PDB-1pn0 as the model template. All the manipulations were performed using PdbViewer.
3. Results and discussion
General comparison of the carotenoids biosynthetic genes from cyanobacteria
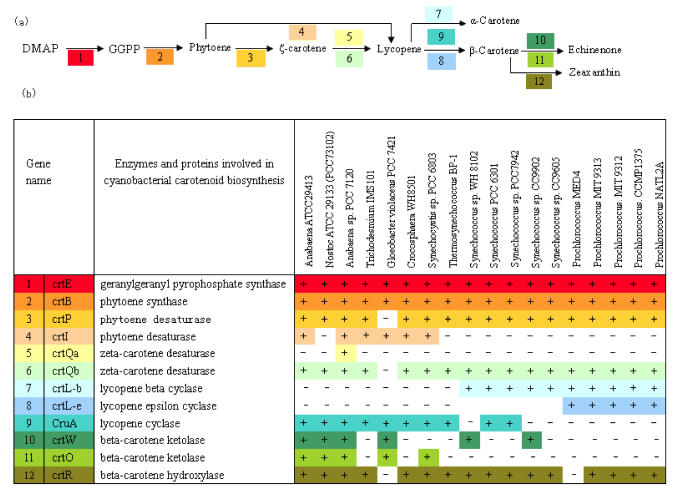
Similarity search between query sequences and cyanobacterial genomes were performed by BLASTP program. The distribution of genes involved in carotenoid biosynthesis across 18 cyanobacterial genomes is summarized in Figure 2. We can see geranylgeranyl pyrophosphate synthase (crtE/GGPS) and phytoene synthase genes (crtB/pys) are widely distributed among all the species. The cyanobacteria share the same carotenoid biosynthetic pathway to lycopene except for G.violaceus 7421.
Fig 2.
Presumed enzymes involved in the biosynthetic pathway of carotenoids among different cyanobacterial species. Some enzymes have been functionally identified, while others are just suggested by sequence homology. Database searches were carried out with the BLASTP program. (a) Putative carotenoid synthetic pathway diagram. The shading reflects correspondence to specific genes, numbers and colors are the same as in (b). (b) A list of major enzymes and proteins involved in cyanobacterial carotenoid biosynthesis. Presence or absence of putative orthologs in a given genome is indicated by '+' or '-', respectively. The gene IDs of putative othologous genes are listed in supplemental material.
Multiple alignments of the predicted amino acid sequences from the homologous carotenoid biosynthetic genes from cyanobacteria were constructed. A similar degree of difference in these proteins among cyanobacteria is noted here. Consistent with the hypothesis that the early reactions of carotenoid biosynthesis are conserved 8, the present study also reveals the enzymes are more conserved in the upstream pathway. In spite of the difference in the lycopene biosynthetic pathway between G. violaceus PCC 7421 and other species, the enzymes in the formation of phytoene have the close phylogenetic relationship supported by more conserved domain. Aside from G. violaceus PCC 7421, the crtP shares more than 60% amino acid identity across different species. With exception of the crtQa from Anabaena sp. PCC 7120, ζ-carotene desaturase also have highly similarity with the amino acid identity from 55% to 99.3% among various cyanobacteria. While the carotene ketolase and carotene hydroxylase in the late steps are significantly less conserved than other enzymes in the pathway.
The diversity of enzyme involved in the desaturation step
Phytoene is converted to lycopene by four-step desaturation and use two related enzymes phytoene desaturase (CrtP/Pds) and ζ-carotene desaturase (CrtQ/Zds) in the most of cyanobacteria; However, G.violaceus PCC7421, like most bacteria and fungi, uses only one enzyme, phytoene desaturase (CrtI)6, catalyzing four-step in this pathway. Surprisingly, homologues of CrtI from G. violaceus PCC7421 are also found in Anabaena variabilis ATCC 29413 (e=0/Identities=57%), Anabaena sp. PCC 7120 (e=0/Identities57%), Trichodesmium erythraeum IMS101 (e=0/Identities=57%), Crocosphaera wastsonii WH8501 (e=0/Identities=55%), Synechocystis sp.PCC6803 (e=0/Identities=56%), which are not involved in the lycopene biosynthetic pathway. Thus, although the crtI homologs in these cyanobacteria appear to be involved in carotenoid biosynthesis, their functions are different from that of crtI in G. violaceus PCC 7421 and bacteria. We therefore propose that these enzymes originated in a same ancestor and then evolved into a different enzyme in different cyanobacteria that produces novel carotenoids that acquire new physiological function. The carotenoid biosynthetic pathway in G. violaceus PCC 7421 is unique contrast to other cyanobacteria. The molecular phylogenetic analysis based on 16S rRNA also demonstrated an isolated position away from other groups of cyanobacteria for G. violaceus 24. This organism is thought to retain traces of the ancestral properties of cyanobacteria.
crtQa from Anabaena sp. PCC 7120 which had been functionally identified to convert ζ –carotene to lycopene 13 while crtQb is involve in this desaturation step in other species 14. By BLASTP program, we also found homologue of crtQb from Anabaena sp. PCC7120, but the information on its function is not available yet. crtQa was found no homologues in other species. Nevertheless both crtQa and crtQb convert ζ –carotene to lycopene, they have no similarity in sequence, and only crtQb displays high conservation with the plant counterparts. The crtQb and crtP from cyanobacteria show high similarities in their amino acid sequence and both contained partial amine oxidoreductase domain. It is very likely that they evolved from the same ancestor. Surprisingly, crtQa is share little sequence similarity to the 'plant-type' phytoene desaturase (crtP gene product), but it has considerable conserved with the bacterial-type enzyme (crtI gene product). It is possible that the cyanobacterial crtQa gene and crtI gene of other microorganisms originated in evolution from a common ancestor.
The evolutionary analysis of crtL- type cyclase and its absence in some species
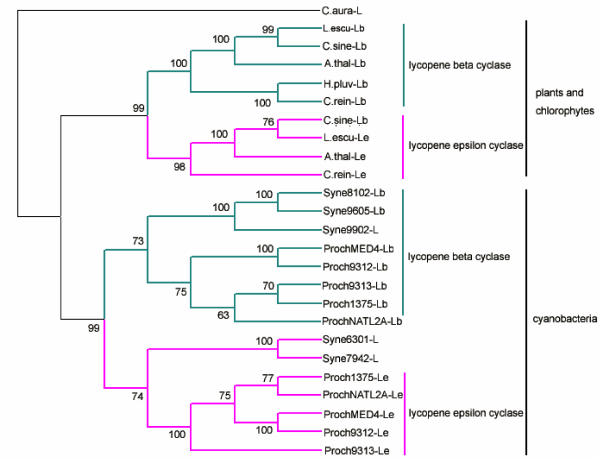
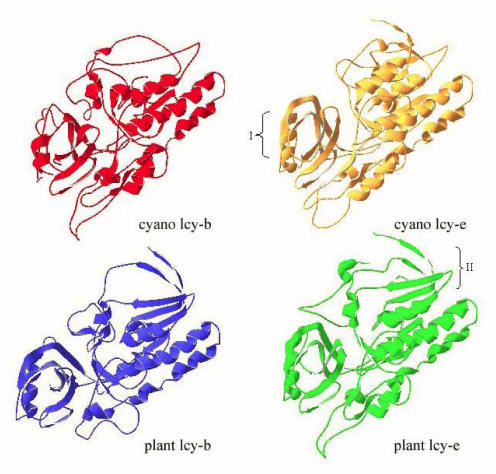
The cyclization reaction of lycopene to β-carotene is also related to different enzymes. The ends of the resulting acyclic lycopene may be cyclized to β-ionone, or ε-ionone rings. The formation of β-ionone rings and of ε-ionone rings in plants is catalyzed by two different enzymes, the β-cyclase and the ε-cyclase. The same case is in some cyanobacteria. Both enzymes show high similarities in their amino acid sequence and it is very likely that they evolve from the same ancestor 25. The phylogenetic relationship among the crtL from cyanobacteria, green algae and higher plants is depicted in Fig.3. From the phylogenetic tree, we can see the enzymes fall into two groups. The cyclase from cyanobacteria separating from other cyclase formed monophyletic group divided into two subclusters containing the β-cyclase and the ε-cyclase. So we suppose that gene may be duplicated after the speciation of the cyanobacteria, chlorophytes and plants. In order to well understand the evolution of the β-cyclase and the ε-cyclase, we examined the tertiary structure using the lycopene cyclases from Arabidopsis thaliana and Prochlorococcus marinus str. MIT 9312 as an example (Fig. 4). A comparable analysis for the tertiary structure of cyclase from cyanobacteria and plants reveal β-cyclase and ε-cyclase have similar structure folds from the same organisms. A single loop formed with five β-strands and one α-helix has conserved in four models, which may be related to binding domain. Several antiparallel β-strands both contained in the tertiary structure of plant-type β-cyclase and ε-cyclase are lacking in that of the cyanobacteria. We supposed lycopene cyclase in a given lineage may evolve through gene duplication that happened after cyanobacteria and chlorophytes/plants speciation event.
Fig 3.
Phylogenetic tree analysis of lycopene cylase based on the amino acid sequences of cyanobacteria, algae and plants by the neighbor-joining method. The Chloroflexus aurantiacus sequence, a distinct ortholog, was chosen to root the tree. Numbers on branches indicate the percentage of 1000 bootstrap replicates that support the adjacent node; Accession numbers: ProchMED4-Lb, NP_893181; Proch9312-Lb, YP_397570; Proch1375-Lb, NP_875528; ProchNATL2A-Lb, YP_291882; Proch9313-Lb, NP_894954; Syne9605-Lb, YP_382237; Syne8102-Lb, NP_896821; Syne7942-L, ZP_00165074; Syne6301-L, YP_172741; Syne9902-Lb, YP_376736; ProchNATL2A-Le, YP_291268; Proch1375-Le, NP_875182; Proch9313-Le, NP_895600; ProchMED4-Le, NP_892751; Proch9312-Le, YP_397130; H.pluv-Lb, AO64977; C. rein-Lb, AAX54906; C. rein-Le, AAT46065; A.thal-Lb, NP_187634; A.thal-Le, NP_200513; L. escu-Le, CAA74745; L.escu-Lb, CAA60170; C.sien-Lb, AAU05146; C.sien-Le, AAS48096, C.auran-L, ZP_00766039.
Fig 4.
Model of structure of lycopene cyclases from cyanobacteria and plants. A, The structure-model of β-lycopene cyclase from Prochlorococcus MIT9312; B, The structure-model of ε-lycopene cyclase from Prochlorococcus MIT9312; C, The structure-model of β-lycopene cyclase from Arabidopsis thaliana; D, The structure-model of ε-lycopene cyclase from Arabidopsis thaliana.
However, it is interesting that only in genus Prochlorococcus, both of lycopene β-(crtL-b) and ε-cyclase (crtL-e) enzymes were found, while, in Synechococcus only one enzyme has good hit with the query sequence. Although there is not only no detectable crtL-e- but also no crtL-b-like lycopene cyclase gene in the genomes of Synechocystis sp. 6803, Thermosynechococcus elongatus, Trichodesmium erythraeum, Gloeobacter, Crocosphaera wastsonii WH8501, Nostoc punctiforme and Anabaena, the related carotenoids had been detected in some species5-7. It would be of interest to know which enzymes converting lycopene to β-carotene in these cyanobacteria. Recently, Takaichi et al (2005) 7 found Anabaena sp. PCC 7120 alr3524 has sequence homology to a new type lycopene cyclase CruA from Chlorobium tepidum 26. Then we used alr3524 from Anabaena sp.7120 and CruA from C. tepidum as query sequence, it is interesting to found homologous enzymes were identified in Thermosynechococcus elongates, Anabaena, Nostoc, Synechocystis sp. PCC 6803, Trichodesmium, C.watsonii WH 8501, G.violaceus PCC7421 other than in Prochlorococcus and Synechococcus (Table1), but their functions have yet to be investigated.
Table 1.
Putative lycopene cylase and gene number of cyanobacteria in which lacking homolog of crtL.
| Species | Query sequence for BLASTP All3524 from Anabaena sp.7120/ CT0456 from C. tepidum | ||
|---|---|---|---|
| ORF | Identity (%) | E value | |
| Anabaena sp. PCC 7120 | Alr3524 Alr0920 | Query/35 31/28 | Query/ 5-67 1e-57/4e-37 |
| A. variabilis ATCC 29413 | Ava3214 Ava4521 | 94/38 31/27 | 0/2e-66 1e-56/5e-36 |
| N. punctiforme PCC 73102 | NpR4002 NpR4207 | 79/38 31/30 | 0/1e-65 7e-54/3e-42 |
| T. erythraeum IMS 101 | Or4079 Or2980 | 61/35 34/30 | 0/5e-65 5e-59/6e-46 |
| C.watsonii WH8501 | Or6211 Or1341 | 64/34 34/30 | 0/9e-66 5e-59/4e-43 |
| Synechocystis sp.PCC 6803 | Sll0147 Sll0659 | 62/34 32/31 | 0/5e-65 2e-67/1e-44 |
| G. violaceus PCC 7421 | Gll3598 Gll2484 | 55/34 33/30 | 0/5e-64 1e-54/9e-42 |
| Thermosynechococcus elongatus | Tlr1139 | 60/34 | 0/2e-66 |
| Synechococcus sp. PCC 6301 | Syn0876_d | 33/28 | 2e-58/2e-38 |
| Synechococcus sp. PCC 7942 | Syn_PCC79420625 | 33/38 | 2e-58/2e-38 |
Note: Database searches were carried out with the BLAST program. The value before virgule is the subject with Alr3524 as the query sequence; the value behind virgule is the subject with CT0456.
Conserved domain between crtW-type ketolase and crtR-type hydroxylase
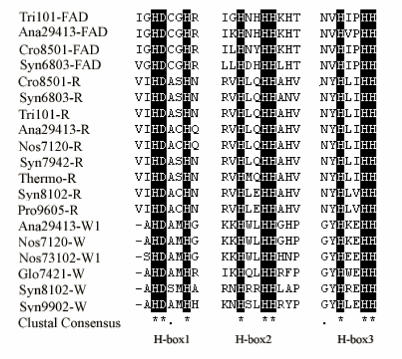
Two distinct β-carotene ketolase genes, crtW and crtO, were found in the genome sequences of cyanobacteria. Anabaena sp. PCC 7120, N.punctiforme PCC 73102, Anabaena ATCC 29413 and G.violaceus PCC 7421 and Synechocystis sp. PCC 6803 were found contain crtO homologous gene, Synechococcus WH8102 and Synechococcus sp. CC9902 were found contain crtW homologous genes. Although these two enzymes involve the same β-carotene ketolation, the characteristics of enzymes are different. CrtO and crtW do not share significantly amino acid sequence homology. CrtOs have six conserved regions including the FAD binding motif 27 and show partial amino oxidase domain, while crtWs sharing three typical histidine rich motifs (Table2) show some characters of fatty acid desaturase. Carotenoid hydroxylases (crtR) in cyanobacteria bears little or no relationship to the carotennoid hydroxydrases from plants and bacteria. It shows some similarity to crtW-type ketolase, especially conserved in the three H-Boxes (Fig.5), which reveal the crtR and crtW might have a common ancestor and acquire the different function during the evolution. The origin and phylogenetic position of crtR and crtW relative to other members of the three H–boxes FA protein family is of considerable interest.
Table 2.
Conserved motifs in crtW-type ketolase.
| Name | H-box1 | H-box2 | H-box3 |
|---|---|---|---|
| Ana29413W1 | TAHDAMH | KHXLHH | CYHFGYHXEHH |
| Ana7120-W | TAHDAMH | KHXLHH | CYHFGYHXEHH |
| Ana29413-W2 | TAHDAMH | KHXLHH | CYHFGYHXEHH |
| Nos73102W1 | TSHDAMH | KHXLHH | CYHFGYHXEHH |
| Nos73102W2 | TAHDAMH | NHXLHH | CYHFGYHXEHH |
| Glo7421-w | TAHDAMH | KHXLHH | CYHFGYHXEHH |
| Syn8102-w | VAHDAMH | NHXRHH | CYNFGYHXEHH |
| Syn9902-w | VAHDAMH | NHXLHH | CYHFGYHXEHH |
Fig 5.
Comparison of the amino acid sequence of fatty acid desaturases, β-hydroxylase and β-ketolase. The conserved H-boxes were in black. 'FAD' represents fatty acid desaturase; 'R' represents crtR; 'W' represents crtW. The sequence sources are as follow: Tri101-FAD ZP_00675708; Ana29413-FAD YP_324705; Syn6803-FAD NP_441489; Cro8501-R ZP_00514501; Syn6803-R NP_440788; Ana29413-R YP_322210; Ana7120-R, NP_488049; Syn7942-R, YP_401456; Thermo-R NP_682690; Pro9605-R YP_380617; Ana29413-W1 YP_322565; Ana7120-W NP_487229; Nos73102-W1 ZP_00111258; Glo7421-W NP_924674; Syn8102-W NP_897461; Syn9902-W YP_376982.
Structure of crt gene cluster in the cyanobacterial chromosomes
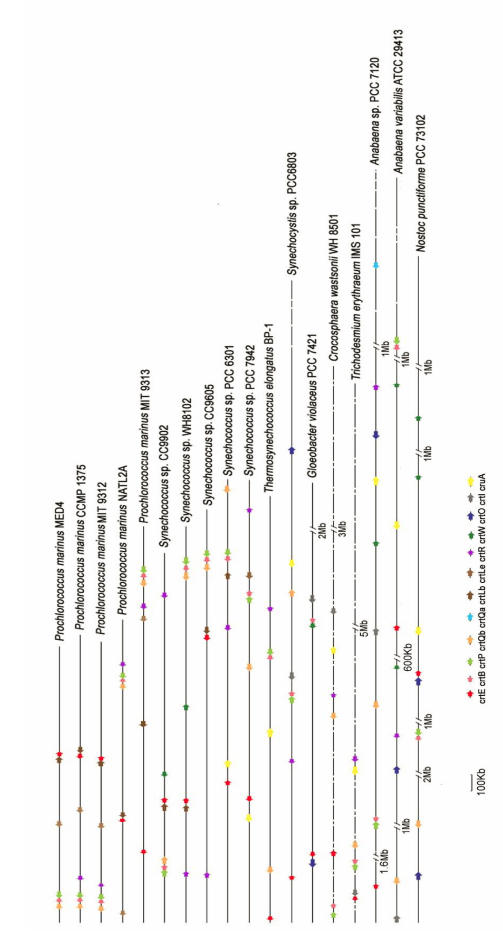
To elucidate the complete genomic structure of the crt genes, we mapped them onto cyanobacterial genomes (Fig. 6). The structure of the crt gene clusters varies greatly among species. In Prochlorococcus, crtB, crtP and crtQb often clustered and transcribed in the same direction. Actually, in many cases, crtB and crtP are directly adjacent to each other on the chromosome and may form an operon. crtE and crtL-b formed an operon in Prochlorococcus NATL, Prochlorococcus MED4, Synechococcus WH8102 and Synechococcus sp. CC9902. While other crt genes are arranged in random in the genome and they are not always transcribed in the same direction. It is interesting cyanobacteria is distinct from other eubacteria in the organization of crt clusters although forming a coherent systematic group, genes for carotenoid biosynthetic enzymes are frequently clustered into large operons 28-30 in typical bacteria, but this does not appear to be the case in cyanobacteria. Although the cartoenoid biosynthetic pathway in G. violaceus PCC 7421 is similar with other eubacteria, the genomic structure of crt genes is not distinct from other cyanobacteria.
Fig 6.
Genomic organization of the crt genes potentially encoding enzymes involved in the carotene biosynthetic pathway of various cyanobacterial species. Long horizontal line indicates the chromosome, whereas short horizontal lines denote the extranuclear plasmids. Deduced chromosomal positions of the crt genes are marked by arrows with different colors. Arrows represent the direction of translation and the relative sizes of ORFs deduced from analysis of the nucleotide sequence. Note that C.watsonii WH8501 and T. erythraeum IMS 101 genomes were still in draft format, and crt genes were mapped onto the chromosome evenly. Gene names are given at the bottom of the figure.
The crt genes are arranged in random in the cyanobacteria chromosomes. These loosely organized operon structures are sometimes considered ''destructed'' due to genome rearrangement, and secondary in origin 31. While genome rearrangement and even gene displacement can be common during operon evolution 32, fragmentation of a well adapted operon will at least require the evolution of regulatory elements for newly generated gene clusters. The crt genes will acquire the new regulatory elements respectively to adapt for new environments.
Each of these enzymes is a single-gene produce in most cases. Multiple copies of ketolases were only identified in the filamentous species. Actually, two carotenoid ketolase genes crtW38 and crtW148 were cloned from the cyanobacterium, Nostoc punctiforme PCC 73102 and functionally characterized 17. Scanning the genomics of all species for crt genes by the similarity search we also found two crtO ketolases and two crtW existed in Nostoc punctiforme PCC 73102 and Anabaena ATCC 29413 respectively. There are no paralogous copies of crt genes other than in filamentous cyanobacteria. Most of filamentous cyanobacteria exhibit a wide range of ecological tolerance and are found in freshwater, marine and terrestrial habitats. The increased number of isozymes associated with pigment biosynthesis in filamentous cyanobacteria relative to unicellular species may be related to increased regulatory demands and perhaps also to different local environments.
Table 3.
The gene IDs of putative othologous genes for Fig.2.
| Species | Gene product | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| crtE | crtB | CrtP | crtI | crtQ | crtL | cruA | crtW | crtO | crtR | ||
| Anabaena sp. 7120 | 4211070 | 4227390 | 4227380 | 4237060 | 4265750(a) 4233000 | 4244510 | 4241130 | 4246730 | 4249390 | ||
| Nostoc punctiforme ATCC 29133 | 402037140 | 402026830 | 402026840 | 402003840 | 402039490 | 402047580 402059010 | 402036890 402001100 | 402042230 | |||
| Trichodesmium erythraeum IMS101 | 403261180 | 403262620 | 403262610 | 403225420 | 403263060 | 403259560 | 403223080 | ||||
| Gloeobacter violaceus PCC 7421 | 4779990 | 4793460 | 4795260 | 4812220 | 4793300 | 4779770 | |||||
| Crocosphaera watsonii WH8501 | 400849100 | 400840970 | 400840960 | 400864170 | 400880630 | 400887960 | 400880820 | ||||
| Synechocystis sp. PCC 6803 | 4274680 | 4286390 | 4286380 | 4287260 | 4291990 | 4293600 | 4299880 | 4282550 | |||
| Thermosynechococcus BP-1 | 4667100 | 4682740 | 4682750 | 4670320 | 4678460 | 4686180 | |||||
| Synechococcus sp. WH 8102 | 5175220 | 5190760 | 5190770 | 5190320 | 5175080 | 5181560 | 5170660 | ||||
| Synechococcus sp. PCC6301 | 610215650 | 610229570 | 610229580 | 610233440 | 610228750 | 610216840 | 610225020 | ||||
| Synechococcus sp. 7942 | 403097570 | 403109760 | 403109750 | 403105020 | 403110550 | 403096300 | 403114410 | ||||
| Synechococcus sp. CC9902 | 403151250 | 403146750 | 403146740 | 403147120 | 403151100 | 403153610 | 403164780 | ||||
Acknowledgments
This research was supported by funds named Key Innovative Project of Chinese Academy of Sciences (KZCX3-SW-215).
References
- 1.Olson JA, Krinsky NI. The colorful, fascinating world of the carotenoids: important physiologic modulators. FASEB J. 1995;9:1547–1550. doi: 10.1096/fasebj.9.15.8529833. [DOI] [PubMed] [Google Scholar]
- 2.Altermann W, Kazmierczak J. Archean microfossils: a reappraisal of early life on Earth. Res Microbiol. 2003;154:611–617. doi: 10.1016/j.resmic.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Schopf JW. Microfossils of the early Archean apex chert: new evidence of the antiquity of life. Science. 1993;260:640–646. doi: 10.1126/science.260.5108.640. [DOI] [PubMed] [Google Scholar]
- 4.Bryant DA. The molecular biology of cyanobacteria. Kluwer Academic publishers. 1996. pp. 559–579.
- 5.Steiger S, Jackisch Y, Sandmann G. Carotenoid biosynthesis in Maresca Gloeobacter violaceus PCC7421 involves a single crtI-type phytoene desaturase instead of typical cyanobacterial enzymes. Arch Microbiol. 2005;184:207–214. doi: 10.1007/s00203-005-0004-5. [DOI] [PubMed] [Google Scholar]
- 6.Takaichi S, Maoka T, Masamoto K. Myxoxanthophyll in Synechocystis sp PCC 6803 is Myxol 2'-Dimethyl-Fucoside, (3R,2'S)-Myxol 2'-(2,4-di-O-Methyl-α-L-Fucoside), not rhamnoside. Plant Cell Physiol. 2001;42:756–762. doi: 10.1093/pcp/pce098. [DOI] [PubMed] [Google Scholar]
- 7.Takaichi S, Mochimaru M, Maoka T, Katoh H. Myxol and 4-ketomyxol 2'-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol. 2005;46:497–504. doi: 10.1093/pcp/pci049. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong GA, Alberti M, Hearst JE. Conserved enzymes mediate the early reactions of carotenoid biosynthesis in nonphotosynthetic and photosynthetic prokaryotes. Proc Natl Acad Sci USA. 1990;87:9975– 9979. doi: 10.1073/pnas.87.24.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong GA. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu Rev Microbiol. 1997;51:629– 659. doi: 10.1146/annurev.micro.51.1.629. [DOI] [PubMed] [Google Scholar]
- 10.Phadwal K. Carotenoid biosynthetic pathway: molecular phylogenies and evolutionary behavior of crt genes in eubacteria. Gene. 2005;345:35– 43. doi: 10.1016/j.gene.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Sandmann G. Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiol Plant. 2002;116:431–572. [Google Scholar]
- 12.Sandmann G. Phytoene desaturases: genes, enzymes and phylogenetic aspects. J Plant Physiol. 1994;143:444–447. [Google Scholar]
- 13.Linden H, Misawa N, Saito T, Sandmann G. A novel carotenoid biosynthesis gene coding for ζ-carotene desaturase: functional expression, sequence and phylogenetic origin. Plant Mol Biol. 1994;24:369–379. doi: 10.1007/BF00020174. [DOI] [PubMed] [Google Scholar]
- 14.Breitenbach J, Fernández-González B, Vioque A, Sandmann G. A higher-plant type ζ-carotene desaturase in the cyanobacterium Synechocystis PCC 6803. Plant Mol Biol. 1998;36:725–732. doi: 10.1023/a:1005997405431. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Gonzalez B, Sandmann G, Vioque A. A new type of asymmetrically acting β-carotene ketolase is required for the synthesis of echinenone in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1997;272:9728–9733. doi: 10.1074/jbc.272.15.9728. [DOI] [PubMed] [Google Scholar]
- 16.Misawa N, Satomi Y, Kondo K, Yam AY, Kajiwara S, Saito T, Ohtani T, Miki W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. 1995;177:6575–6584. doi: 10.1128/jb.177.22.6575-6584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiger S, Sandmann G. Cloning of two carotenoid ketolase genes from Nostoc punctiforme for the heterologous production of canthaxanthin and astaxanthin. Biotech Lett. 2004;26:813–817. doi: 10.1023/b:bile.0000025880.91269.cf. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequences alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: exible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. J Mol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Tamura K, Jakobsen IB. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 24.Honda D, Yokota D, Sugiyama J. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J Mol Evol. 1999;48:723–739. doi: 10.1007/pl00006517. [DOI] [PubMed] [Google Scholar]
- 25.Krubasik P, Sandmann G. Molecular evolution of lycopene cyclases involved in the formation of carotenoids with ionone end groups. Biochemical Society Transactions. 2000;28:806–810. [PubMed] [Google Scholar]
- 26.Maresca JA, Frigaard NF, Bryant DA. Identification of a novel class of lycopene cyclases in photosynthetic bacteria. In: van der Est A, Bruce D, editors. Photosynthesis: Fundamental Aspects to Global Perspectives. Allen Press; 2005. [Google Scholar]
- 27.Mochimaru M, Masukawa H, Takaichi S. The cyanobacterium Anabaena sp. PCC 7120 has two distinct β-carotene ketolases: CrtO for echinenone and CrtW for ketomyxol synthesis. FEBS Letters. 2005;579:6111–6114. doi: 10.1016/j.febslet.2005.09.081. [DOI] [PubMed] [Google Scholar]
- 28.Krubasik P, Sandmann G. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol Gen Genet. 2000;263:423–432. doi: 10.1007/s004380051186. [DOI] [PubMed] [Google Scholar]
- 29.Viveiros M, Krubasik P, Sandmann G, Houssaini-Iraqui M. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+ FEMS Microbiol Lett. 2000;187:95–101. doi: 10.1111/j.1574-6968.2000.tb09143.x. [DOI] [PubMed] [Google Scholar]
- 30.Sedkova N, Tao L, Rouvière PE, Cheng Q. Diversity of carotenoid synthesis gene clusters from environmental Enterobacteriaceae strains. Appl Environ Microb. 2005;17:8141–8146. doi: 10.1128/AEM.71.12.8141-8146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol Biol Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 32.Omelchenko MV, Makarova KS, Wolf YI, Rogozin IB, Koonin EV. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 2003;4:R55. doi: 10.1186/gb-2003-4-9-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]