Abstract
CTCF (CCCTC-binding factor) binds sites around the mouse β-globin locus that spatially cluster in the erythroid cell nucleus. We show that both conditional deletion of CTCF and targeted disruption of a DNA-binding site destabilize these long-range interactions and cause local loss of histone acetylation and gain of histone methylation, apparently without affecting transcription at the locus. Our data demonstrate that CTCF is directly involved in chromatin architecture and regulates local balance between active and repressive chromatin marks. We postulate that throughout the genome, relative position and stability of CTCF-mediated loops determine their effect on enhancer–promoter interactions, with gene insulation as one possible outcome.
Keywords: CTCF, globin, insulator, higher-order chromatin structure, looping, transcription
Chromatin insulators are DNA sequences that confer autonomous expression on genes by protecting them against inadvertent signals coming from neighboring chromatin. CTCF (CCCTC-binding factor) is the prototype vertebrate protein exhibiting insulator activity (West et al. 2002) that can act as an enhancer blocker or as a barrier against repressive forces from nearby heterochromatin in vitro (Defossez and Gilson 2002; Recillas-Targa et al. 2002). In vivo, CTCF binds to the imprinting control region of the H19/insulin-like growth factor (Igf2) locus, where it acts as a methylation-sensitive enhancer blocker (Bell and Felsenfeld 2000; Hark et al. 2000). Moreover, CTCF-binding sites have been found—and its insulator activity has been anticipated—at the imprinting center that determines choice of X inactivation (Chao et al. 2002), at boundaries of domains that escape X inactivation (Filippova et al. 2005), and at sites flanking CTG/CAG repeats at the DM1 locus (Filippova et al. 2001). CTCF was first defined as an insulator protein when it was found to be required for the enhancer-blocking activity of a hypersensitive site 5′ of the chicken β-globin locus (5′HS4) (Bell et al. 1999). A similar CTCF-dependent insulator site was subsequently found at the 3′ end of the locus and both sites coincide with erythroid-specific transitions in DNase I sensitivity of chromatin (Saitoh et al. 2000). Such observations suggested that CTCF partitions the genome in physically distinct domains of gene expression. The molecular mechanism underlying CTCF's insulating activity is still unknown.
CTCF-binding sites also flank the human and mouse β-globin locus (Fig. 1A), which contains a number of developmentally regulated, erythroid-specific β-globin genes and an upstream locus control region (LCR) required for high β-globin expression levels. In mice, three CTCF-binding sites have been identified upstream (HS-85, HS-62, and HS5) and one downstream (3′HS1) of the locus (Farrell et al. 2002; Bulger et al. 2003). Previously, we applied chromosome conformation capture (3C) technology (Dekker et al. 2002) to study long-range DNA interactions between these and other sites in the β-globin locus. In erythroid cells, the CTCF-binding sites (including HS-85; see below) were found to participate in spatial interactions between the LCR and the active β-globin genes, and collectively form an active chromatin hub (ACH) (Tolhuis et al. 2002). No such long-range DNA interactions were detected in nonerythroid cells. However, in established I/11 erythroid progenitor cells that do not yet show activated β-globin gene expression, contacts between the LCR and the genes are absent, but long-range DNA interactions already exist between the hypersensitive sites that contain CTCF-binding sites (Palstra et al. 2003). Here, we investigated the involvement of CTCF in the formation of these loops.
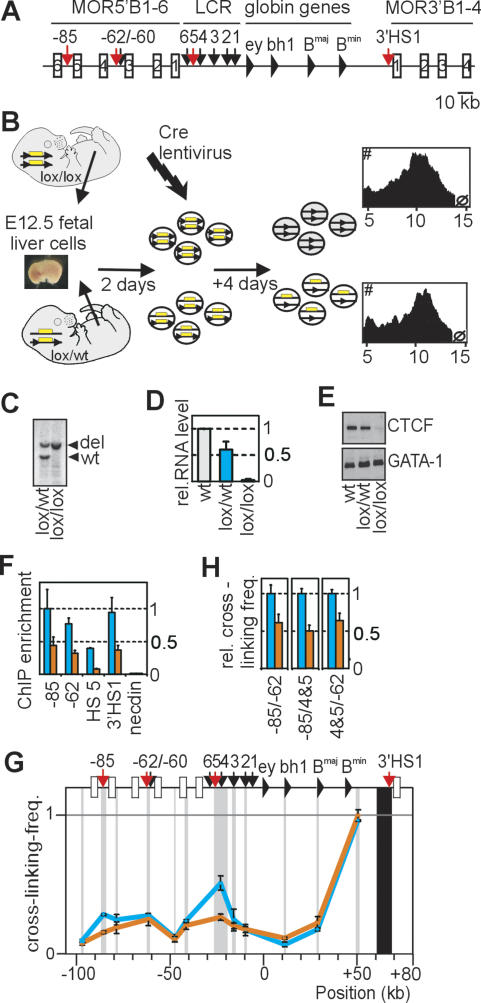
Figure 1.
Deleting CTCF in primary erythroid progenitors reduces the frequency of interactions between cognate binding sites in the β-globin locus. (A) Schematic presentation of the mouse β-globin locus. DNase I hypersensitive sites (arrows) and CTCF-binding sites (red) are indicated. (B) Strategy to delete CTCF. Plots show a similar cellular size distribution for homozygous and heterozygous conditional knockout cells. (C) Southern blot analysis showing complete deletion of CTCF conditional knockout alleles. (D) CTCF mRNA levels (as determined by Q-RT–PCR) in untreated wild-type (level set to 1) and Cre-recombined lox/wt and lox/lox cells. Standard deviation is indicated. (E) Western blot analysis of CTCF protein and GATA-1 protein (control, stripped, and rehybridized blot). (F) CTCF ChIP analysis. Lox/lox (orange) vs. control cells (blue). (G) 3C analysis demonstrating reduced interaction frequencies between 3′HS1, HS4/5, and HS-85 in lox/lox cells (orange), as compared with control cells (blue). (H) 3C analysis demonstrating reduced interaction frequencies in lox/lox cells between the other CTCF-binding sites in the β-globin locus.
Results and Discussion
β-globin locus conformation in erythroid cells with reduced levels of CTCF protein
To investigate the role of CTCF in the formation of chromatin loops, we analyzed β-globin DNA contacts in cells lacking the CTCF protein. Analysis was focused on embryonic day 12.5 (E12.5) erythroid progenitor cells because they can be expanded ex vivo (Dolznig et al. 2001) and lack stable LCR–gene contacts, and therefore best reveal the interactions between outer hypersensitive sites. Chromatin immunoprecipitation (ChIP) experiments revealed that CTCF was bound in vivo to cognate sites in the β-globin locus in these cells (Fig. 1F), while the protein was absent from HS5 and 3′HS1 in brain cells not showing these loops (Supplementary Fig. 1). Since CTCF-null mice die early during embryogenesis, a conditional knockout mouse model was generated by inserting two lox sites upstream of and downstream from the first and last coding exon of CTCF, respectively. To delete CTCF, fetal liver cells were isolated from lox/lox E12.5 embryos, cultured under conditions that select erythroid progenitors, and infected with a replication-deficient lentivirus expressing Cre recombinase (Fig. 1B). Heterozygous (lox/wt) cells from littermate embryos underwent the same treatment and served as controls. Cre recombination resulted in nearly 100% deletion of targeted CTCF alleles, with a reduction in mRNA and protein levels to 2%–3% and 10%–25%, respectively, in lox/lox as compared with wild type (Fig. 1C–E). CTCF binding to cognate sites in the β-globin locus was reduced but not completely abolished in lox/lox cells, as demonstrated by ChIP (Fig. 1F). To investigate β-globin locus conformation in these cells by 3C technology, we used a novel TaqMan probe-based quantitative PCR (Q-PCR) strategy to accurately quantify 3C ligation efficiencies (Supplementary Fig. 2). We found that the structure of the β-globin locus in wild-type and lox/wt E12.5 progenitor cells was essentially the same as previously observed in I/11 progenitor cells (data not shown), with long-range interactions between the CTCF-binding sites HS-85, HS-62/60, HS4/5, and 3′HS1 (Fig. 1G). In lox/lox cells containing lower levels of CTCF protein, however, clearly reduced DNA–DNA interaction frequencies were observed specifically between the sites that normally bind CTCF (Fig. 1G,H). This is true for all combinations of binding sites, except for the interaction between 3′HS1 and HS-62 (see below). The results demonstrated that CTCF is required for long-range DNA–DNA interactions between cognate binding sites in the β-globin locus. Gene expression analysis revealed the same low levels of expression for all β-globin genes in wild-type versus lox/lox progenitor cells (Supplementary Fig. 3). Moreover, we did not find activation of any of the mouse olfactory genes immediately surrounding the β-globin locus (MOR5B1-3 and MOR3B1-4) (data not shown). Hence, the reduction of CTCF protein to low levels had no appreciable effect on gene expression at or around the β-globin locus in erythroid cells, representing a differentiation stage prior to LCR-mediated gene activation.
Long-range interactions of 3′HS1 containing nucleotide changes that disrupt CTCF binding
The structural changes in the β-globin locus that we observed in cells with deleted CTCF may be a direct consequence of reduced protein binding to the locus, or could be caused by secondary pathways that fail to act on the locus in the absence of sufficient CTCF. To investigate this, we disrupted CTCF binding locally by changing four conserved nucleotides in the core CTCF-binding site of the endogenous 3′HS1 (Supplementary Fig. 4). Bandshift assays confirmed that these alterations completely abolished CTCF binding in vitro (Supplementary Fig. 5). Targeting was performed in embryonic stem (ES) cells that were established from a cross between the two inbred strains 129 and C57BL/6 (B6) and was directed to the B6 allele. Two additional, nonconserved nucleotides were changed 70 base pairs (bp) downstream from the core CTCF-binding site to allow allele-specific analysis of CTCF binding to 3′HS1 by ChIP. Moreover, an extra HindIII restriction site was introduced ~850 bp downstream from the CTCF-binding site, which enabled us to exclusively analyze DNA interactions of the targeted 3′HS1 by 3C. An independent control ES line was generated containing the extra HindIII site with the normal 3′HS1. In each cell line, the neomycin selection cassette was removed by transient expression of Cre recombinase, leaving behind a single lox site immediately downstream from the newly introduced HindIII site (Supplementary Fig. 4).
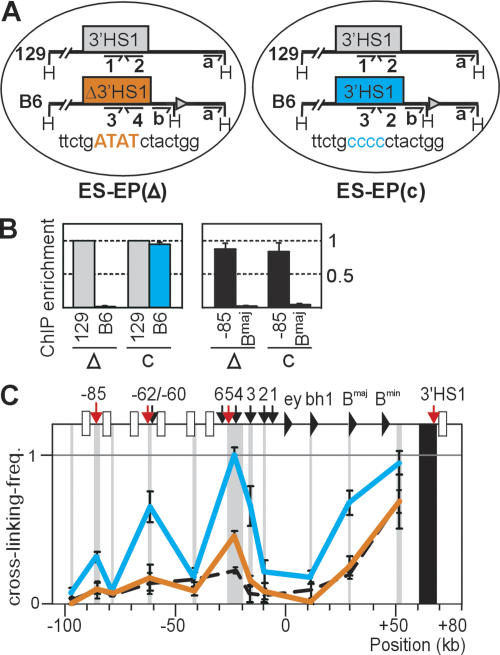
Definitive erythroid progenitors were generated from the ES cells in vitro to analyze the consequences of the targeted nucleotide changes in erythroid cells. We established two such ES-EP cell lines, ES-EP(Δ3′HS1) (or Δ) and the control line ES-EP(c) (or c) (Fig. 2A). After validation of the cells as a model system for erythroid differentiation (Supplementary Fig. 6; Carotta et al. 2004), we analyzed CTCF binding to mutated and wild-type 3′HS1 in vivo. In the control line ES-EP(c), CTCF bound strongly and equally well to 3′HS1 on both alleles. In ES-EP(Δ3′HS1), however, binding to 3′HS1 on the non-targeted 129 allele was the same as in ES-EP(c), but binding to the mutated 3′HS1 on the targeted B6 allele was completely abolished (Fig. 2B). Thus, the change of 4 nucleotides (nt) in the core CTCF-binding site also prevented binding of CTCF to 3′HS1 in vivo. Next we analyzed whether disruption of CTCF binding at 3′HS1 affected its long-range DNA interactions in the β-globin locus. The extra HindIII restriction site introduced downstream from 3′HS1 was used to focus 3C analysis exclusively on the targeted B6 allele. In undifferentiated ES-EP(c), the wild-type β-globin B6 allele formed a chromatin hub typically observed in normal erythroid progenitor cells, with 3′HS1 interacting with HS4/5, HS-62, and HS-85 (Fig. 2C). In undifferentiated ES-EP(Δ3′HS1), however, the mutated 3′HS1 showed a dramatic drop in interaction frequencies with all these DNA elements, to levels similar to those observed in nonexpressing fetal brain cells (Fig. 2C). Thus, disruption of CTCF binding to 3′HS1 severely destabilized the large chromatin loop containing the LCR and the globin genes in erythroid progenitor cells. The fact that the interaction with HS-62 was lost upon targeted disruption of CTCF binding to 3′HS1, but not in our conditional CTCF knockout experiments, suggests that this interaction is more resistant than others to the reduction of levels of CTCF protein.
Figure 2.
Targeted nucleotide changes in 3′HS1 disrupt CTCF binding and reduce frequency of long-range 3′HS1 interactions. (A) Erythroid progenitor cell lines derived in vitro from ES cells. ESEP(Δ3′HS1) harbors four targeted nucleotide changes in the core CTCF-binding site of 3′HS1 on the B6 allele (orange). ES-EP(c) contains wild-type 3′HS1 on the B6 allele (blue). The nontargeted, intact 129 allele is in gray. For ChIP, each 3′HS1 CTCF-binding site can be analyzed with a unique primer pair (#1–4). An extra HindIII site targeted downstream from 3′HS1 allows exclusive 3C analysis of B6 allele (with 3C-primer b). (B) ChIP on undifferentiated ES-EP cell lines, with antibody against CTCF. (Left) 3′HS1 alleles in the two ES-EP lines (Δ and c). (Right) Positive (HS-85) and negative (βmajor) controls. (C) 3C analysis with primer b (see above). Note that interaction frequencies with mutated 3′HS1 (orange) are reduced compared with wild-type 3′HS1 (blue). (Black dashed line) 3′HS1 interactions in fetal brain, analyzed with primer a and plotted for comparison.
Expression of β-globin and surrounding olfactory receptor genes in the absence of CTCF-mediated chromatin loops
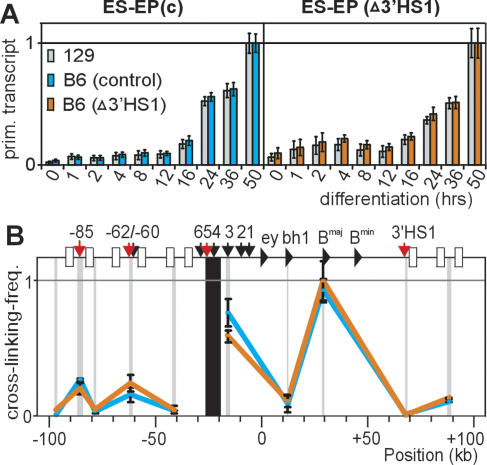
Since the large, CTCF-dependent loops are formed only in human or mouse cells that are committed to, or highly express, the β-globin genes (Palstra et al. 2003), we investigated the relationship between these loops and transcriptional regulation in detail. First, we analyzed whether CTCF at 3′HS1 serves as an enhancer blocker that prevents the inappropriate activation of downstream mouse olfactory receptor genes (MORs) by the β-globin LCR in erythroid cells, as suggested previously (Farrell et al. 2002). For this, we compared mRNA levels of the MOR3′B1-4 genes between differentiated ES-EP(c) and ES-EP(Δ3′HS1) cells when the LCR is fully active. We found no inappropriate activation of any of the downstream MORs, or of MOR5′B3, in the differentiated ESEP(Δ3′HS1) cells (data not shown), and we concluded that insulator activity of CTCF at 3′HS1 is not required for blocking LCR-mediated activation of downstream MOR genes in ES-EP cells. Noteworthy, it was previously found that deletion of the complete HS5 from the endogenous locus also had no effect on expression of the surrounding MOR genes (Bulger et al. 2003). We envision that the transcription factor environment in erythroid cells does not support the activation of olfactory receptor genes. Next, we analyzed whether the CTCF-dependent loops influence β-globin gene expression. Upon erythroid differentiation, the LCR forms stable contacts with the active β-globin genes and strongly up-regulates their transcription rate (Carter et al. 2002; Tolhuis et al. 2002). We reasoned that a shared presence on one chromatin loop anchored by CTCF in progenitor cells would decrease the spatial distance between the LCR and genes, which may facilitate their productive interaction later during differentiation. If true, the absence of such a pre-existing loop could possibly result in a delay of full β-globin gene activation. To test this, we compared the kinetics of LCR-mediated gene activation between the individual alleles of differentiating ES-EP(c) and ESEP(Δ3′HS1) cells. Two sets of βmajor intron primers were designed that allowed independent analysis of ongoing transcription from the B6 allele and 129 allele (Supplementary Fig. 7). ES-EP(c) and ES-EP(Δ3′HS1) cells were induced to undergo synchronous differentiation, and RNA was collected at various time intervals. As expected, βmajor transcription rates increased strongly upon differentiation. However, at each given stage of differentiation, we detected the same gene activity between the 129 allele and B6 allele, both in ES-EP(c) and ESEP(Δ3′HS1) cells (Fig. 3A). Thus, the CTCF-dependent chromatin loop with 3′HS1 that topologically defines the β-globin locus in erythroid cells does not detectably influence the expression kinetics or levels of the β-globin and nearby MOR genes. This was also true for the embryonic β-globin genes εy and βH1, which were off in both cell lines before and after differentiation (data not shown).
Figure 3.
Targeted nucleotide changes in 3′HS1 do not affect β-globin gene expression. (A) Ongoing βmajor transcription as measured by Q-RT–PCR, using 129- and B6-specific primers against intron 2 of the βmajor gene. (X-axis) Hours after induction of differentiation. Error bars represent standard error of mean. (B) Locus-wide, B6-specific analysis of interaction frequencies with HS4/5 after differentiation. Note that primer a was used near 3′HS1, which on the B6 allele analyzes a small (0.5-kb) fragment downstream from (i.e., not containing) 3′HS1, whereas on the 129 allele this primer would analyze an ~8-kb fragment encompassing 3′HS1. The dramatic drop in interaction frequencies shows that analysis is restricted to the B6 allele.
Establishment of LCR–gene contacts in the absence of a pre-existing loop with 3′HS1
The unaltered β-globin gene expression patterns from the targeted allele in ES-EP(Δ3′HS1) cells suggested that in the absence of a pre-existing chromatin loop, LCR–gene contacts can still be established normally upon erythroid differentiation. To test this, we searched for 129/ B6 polymorphisms near restriction sites in the LCR that would allow allele-specific 3C analysis. This resulted in the design of a TaqMan probe for a HindIII fragment encompassing HS4 and HS5 that signals exclusively from the B6 allele (Supplementary Fig. 8). Although HS4 and HS5 are not prime candidates in the LCR to directly contact the genes, this relatively large HindIII fragment was previously shown to be representative of the complete LCR, since it displayed a prominent peak of interaction with the βmajor gene upon erythroid differentiation (Tolhuis et al. 2002). In both differentiated ES-EP(c) and ES-EP(Δ3′HS1) cell lines, we found identical locus-wide interaction frequencies for HS4/5 between the B6 alleles containing either wild-type or mutated 3′HS1, and both showed a strong peak of interaction with the βmajor gene (Fig. 3B). This demonstrated that a pre-existing loop between upstream sites and 3′HS1 is dispensable for the establishment of stable LCR–gene contacts later during erythroid differentiation. Such a conclusion is in agreement with transgenic experiments showing full β-globin expression from constructs lacking 3′HS1 (Strouboulis et al. 1992).
Histone modifications in the absence of CTCF binding
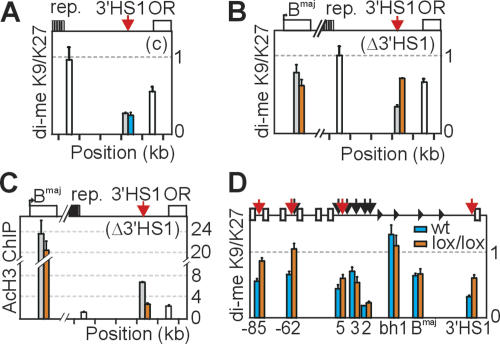
3′HS1 was previously shown to be present in, and close to, the 3′ border of an open chromatin domain spanning ~145 kb around the β-globin locus in erythroid cells (Bulger et al. 2003). Within this domain, a large (~15 kb) region of highly repetitive DNA is present ~3 kb upstream of 3′HS1 that cannot be analyzed for nuclease sensitivity, but likely is packed into compact chromatin. To further investigate this, we analyzed histone modifications at and directly around 3′HS1 in ES-EP cells. Using an antibody that recognizes both dimethyl H3K9 and dimethyl H3K27, we found that these repressive marks were abundantly present on both sides of 3′HS1, but not inside 3′HS1 (Fig. 4A). Conversely, acetylation of histone H3, a mark for open chromatin, was clearly enriched at 3′HS1 but not, or much less, at sites surrounding the hypersensitive site (Fig. 4C). These data argued against the existence of a large open chromatin domain extending across 3′HS1 and suggested that 3′HS1 forms an isolated entity of open chromatin. To address whether CTCF plays a role in the establishment of this pattern, we performed ChIP on ES-EP(c) and ES-EP(Δ3′HS1) cells and used allele-specific primer pairs to compare modifications at 3′HS1 on targeted versus nontargeted alleles. In the control cell line, we found identical levels of di-meH3K9/K27 at 3′HS1 on the two alleles. In ES-EP(Δ3′HS1), however, loss of CTCF binding was accompanied by an increase of di-meH3K9/K27 and concomitant decrease of AcH3 at 3′HS1 (Fig. 4B,C). We found no indication for spreading of the methyl mark into the locus, either locally (Fig. 4, cf. levels of enrichment in A and B) or at the βmajor gene, which locates more inside the locus (Fig. 4B, analyzed by allele-specific primers). In fact, AcH3 levels at βmajor were also similar for the targeted and nontargeted allele in ES-EP(Δ3′HS1) cells—two observations that were fully in agreement with our finding that βmajor gene expression was not affected by disrupted CTCF binding to 3′HS1 (Fig. 3A).
Figure 4.
Histone modifications in the absence of CTCF binding. (A) ChIP enrichment for di-meH3K9/K27 in undifferentiated control ES-EP(c) cells on the 129 allele (gray), B6 allele (blue), or on both alleles (white). Note that values in A–C were normalized to input, and therefore white bars also represent enrichment per allele. (B,C) ChIP enrichment for di-meH3K9/K27 (B) and acetylated histone H3 (C) in undifferentiated ES-EP(Δ3′HS1) cells. (Orange bars) B6 allele; (gray bars) 129 allele. Note increased methylation (B) and decreased acetylation (C) only at mutated 3′HS1 on B6 allele. (D) ChIP enrichment for di-meH3K9/K27 at the β-globin locus in control (blue) and conditional CTCF knockout (lox/lox) (orange) E12.5 erythroid progenitors cells. Note that reduced levels of CTCF cause an increase in di-meH3K9/K27 specifically at the CTCF-binding sites HS-85, HS-62, HS5, and 3′HS1.
We considered the possibility that the spreading of dimeH3K9/K27 into the locus requires the disruption of CTCF binding to more sites than just 3′HS1. To investigate this, we compared di-meH3K9/K27 levels in E12.5 wild-type versus lox/lox conditional CTCF knockout progenitor cells, the latter containing reduced amounts of CTCF (Fig. 1). We found that loss of CTCF binding to 3′HS1, HS5, HS-62, and HS-85 coincided with locally increased amounts of di-meH3K9/K27, while modification levels elsewhere in the locus appeared unaffected (Fig. 4D). Since CTCF binding to β-globin sites was reduced but not absent in lox/lox cells, this leaves open the possibility that residual CTCF amounts prevent spreading of di-meH3K9/K27 into the locus. We concluded that CTCF regulates the balance between active and repressive chromatin modifications at its binding sites, and we propose that CTCF-mediated acetylation of histones prevents their methylation. Mechanistically, CTCF may directly attract histone acetyl-transferases (HATs), although current evidence for this interaction is lacking. Alternatively, CTCF-mediated chromatin looping brings binding sites into spatial proximity with HATs bound elsewhere to the DNA (de Laat and Grosveld 2003). The observation that CTCF binding was required for histone acetylation is interesting because these two events were previously claimed to be uncoupled (Recillas-Targa et al. 2002). Our data do not support the generality of boundaries demarcating expression domains, but fit better with the concept that genes maintain autonomous expression profiles mostly through their unique ability to productively interact with positive regulatory elements (Dillon and Sabbattini 2000; de Laat and Grosveld 2003).
CTCF organizes higher-order chromatin structure
We have presented two independent lines of evidence that together firmly establish that CTCF functions in the formation of chromatin loops; removal of most CTCF protein, as well as targeted disruption of a CTCF-binding site, resulted in destabilization of long-range contacts between cognate binding sites in the β-globin locus. CTCF is critical for the looped conformation present in erythroid progenitor cells, but is dispensable for LCR–gene contacts established later during differentiation. We, and others, have shown previously that the latter contacts depend on the transcription factors EKLF and GATA-1 (Drissen et al. 2004; Vakoc et al. 2005). Together, these studies begin to delineate the factors that act sequentially to form a functional β-globin ACH in differentiated erythroid cells where β-globin genes are fully expressed. Based on the observations that CTCF-dependent chromatin loops are tissue-specific and evolutionarily conserved between mouse and man, it seemed reasonable to expect that these loops would play a role in gene expression. Such function may exist but is beyond our current detection limits. An alternative view is that evolutionary selection against sites forming chromatin loops within a gene locus positions them outside the β-globin locus, without necessarily being selected to act, positively or negatively, on gene expression (Dillon and Sabbattini 2000).
We hypothesize that CTCF also organizes higher-order chromatin structure at other gene loci, and we predict that such chromatin loops facilitate communication between genes and regulatory elements but can also lead to the exclusion of interactions between elements. In terms of transcriptional regulation, the final outcome of such chromatin loops will depend on the position of CTCF-binding sites relative to other regulatory elements and the genes, the concentration of the trans-acting factors involved, and the affinities of the (long-range) interactions. In Drosophila, a limited 3C analysis previously provided indications for a loop between the scs and scs’ enhancer-blocking elements (Blanton et al. 2003). Moreover, insulator proteins like suppressor of Hairy-wing [Su(Hw)] and Modifier of mdg4 2.2 [Mod(mdg4)2.2] have been found to coalesce into large foci, called insulator bodies. These bodies preferentially localize at the nuclear periphery and are hypothesized to bring together distant insulator sites, with intervening chromatin fibers looped out to form isolated expression domains (Gerasimova et al. 2000). Our observations made on the CTCF protein provide high-resolution insight into the nature of such loops in mammals. It will be interesting to see if CTCF forms chromatin loops through multimerization of CTCF molecules bound to distinct DNA elements (Yusufzai et al. 2004), or whether this loop formation also involves other factors. Similarly, future experiments should provide insight into whether CTCF-dependent chromatin looping occurs at a defined physical structure in the nucleus (Dunn et al. 2003; Yusufzai and Felsenfeld 2004; Yusufzai et al. 2004), or whether the base of such loops has a more fluid nature.
Materials and methods
Generation of conditional CTCF knockout mice and CTCF antibody
Targeting constructs and strategy for the generation of conditional CTCF knockout mice as well as the polyclonal antibody generated against CTCF will be described in detail as part of a study that addresses the role of CTCF in T-cell development (H. Heath, in prep.).
Lentivirus production and infection
Cre-lentivirus was produced by transient transfection of 293T cells according to standard protocols (Zufferey et al. 1997). Fetal livers were isolated from E12.5 embryos and stored until genotyping of embryos was completed. Cells from three fetal livers of the same genotype were pooled, cultured (Dolznig et al. 2001) for 2 d, and infected (Supplemental Material).
3C analysis, ChIP, and analysis of gene expression
These PCR-based techniques were performed according to standard procedures (Splinter et al. 2004). For details and probe and primer sequences, see the Supplemental Material.
In vitro differentiation of ES cells into ES-EPs and characterization of ES-EPs
Differentiation of ES cells into ES-EPs, expansion of ES-EPs, and in vitro differentiation of ES-EPs into eryhrocytes were performed as described (Epo was a kind gift from Ortho-Biotech; Carotta et al. 2004), except that embryoid bodies were formed in 4000–6000 hanging drop cultures (~200 ES cells/drop) that were pooled and disrupted after 6 d of culturing to generate ES-EPs. After 2–3 wk of cultivation, a homogeneous erythroid progenitor population was obtained (Supplementary Fig. 6).
Acknowledgments
We thank A. de Wit, T. van Dijk, and M. von Lindern for help with experiments. This work was supported by a grant from the division of Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO), through the ESF EuroDYNA programme to N.G., EU grants LSHG-CT-2004-503433 (Epigenetic) and LSHM-CT-2003-504468 (Cells into Organs) and NWO grant 912.03.009 to F.G., and NWO grants 016-006-026 and 912-04-082 to W.d.L.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.399506.
References
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Blanton J., Gaszner M., Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Schubeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Pilat S., Mairhofer A., Schmidt U., Dolznig H., Steinlein P., Beug H. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104:1873–1880. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chao W., Huynh K.D., Spencer R.J., Davidow L.S., Lee J.T. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- de Laat W., Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Gilson E. The vertebrate protein CTCF functions as an insulator in Saccharomyces cerevisiae . Nucleic Acids Res. 2002;30:5136–5141. doi: 10.1093/nar/gkf629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dillon N., Sabbattini P. Functional gene expression domains: Defining the functional unit of eukaryotic gene regulation. Bioessays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Dolznig H., Boulme F., Stangl K., Deiner E.M., Mikulits W., Beug H., Mullner E.W. Establishment of normal, terminally differentiating mouse erythroid progenitors: Molecular characterization by cDNA arrays. FASEB J. 2001;15:1442–1444. doi: 10.1096/fj.00-0705fje. [DOI] [PubMed] [Google Scholar]
- Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes & Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.L., Zhao H., Davie J.R. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 2003;288:218–223. doi: 10.1016/s0014-4827(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Farrell C.M., West A.G., Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd K., Corces V.G. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F., Pikaart M.J., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl. Acad. Sci. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Bell A.C., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G. Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., Grosveld F., de Laat W. 3C technology: Analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- Strouboulis J., Dillon N., Grosveld F. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes & Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- West A.G., Gaszner M., Felsenfeld G. Insulators: Many functions, many mechanisms. Genes & Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Yusufzai T.M., Felsenfeld G. The 5′-HS4 chicken β-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T.M., Tagami H., Nakatani Y., Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]