Abstract
The Schizosaccharomyces pombe CLIP170-associated protein (CLASP) Peg1 was identified in a screen for mutants with spindle formation defects and a screen for molecules that antagonized EB1 function. The conditional peg1.1 mutant enabled us to identify key features of Peg1 function. First, Peg1 was required to form a spindle and astral microtubules, yet destabilized interphase microtubules. Second, Peg1 was required to slow the polymerization rate of interphase microtubules that establish end-on contact with the cortex at cell tips. Third, Peg1 antagonized the action of S. pombe CLIP170 (Tip1) and EB1 (Mal3). Fourth, although Peg1 resembled higher eukaryotic CLASPs by physically associating with both Mal3 and Tip1, neither Tip1 nor Mal3 was required for Peg1 to destabilize interphase microtubules or for it to associate with microtubules. Conversely, neither Mal3 nor Tip1 required Peg1 to associate with microtubules or cell tips. Consistently, while mal3.Δ and tip1.Δ disrupted linear growth, corrupting peg1 + did not. Fifth, peg1.1 phenotypes resembled those arising from deletion of the single heavy or both light chains of fission yeast dynein. Furthermore, all interphase phenotypes arising from peg1 + manipulation relied on dynein function. Thus, the impact of S. pombe CLASP on interphase microtubule behavior is more closely aligned to dynein than EB1 or CLIP170.
Keywords: CLASP, dynein, S. pombe, fission yeast, CLIP170, EB1
Microtubules grow by incorporating αβ-tubulin heterodimers into the dynamic plus ends while their minus ends are often anchored at microtubule organizing centers. Plus ends exhibit a stochastic “dynamic instability,” so that neighboring microtubules oscillate between phases of growth and shrinkage independently of one another. The frequency with which microtubules change from growth to shrinkage is influenced by a variety of stimuli so that the distribution of the microtubule cyto-skeleton changes in response to developmental or positional cues (Kirschner and Mitchison 1986). The class of microtubule-associated proteins (MAPs) known as +TIPs plays a critical role in regulating microtubule dynamics. +TIPs associate with the dynamic, plus ends to modulate microtubule dynamics to mediate interactions between microtubule ends and specific docking sites within the cell (Akhmanova and Hoogenraad 2005; Galjart 2005). All major metazoan +TIPs studied so far including the dynactin component p150glued, Lis1, CLASP (CLIP170-associated protein), and EB1 are physically or functionally linked with the founder +TIP CLIP170 (Coquelle et al. 2002; Goodson et al. 2003; Lansbergen et al. 2004; Mimori-Kiyosue et al. 2005). Although there are functional distinctions between different +TIPs, they do physically interact with one another and so presumably influence each other's function. Importantly, an individual +TIP can interact with two partner +TIPs by distinct mechanisms (Coquelle et al. 2002; Goodson et al. 2003; Mimori-Kiyosue et al. 2005; Wolyniak et al. 2006), highlighting the complexity of the integration of +TIP function at microtubule ends.
CLASP molecules were first identified by genetic approaches (Pasqualone and Huffaker 1994; Inoue et al. 2000) but subsequently gained their name by virtue of their physical association with CLIP170 and CLIP115 (Akhmanova et al. 2001). CLASPs are required to maintain a stem cell component in Drosophila and for ordered cell migration in vertebrates (Mathe et al. 2003; Lee et al. 2004) and can be critically important for localized modulation of microtubule dynamics (Inoue et al. 2004). Mammalian CLASPs stabilize the association of microtubules and the cell cortex (Mimori-Kiyosue et al. 2005).
The simple fission yeast cytoskeleton offers unique opportunities for the analysis of conserved aspects of microtubule function. A differentiated interphase cytoskeleton, in which the plus ends extend away from a centrally placed nucleus, distributes mitochondria, maintains the central position of the nucleus, and delivers cell polarity determinants such as the Kelch domain protein Tea1 to the cell tips (Mata and Nurse 1997; Hagan 1998). Tea1 is transported to cell tips as a result of its association with the +TIPs Tea2 (kinesin-related protein), Tip1 (CLIP170), and Mal3 (EB1) (Browning et al. 2000, 2003; Brunner and Nurse 2000). Exactly how they mediate Tea1 transport remains to be established; however, all four molecules move along the microtubule lattice as well as associating with the growing plus ends. Although Tip1 can associate with microtubules in cells from which the tea2 + gene has been deleted (Busch and Brunner 2004), a large part of the movement of the Tea1/ Tip1/Mal3 complex along the microtubule lattice is mediated by Tea2 activity, and the physical association between Mal3 and Tea2 stimulates the ATPase activity of the kinesin (Browning et al. 1998, 2000; Browning and Hackney 2005).
We now show that the Schizosaccharomyces pombe CLASP Peg1 destabilized microtubules in interphase and yet promoted microtubule stability in mitosis. Focusing on interphase, we did not find a strong link between the function of Peg1 and that of Mal3 or Tip1; rather, we revealed a strong functional link between Peg1 and dynein.
Results
CLASP executes an essential role in spindle formation
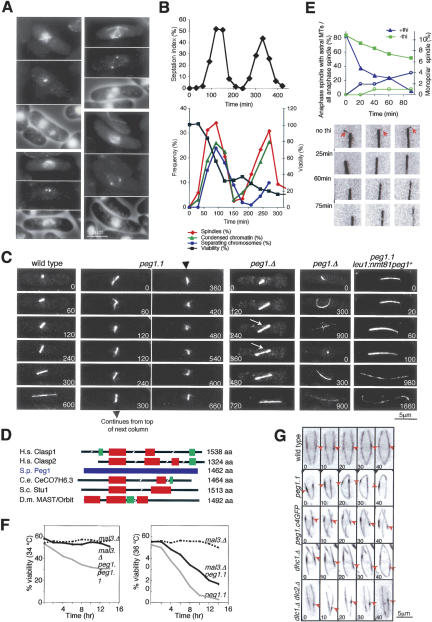
A screen for mutants that could not form a bipolar spindle at 36°C identified an L747F mutation in S. pombe SPAC3G9.12 (Fig. 1 A). As SPAC3G9.12 encodes S. pombe CLASP (Fig. 1D) and a peg is a type of clasp, we named SPAC3G9.12 “Peg1.” At 36°C, peg1.1 mutants exhibited spindle formation defects ranging from star-shaped monopolar spindles through to separated spindle pole bodies (SPBs) on the opposing sides of an undivided nucleus that were connected by very few, or no, micro-tubules (Fig. 1A). Time-lapse imaging of GFP-tubulin-expressing peg1.1 cells (see below) established that individual mutant cells repeatedly formed short, very weak spindles that collapsed (Fig. 1C; Supplementary Movies 2A,B). Very occasionally a robust spindle spanned the nucleus and the nucleus then divided, suggesting that Peg1 function was less critical within the central spindle if cells passed the metaphase/anaphase transition (Supplementary Movie 2B, right cell). The drop in viability in a peg1.1 population that had been synchronized with respect to cell division by size selection coincided with the defective mitosis (Fig. 1B).
Figure 1.
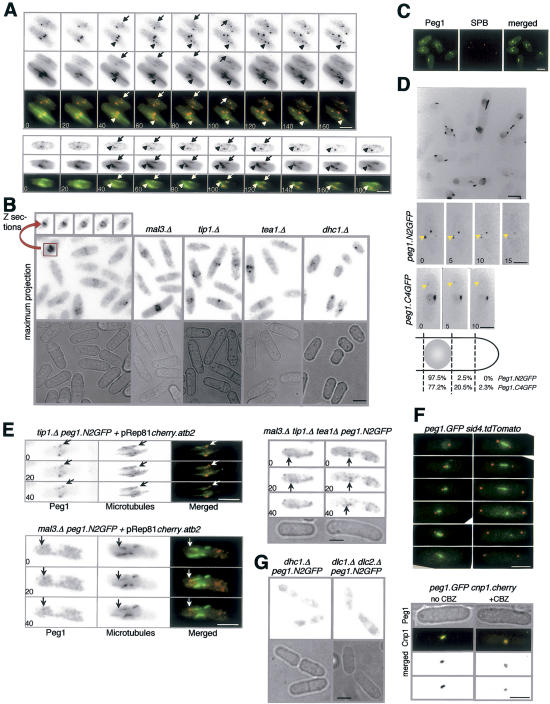
Peg1 and the formation and function of the mitotic spindle and the architecture of interphase microtubule bundles. (A,B) Temperature-sensitive peg1.1 cells were synchronized with respect to cell cycle progression by elutriation, and the majority of the culture was shifted to 36°C immediately after isolation. Samples were processed for immunofluorescence microscopy and viability at 36°C, and to count cell plates with calcofluor at 25°C. (A) Cells 120 min after shift: microtubules (top), SPBs (middle), and chromatin cell periphery (bottom). (B) The top graph shows the septation index at the indicated times at 25°C, while the bottom graph shows the criteria indicated in the legend. (C) Imaging atb2.nmt81GFP cells. peg1.1 cells repeatedly formed short spindles that then collapsed. peg1.Δ cells in which the interphase microtubule phenotype was moderate had weaker central spindles than wild-type controls (arrow) or formed bent spindles. (D) A cartoon comparing the peg1 + predicted amino acid sequence to those of CLASPs from different species. The frequency of shared identity between CLASPs from different species and Peg1 sequences (blue), as derived from the BlastP algorithm: (red) 21%–24% amino acid identity to Peg1; (green) 27%–31% identity to Peg1. (H.s.) Homo sapiens; (S.p.) S. pombe; (C.e.) Caenorhabditis elegans; (S.c.) S. cerevisiae; (D.m.) Drosophila melanogaster. (E) Repressing the expression of peg1 + from the nmt81 promoter in peg1.1 cells at the same time (t = 0) as cells were shifted from 25°C to 36°C resulted in a greater reduction in the number of anaphase B spindles that had astral microtubules (left axis, blue triangles) and bent spindles (C) than in strains where peg1 + expression was maintained by the exclusion of thiamine (left axis, green squares), while in both cases the number of cells that could not form a spindle at all was minimal (right axis, circles). The images below the graph show ends of anaphase B spindles at the indicated times. (F) Viability assays. Cells were shifted from 25°C to 34°C (left) or 36°C (right) at the start of the experiment, and aliquots were spread on solid medium to determine their ability to form colonies. (G) The bright “overlap” zone in peg1.1, peg1.C4GFP dhc1.Δ, dlc1.Δ dlc2.Δ undergoes more extreme excursions than in wild-type cells. Numbers in the corner of panels indicate relative timing in seconds.
Generation of a heterozygous peg1.1/peg1 + established that peg1.1 was a recessive mutant, while tetrad analysis of a diploid in which one copy of the peg1 + ORF had been replaced with a hygr R marker gene, showed that peg1 + is an essential gene (data not shown). Germinating spores from such a peg1.Δ/peg1+ diploid (peg1.Δ is a deletion of peg1 +) in which both atb2 + (α-tubulin) genes were fused to GFP established that complete ablation of Peg1 could produce the same spindle formation defects as peg1.1 at 36°C (data not shown). However, the variable amounts of Peg1 inherited from the parental zygote by different spores meant that some germinating spores completed their first mitosis yet underwent a cut division with a fully defective spindle in the next cycle. The central spindles of these functional divisions were often noticeably weakened until cells entered anaphase B (Fig. 1C, third data set, white arrow), whereupon some anaphase B spindles bent to form an arch (Fig. 1C, fourth data set). The lack of astral microtubules on such bent peg1.Δ spindles suggested that Peg1 function was required for astral microtubule stability and that the bending might arise from abnormal interactions with the cortex. We therefore generated a peg1.1 strain that harbored an integrated version of the peg1 + gene whose expression was under the control of a repressible (nmt81) promoter. Simultaneous repression of peg1 + transcription, by the addition of thiamine, and switch from 25°C to 36°C to inactivate the peg1.1 product, led to a progressive loss of anaphase B spindles that had astral microtubules 5% of which were bent into an arch (Fig. 1C, fifth data set), while there was a very modest increase in the number of cells that were unable to form a spindle (Fig. 1E), indicating that Peg1 was required for the generation or stability of astral microtubules.
Peg1 (CLASP) function antagonizes Mal3 (EB1)
A separate screen for genomic sequences that would suppress the sensitivity to the anti-microtubule drug thiabendazole (TBZ) of strains in which the EB1 homolog Mal3 was defective identified a plasmid containing the 3′ third of peg1 + (data not shown). We therefore asked whether removing EB1 function with mal3.Δ alleviated the impact of the peg1.1 mutation and found clear evidence for such compensation at 34°C (Fig. 1F). Thus, S. pombe CLASP (Peg1) counteracts the function of its EB1 (Mal3).
Selection of GFP/tubulin construct
Before embarking on a detailed characterization of the impact of Peg1 on microtubule dynamics, we assessed different GFP/α-tubulin fusion proteins that have been used to image microtubule dynamics in fission yeast (Ding et al. 1998; Drummond and Cross 2000; Garcia et al. 2001; Glynn et al. 2001) in order to find the approach that would have the least impact on microtubule function. All strains were processed for anti-tubulin immunofluorescence microscopy (Hagan and Hyams 1988) and the length and number of microtubule bundles recorded. Only microtubules of an atb2.nmt81GFP strain in which expression of the fusion protein had been fully induced by removal of thiamine were indistinguishable from wild-type cells. This atb2 allele has GFP coding sequences fused to the 5′ end of the native atb2 + α-tubulin ORF, and the chimeric gene is under the control of the nmt81 promoter (Garcia et al. 2001). Unless otherwise indicated, like Sawin, Toda, and colleagues (Garcia et al. 2001; Sawin et al. 2004), we use atb2.nmt81GFP throughout this study.
Loss of Peg1 produces more intense interphase bundles that contain more stable microtubules
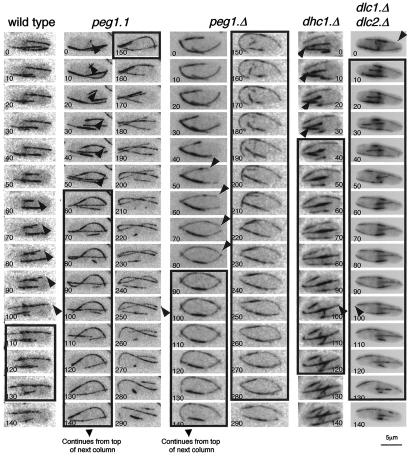
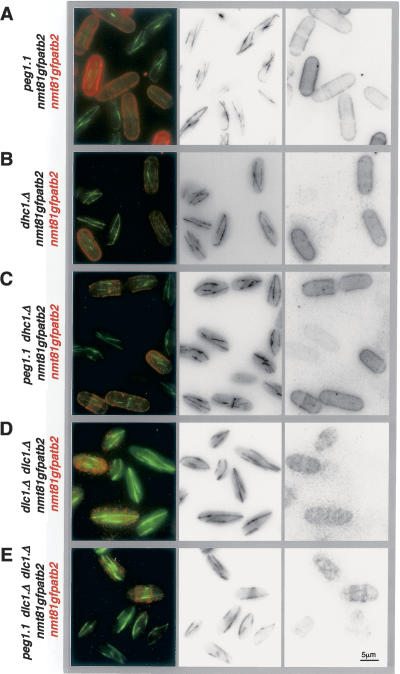
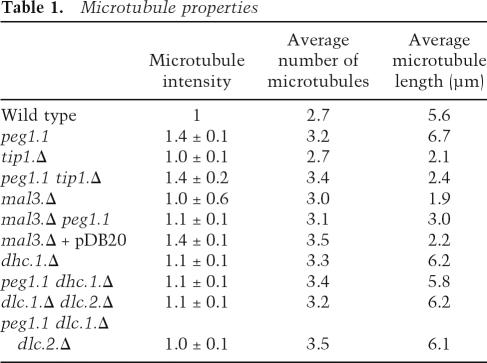
The microtubule bundles of atb2.nmt81GFP peg1.1 (Supplementary Movie 3) were brighter and longer than those of atb2.nmt81GFP peg1 + (Supplementary Movie 4). Furthermore, peg1.1 cells differed from wild-type counterparts as 13% of microtubules curled around the cell tips (Fig. 2). Labeling peg1 + cells with red fluorescent lectin distinguishing them from isogenic peg1.1 cells in the same field of view established that microtubule bundles of peg1.1 cells were 1.4 ± 0.1-fold brighter and 19% longer than in peg1 + (Fig. 3; Table 1). Most significantly, even though the growth and shrinkage rates were unaffected by the peg1.1 mutation, microtubule ends spent 1.8 times longer at the tips of peg1.1 cells than in peg1 + control cells (Fig. 2; Table 2A).
Figure 2.
Microtubules persisted at cell tips when Peg1 or dynein heavy-chain functions were compromised. GFP α-tubulin was expressed by the atb2.nmt81GFP allele in the indicated strains at 36°C. Each panel shows, from the top, a 10-sec-interval series of maximal projections of the indicated strains. The arrows highlight microtubule tips that will pause at the cell end for the period indicated by the boxes around the frames. Thus, the microtubule in the example of a wild-type cell pauses at the right tip for 30 sec, while in peg1.1, the microtubule resides at the cell tip for 100 sec. Numbers in the corner of panels indicate relative timing in seconds.
Figure 3.
Compromising Peg1 function results in more and brighter microtubules. atb2.nmt81GFP cells were briefly resus-pended in TRITC-conjugated lectin before being washed twice in filter-sterilized minimal medium and imaged alongside atb2.nmt81GFP peg1.1 (A), atb2.nmt81GFP dhc1.Δ (B), atb2.nmt81GFP dhc1.Δ peg1.1 (C), dlc1.Δ dlc2.Δ (D), or peg1.1 dlc1.Δ dlc2.Δ (E) cells (that had not been stained with TRITC lectin).
Table 1.
Microtubule properties
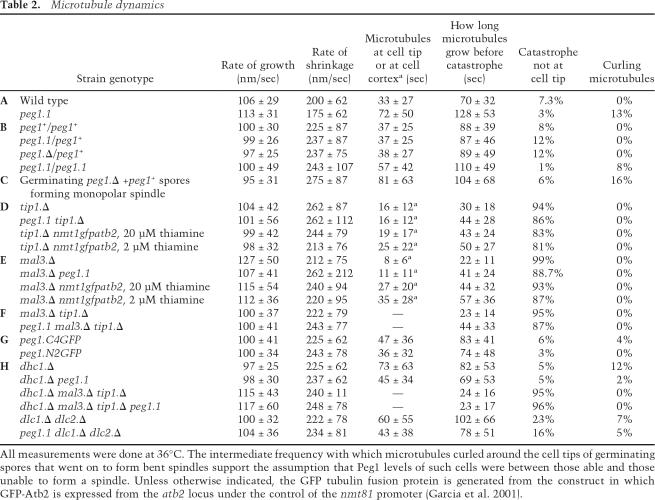
Table 2.
Microtubule dynamics
To understand the impact of the peg1.1 mutation on microtubule dynamics, it was necessary to establish whether peg1.1 led to a loss or modification of Peg1 function. We therefore compared microtubule dynamics, dwell times, and curling around cell tips at 36°C in atb2.nmt81GFP/atb2.nmt81GFP diploids in which the peg1 loci were either peg1.1/peg1.1, peg1 +/peg1 +, peg1.1/ peg1 +, or peg1 + /peg1.Δ. Heterozygote mutants were indistinguishable from peg1 +/peg1 + homozygotes. In contrast, the microtubules of peg1.1/peg1.1 homozygotes mimicked those in haploid peg1.1 cells in spending longer at the cell tips (Table 2B). The recessive nature of the impact of the peg1.Δ and peg1.1 mutations on microtubule dynamics indicated that peg1.1 is a loss-of-function mutation. Thus, in contrast to its stabilizing impact on cytoplasmic astral microtubules in mitosis, Peg1 destabilizes cytoplasmic microtubules in inter-phase.
To gauge the severity of the defect imposed by the peg1.1 mutation, we compared it with that of a complete deletion of the gene by imaging microtubules in peg1.1 cells and germinating peg1.Δ spores at 36°C. As the persistence of some parental gene product in the spores meant that the first mitosis was sometimes successful (Fig. 1C), cells were followed into mitosis after assessment of interphase dynamics to study only those that were unable to form a spindle. The dwell times of microtubule ends at the tips of these cells were similar to those in peg1.1 cells (Table 2C; Fig. 2) indicating that peg1.1 severely compromised the interphase as well as the mitotic function of Peg1 at 36°C.
Peg1 (CLASP) associates with Tip1 (CLIP170) and Mal3 (EB1) but not Tea1
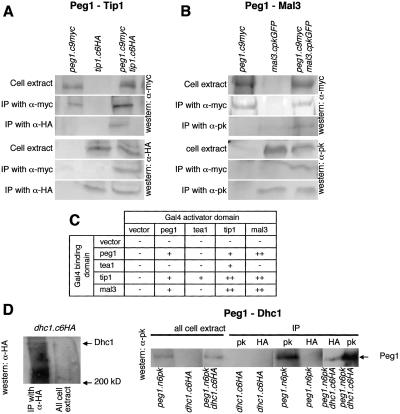
To address how closely the function of S. pombe CLASP may resemble those of higher eukaryotic CLASPs, we asked whether it mimicked these CLASPs from more complex systems in physically associating with CLIP170 (Tip1) and EB1 (Mal3) (Akhmanova et al. 2001; Mimori-Kiyosue et al. 2005). Peg1 clearly associated with Tip1 (CLIP170) and Mal3 (EB1) in two hybrid and coimmuno-precipitation assays (it showed no affinity for another +TIP, Tea1, in either assay) (Fig. 4A–C).
Figure 4.
Peg1 physically associated with Tip1 (CLIP 170), Mal3 (EB1), and dynein (Dhc1), but not Tea1. (A,B,D) Immuno-precipitation reactions using the indicated antibodies on the indicated strains established that CLASP (Peg1) bound S. pombe CLIP170 (Tip1), EB1 (Mal3), and dynein heavy chain (Dhc1). (C) Summary of two-hybrid assays.
The impact of Tip1 and Mal3 on microtubule dynamics
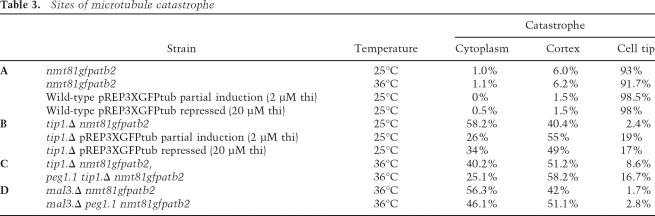
It has been reported that the life span of microtubules that contact the cortex is shorter than those in the cytoplasm after the tip1 + gene has been deleted (tip1.Δ) (Brunner and Nurse 2000). However, there was no context specificity to the instability in tip1.Δ in our assay conditions (Tables 2D, 3B,C; Supplementary Movie 5; Supplementary Fig. S1). We therefore switched to the higher expression conditions of the previous study (multicopy pREP3X nmt1 + promoter-driven GFP tubulin plasmid) and marked the cell cortex with TRITC lectin. We then found context specificity when tubulin levels were changed by some leak through expression of the GFP/tubulin fusion protein from this plasmid when 20 μM thiamine was included to reduce transcription to a minimum. The lifetime of cytoplasmic microtubules (the period from the initiation of a phase to growth to the point where shrinkage is initiated) now increased 43% relative to the lower expressing tip1.Δ atb2.nmt81GFP cells, whereas that of microtubules touching the cortex increased by only 20% (Tables 2D, 3B; Supplementary Movie 6A,B). This differential impact was mild and microtubules frequently slid along the cell cortex (Supplementary Fig. S2). Increasing tubulin levels even further by the inclusion of just 2 μM thiamine repressor in the medium obscured the effect (Table 2D), indicating that a narrow range of microtubule stabilization conditions is required to reveal the context-dependent impact of tip1.Δ. Compromising Peg1 function in atb2.nmt81GFP tip1.Δ cells, on the other hand, resulted in a much greater context specificity to microtubule lifetime in tip1.Δ cells as the life span of microtubules that did not touch the cortex increased by 50%, whereas those that did remained unchanged (Tables 2D, 3C; Supplementary Movie 7). As tubulin levels were unaffected by the peg1.1 mutation (Supplementary Fig. S3), we conclude that the fundamental consequence of abolishing Tip1 (CLIP170) function is microtubule instability irrespective of the local environment; however, as reported by Brunner and Nurse (2000), microtubule ends gain the potential to behave in a context-dependent manner when microtubule stability is increased by increasing tubulin gene dosage or (to a much greater extent) compromising Peg1.
Table 3.
Sites of microtubule catastrophe
Addressing the impact of tubulin gene dosage and peg1.1 mutation on cells from which the EB1 homolog had been deleted (mal3.Δ) revealed further complexity in microtubule cortex interactions (Tables 2E, 3D; Supplementary Figs. S1, S4; Supplementary Movies 8, 9). Increasing tubulin gene dosage doubled the life span of cytoplasmic microtubules, and yet the residence time at the cortex increased by a much greater 3.4-fold. Thus, either, Mal3 is required for stability of microtubules in the cytoplasm, or Mal3 counteracts a protective impact of proteins on the cortex that otherwise stabilizes micro-tubules. Corrupting Peg1 function in mal3.Δ with the peg1.1 mutation doubled the life span of cytoplasmic microtubules, and yet the residence time of microtubule tips at the cortex increased by only 37% (Table 2E). Thus the impact of altering Peg1 function is distinct from that of driving microtubule polymerization by simply increasing the number of subunits available for polymerization. Rather, it is clear that, as the existence of so many distinct +TIPs might predict, a complex interplay between different +TIPs influences microtubule stability in distinct environments.
Peg1 (CLASP) depolymerized microtubules in the absence of Tip1 (CLIP170) or Mal3 (EB1)
The most important observation arising from our analysis of +TIP mutants was that the duration of microtubule polymerization periods in mal3.Δ tip1.Δ peg1.1 triple mutants at 36°C was double those of mal3.Δ tip1.Δ double-mutant cells (Table 2F; Supplementary Figs. S1, S4). Thus, contrary to models in which CLASP modulates or works alongside EB1 and CLIP170, Peg1 function can have an impact on microtubule dynamics in the complete absence of CLIP170 and/or EB1.
S. pombe CLASP, Peg1, did not control cell polarity
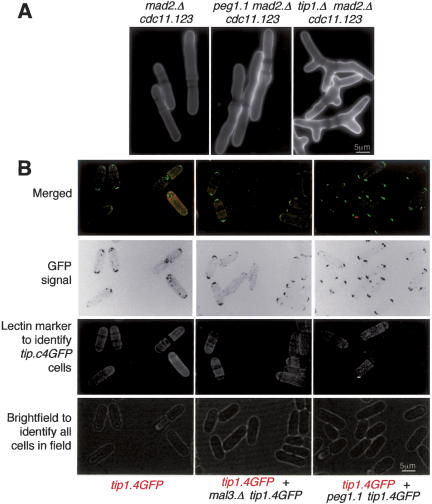
Because S. pombe +TIPs Tea1, Tip1, and Mal3 play critical roles in controlling cell polarity (Beinhauer et al. 1997; Mata and Nurse 1997; Brunner and Nurse 2000), a functional link between Peg1 and these other +TIPs should be reflected in a morphogenetic phenotype in peg1.1. peg1.1 cells normally die by a lethal mitotic cut event (Fig. 1B) and so do not go through the repeated rounds of new tip growth at the restrictive temperature that enable us to assess morphogenesis in peg1.1. We therefore abolished the spindle checkpoint-induced mitotic arrest with mad2.Δ and blocked cytokinesis with the cdc11.123 mutation. The impact on the selection of the site for new tip growth would then be revealed in the resultant peg1.1 cdc11.123 mad2.Δ triple mutant as it underwent repeated rounds of S phase and repolarization of growth at 36°C. Unlike the control tip1.Δ cdc11.123 mad2.Δ strain, which exhibited severe morphology defects, peg1.1 cdc11.123 mad2.Δ cells maintained polarized growth (Fig. 5A). Furthermore, the recruitment of the +TIP polarity determinants to the cell tips was not altered after Peg1 function was compromised by incubating peg1.1 at 36°C (Fig. 5B; Supplementary Fig. S5; data not shown). Thus, unlike these other S. pombe +TIPs, Peg1 function did not influence cell polarity and so showed no signs of influencing the function of the polarity-determining +TIPs, EB1, CLIP170, Tea2, and Tea1.
Figure 5.
Peg1 was not a polarity determinant and was not required for the recruitment of Tip1 to microtubules or cell tips. (A) Peg1 was not required to maintain linear growth. Indicated strains were grown at 36°C for 5 h in EMM2 medium before being stained with calcofluor. Shifting nitrogen-starved cells from 25°C to 36°C and then following them as they recovered from the starvation also failed to reveal a morphology defect when Peg1 was compromised (data not shown). (B) tip1.4GFP cells were briefly resuspended in TRITC-conjugated lectin before being washed twice in filter-sterilized minimal medium and imaged using spinning disk microscopy as described in the legend for Figure 3. Cells were either viewed in isolation (left panel) or alongside the indicated double/triple mutants that had not been stained with TRITC lectin.
Peg1 distribution is not affected by removal of other +TIPs
Fusion of single or multiple copies of GFP or epitope tags to the 3′ end of the peg1 + gene did not affect cell viability or mitotic progression (data not shown), but compromised the ability of Peg1 to induce microtubule instability in interphase (Table 2G; Supplementary Movie 10A). In contrast, interphase microtubule dynamics were indistinguishable from those of wild-type cells when two GFP genes were fused, in frame, to the 5′ end of the native peg1 + locus (Table 2G; Supplementary Movie 10B). In log phase, faint dots of Peg1.N2GFP both moved along microtubule bundles of cells expressing an Atb2.Cherry fusion protein (Fig. 6A, arrowhead) and associated with the tips of growing microtubules (Fig. 6A, arrow). Peg1.N2GFP dots were seen at the points within microtubule bundles at which the decrease in the intensity of Atb2.Cherry indicated the end of a microtubule within the bundle (Streiblova and Girbardt 1980; Busch and Brunner 2004; Carazo-Salas et al. 2005). As cell density increased to 2 × 107 and nutrient supply became limited, strong Peg1.N2GFP foci associated with microtubule bundles near the nucleus (Fig. 6B; data not shown). These dots, one of which was the SPB (Fig. 6C), rarely strayed to cell ends (in contrast, dots of the defective Peg1.C4GFP were more generally distributed throughout nutrient-limited cells; Fig. 6D). Accumulation around nuclei is a feature of microtubule nucleation factors such as Mod20 that are required for microtubule regrowth following transient depolymerization by cold shock (Sawin et al. 2004). As microtubule depolymerization pheno-types occur within 2 min of a shift of peg1.1 to 36°C (data not shown), peg1.1 microtubules were transiently depolymerized by cold before return to 36°C. Microtubule regrowth was indistinguishable from peg1 + controls, indicating that Peg1 was not required for regrowth (Supplementary Fig. S6). However, the most important observations arising from Peg1.2NGFP localization was that deletion of tip1 +, tea1 +, or mal3 + did not affect either type of Peg1.2NGFP distribution (Fig. 6B,E). Thus, in a reflection of the lack of a requirement for Peg1 for the association of Tip1, Tea1, or Mal3 with microtubules and cell ends, these +TIPs were not required for Peg1 distribution.
Figure 6.
Peg1 recruitment to microtubules of log-phase cells required dynein function but not Tip1, Mal3, or Tea1. Panels, B, C, D (top), and F are maximal projections of a Z-series extending through the cells. For A, D (bottom), E, and G, three consecutive planes were compressed to generate the images. (A) Peg1.N2GFP foci associate with the lattice (arrowhead) and tips (arrows) of Atb2.Cherry-marked microtubules. (B) Peg1.N2GFP fluorescence was imaged in nutrient-limited cells of the indicated genetic backgrounds. The individual panels at the top are consecutive Z-slices through the indicated peg1.N2GFP cell. (C) One of the foci that appears around the nucleus upon nutrient limitation colocalizes with the SPBs of peg1.N2GFP sid4.Tdtomato cells. (D) Peg1.C4GFP shows strong signals in nutrient-limited cells that are less constrained around the nucleus than Peg1.N2GFP. (E) Peg1.N2GFP foci in the indicated strains. (F, top) Peg1.N2GFP associates with the mitotic spindle of peg1.N2GFP sid4.Tdtomato cells throughout mitosis. (Bottom) Peg1.N2GFP signals overlay centromere signals of early mitotic peg1.N2GFP cnp1.Cherry cells, but not those of cells in which mitotic progression has been arrested by microtubule depolymerization with 50 μg/mL CBZ. Bar, 5 μm.
Peg1.2NGFP associated with mitotic spindles throughout mitosis accumulating in an ever-decreasing portion of the mid-zone as anaphase progressed (Fig. 6F, top panel). When microtubules were depolymerized by the addition of 50 μg/mL CBZ, Peg1.2NGFP was not associated with kinetochores (Fig. 6F, bottom panel), although CHIP analysis of untreated log-phase cultures showed that Peg1 did associate with centromeres of dividing cells (data not shown).
Peg1 requires dynein to slow microtubule polymerization at cell tips
Our failure to find links between the function of CLASP and CLIP170 or EB1 led us to examine its relationship with the +TIP dynein. The microtubule dynamics following deletion of either the single dynein heavy chain (dhc1.Δ) or both, redundant, light chains (dlc1.Δ dlc2.Δ) were reminiscent of those in which Peg1 function had been compromised. Microtubules were longer, had increased dwell times at cell tips, and curled around cell tips (Fig. 2; Table 2H; Supplementary Movie 11). A second shared phenotype was the exaggerated movements of the stronger region of fluorescence within microtubule bundles. These brighter regions are assumed to represent the regions within the bundles of multiple micro-tubules that contain most microtubules (Streiblova and Girbardt 1980; Busch and Brunner 2004; Carazo-Salas et al. 2005) and are invariably found in the cell center near the nucleus. Time-lapse imaging of dhc1.Δ, dlc1.Δ dlc2.Δ, peg1.1, or peg1.C4GFP cells revealed highly dynamic overlap regions that moved from one end of the cell to the other (Fig. 1G; Supplementary Movies, 3, 10a, 11). A third link came from the fact that the increased intensity and number of interphase microtubule bundles arising from the peg1.1 mutation was no longer seen when dhc1 + or both dlc1 + and dlc2 + were deleted alongside the peg1.1 mutation, indicating that loss of Peg1 had minimal impact on microtubule dynamics in the absence of dynein (Table 1; Fig. 3; Supplementary Fig. S4). The most striking dependency between peg1.1 pheno-types and dynein function was that the increase in microtubule dynamics that is induced in a mal3.Δ tip1.Δ background by corruption of Peg1 function with the peg1.1 mutation relied on dynein function (Table 2F,H) (dhc1.Δ alone did not increase microtubule life span in this mal3.Δ tip1.Δ background). Thus, the consequences of losing dynein and Peg1 function are similar, and two peg1 − phenotypes are epistatic to loss of dynein. Consistently, Peg1.N2GFP distribution in log phase was disrupted in dhc1.Δ and dlc1.Δ dlc2.Δ (Fig. 6G), although the strong staining around nuclei of starved cells was still seen (Fig. 6B; data not shown).
Peg1 is required to slow microtubule polymerization at cell tips
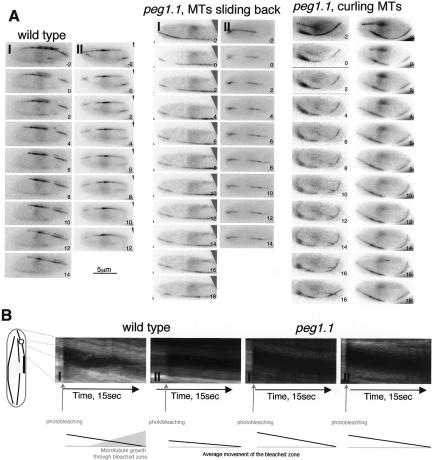
As corruption of Peg1 extended the movements of the intense zones within interphase microtubule bundles (Fig. 1G; Supplementary Movie 3), it was possible that the prolonged dwell time at cell tips after corruption of dynein or Peg1 arose from sliding of preformed microtubules to cell ends rather than from alterations in the dynamics of individual microtubules. We therefore used photobleaching to ask whether microtubule sliding made any contribution to the extended residence time at cell tips we recorded in peg1.1 and dhc1.Δ mutants in Table 2. The rationale was that the distance between the bleached zone and the microtubule tip would diminish if the whole microtubule bundle slid toward the tip, whereas the distance between the bleached zone and the microtubule tip would increase if polymerization accounted for the extended residence time of microtubule ends at the cell tip.
In wild type, dhc1.Δ and peg1.1 bleached bars stayed between the cell tip and the nucleus as microtubules grew toward the tip, and then, once the microtubule end established an end-on association with the cell tip, this bleached zone actually migrated back toward the nucleus, away from the tip (Fig. 7A,B; data not shown). Thus, the increased dwell time of microtubule ends at cell tips in these mutants arose from altered plus-end dynamics rather than sliding.
Figure 7.
Microtubule sliding did not account for the persistence of microtubules at cell tips. Regions of microtubules of the indicated GFP α-tubulin-expressing strains were marked by photobleaching the fluorescence in a microtubule bundle in a zone between the nucleus and the cell tip. (A) Images were taken in a single focal plane at 1-sec intervals. (B) Kymographs showing microtubules from the regions indicated in the cartoon on the left. The cartoon on the left of the kymographs indicates the region of the bleached micro-tubules used to generate the kymographic data. The cartoons below the kymographs summarize the rearward migration of the rear of the bleached zone and the growth of a microtubule through the bleached zone.
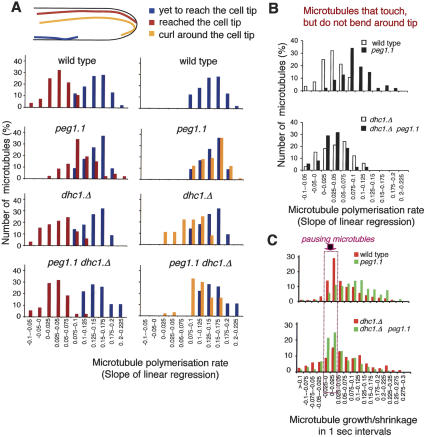
The introduction of the bleached zone also enabled us to compare the dynamics of microtubules after they had established an end-on contact with the cell tip with those that were yet to reach and those that curled around the ends. We calculated the rates of polymerization from plots of the length of the zones of individual microtubules that extended beyond the bleached region against time (Supplementary Fig. S7). The slope of the linear regression of such plots (polymerization rates) was divided into discrete intervals and plotted against the frequency to generate the bar charts shown in Figure 8. Consistent with a previous report (Tran et al. 2001), the polymerization rates of wild-type microtubules that were yet to touch the cell tip (Fig. 8A, blue) were greater than those that had established the transient end-on associations with the cell tip that frequently occur before microtubules start to depolymerize and shrink away from the cell tips (Fig. 8A, maroon). The microtubules also slowed after microtubules established end-on associations with the cortex at cell tips of dhc1.Δ cells. Thus, although dynein was required to induce catastrophe at cell tips (Fig. 2; Table 2H), it was not required to slow the polymerization of microtubules that established end-on associations with cell tips. In contrast, the rate of micro-tubule polymerization did not slow to the same degree as seen in wild-type cells when microtubules established end-on associations with the tips of peg1.1 cells (Fig. 8A,B; Supplementary Fig. S7). Furthermore, they did not slow at all for those microtubules that continued polymerizing after contacting the cortex and the tip to curl around this tip (Fig. 8A; Supplementary Fig. S7). Critically, microtubules that established end-on associations did slow to the same degree as wild type in peg1.1 mutant cells if dynein was also missing alongside the deficiency in Peg1 function (Fig. 8A,B; Supplementary Fig. S7). Thus, the ability of Peg1 to slow the polymerization of microtubules that established end-on contact with the cortex at cell tips depended on dynein.
Figure 8.
Peg1 relied on dynein to slow polymerization of microtubules that establish end-on interactions with the cell cortex at cell tips, and to induce pauses between phases of microtubule polymerization and depolymerization. (A) Following photobleaching of a zone of fluorescence of GFP α-tubulin-expressing strains as in Figure 7, the length of the tip-proximal nonbleached zone was measured and plotted against time (Supplementary Fig. S7), and the linear regression of this slope was calculated. The number of microtubules for which this linear regression (rate of microtubule extension) fell into a particular interval is plotted as bar charts according to whether the microtubules were yet to reach the cell tip (blue), had reached the tip and established an end-on association (maroon), or continued to polymerize to curl around the cell tip (orange). For full data sets, see Supplementary Figure S7. (B) Bar charts that compare the data sets shown in A of the microtubules that continue to polymerize after establishing endon association with the cell tip. (C) The change in the length of the tip-proximal nonbleached zone was scored over discrete 1-sec periods and plotted against the frequency of occurrence for individual classes to generate the plots for the indicated strains. The purple arrow highlights the intervals at −0.025 to 0.0 and 0.0 to 0.025, which correspond to pauses during polymerization.
Peg1 induced pausing of microtubule polymerization depends on Dhc1 function
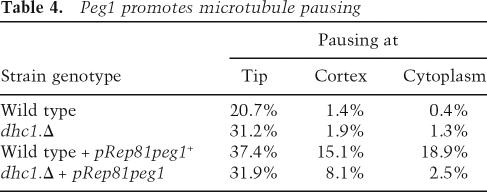
Quantifying the amount of microtubule growth achieved over 1-sec periods and plotting the frequencies with which different degrees of growth were observed as discrete intervals revealed a significant number of pausing events in wild-type cells (Fig. 8C). These pauses were greatly reduced in peg1.1 mutants and considerably extended when Peg1 levels were increased (Fig. 8C; Table 4). Thus, Peg1 mimics human CLASP in promoting pausing events during microtubule growth (Mimori-Kiyosue et al. 2005). Significantly, deletion of dhc1 + alongside peg1.1 restored pausing to peg1.1 mutants (Fig. 8C) and considerably reduced the amount of pausing induced by excessive Peg1 (Table 4). Thus, every interphase consequence of corruption of Peg1 that we monitored depended on dynein function.
Table 4.
Peg1 promotes microtubule pausing
Physical association of Dhc1 with Peg1
The strong links between Peg1 and dynein function prompted us to ask whether the molecules associated with one another. We used 12CA5 antibodies that recognize the HA epitope to generate immunoprecipitates from dhc1 + peg1.N6pK, dhc1.C6HA peg1 +, and dhc1.C6HA peg1.N6pK (peg1.N6pK has normal inter-phase dynamics) (data not shown) and probed with the MAb336 to detect Peg1.N6pK. Consistent with the inability to detect Dhc1 mRNA in vegetative cells (Yamamoto et al. 1999), we did not detect Dhc1.HA in whole-cell extracts (Fig. 4D); however, immunoprecipitation concentrated the protein to a level where it was detected in immunoprecipitates from dhc1.C6HA peg1 +. 12CA5 immunoprecipitates from dhc1.C6HA peg1.N6pK contained Peg1.N6pK, whereas 12CA5 precipitates from dhc1 + peg1.N6pK cells did not (Fig. 4D), indicating that dynein and Peg1 can be part of the same complex in vivo.
Discussion
S. pombe CLASP influences microtubule dynamics in different ways in different cell cycle stages
In accord with data from humans, Saccharomyces cerevisiae, and Drosophila, the S. pombe CLASP Peg1 plays a critical role in regulating mitotic microtubule dynamics (Pasqualone and Huffaker 1994; Inoue et al. 2000, 2004; Maiato et al. 2003, 2004, 2005; Laycock et al. 2006). Progressive depletion of Peg1 led to a progressive loss of cytoplasmic astral microtubules in anaphase B. The reliance on Peg1 for central spindle function appeared to diminish upon transition into anaphase B, as anaphase B spindles were always robust irrespective of whether peg1.1 cells had spent a long time trying to form a spindle or peg1.4CGFP cells had exhibited weakened central spindles prior to anaphase B. The similarity between peg1 − spindle defects and those arising from the loss of CLASP function in budding yeast stu1 and the weakened central spindle of Drosophila ORBIT/MAST mutants suggests that CLASPs may have a conserved role in establishing the integrity of the microtubules of the central spindle (Pasqualone and Huffaker 1994; Inoue et al. 2004).
In interphase, Peg1 destabilized microtubules. This switch in activity is reminiscent of the Drosophila kinesin-related protein KLP67A, which flips from stabilizing microtubules in prometaphase to destabilizing them in anaphase (Gatt et al. 2005), and budding yeast EB1, which stimulates microtubule dynamicity in G1 but not in preanaphase cells (Wolyniak et al. 2006). How such cell cycle-dependent changes in function arise awaits clarification, but phosphorylation is a good candidate. The phosphorylation status of in vitro translated mammalian CLASP appears to modulate its affinity for microtubules, and its in vivo function is influenced by GSK3β kinase activity (Akhmanova et al. 2001; Wittmann and Waterman-Storer 2005).
Distinction between S. pombe CLASP and its EB1 and CLIP170
S. pombe CLASP mimicked CLASPs of higher eukaryotes by associating with CLIP170 or EB1 (Akhmanova et al. 2001; Mimori-Kiyosue et al. 2005). However, the ability to conduct drastic manipulation of the +TIP repertoire in S. pombe enabled us to establish that neither association was required for Peg1 to destabilize inter-phase microtubules. Thus, even though it was the physical association of mammalian CLASP with CLIP170 that gave the name to these molecules (Akhmanova et al. 2001), this association, and that with EB1, may not be critical for CLASP to be able to influence microtubule dynamics. This would be consistent with distinctions that have been recorded between these +TIPs in higher systems. While CLASP is a constitutive kinetochore component that promotes the incorporation of tubulin heterodimers at the kinetochore, CLIP170 leaves the kinetochore upon microtubule attachment (Dujardin et al. 1998; Maiato et al. 2003). Furthermore, EB1 only associates with the kinetochore undergoing anti-poleward migration and is not required for subunit addition at the kinetochores (Tirnauer et al. 2002; Maiato et al. 2003, 2004, 2005). Consistently, depletion of CLASP blocks poleward flux, whereas depletion of EB1 does not (Maiato et al. 2005).
So has the association between CLASPs and CLIP170 and EB1 led to false expectations of the nature of the relationship between CLASP and these two molecules? Localization studies do not dispute this view, as CLASP sits slightly behind the CLIP170 at the very tips of microtubules in migrating axons, behind both EB1 and CLIP170 in migrating fibroblasts, and only partially co-localizes with EB1 in mitotic asters (Akhmanova et al. 2001; Maiato et al. 2003; Lee et al. 2004). So why and when do they associate? Perhaps the association is critical for coordinating the activity of the different +TIPs to generate a concerted action. Alternatively, perhaps the physical association is a transient phase associated with the loading or unloading of +TIPs (Komarova et al. 2005) or the docking or release of target molecules at particular locations.
S. pombe dynein regulates interphase microtubule dynamics
The ability to delete the genes encoding the single dynein heavy and two light chains (dhc1.Δ, dlc1.Δ, dlc2.Δ) from fission yeast cells without affecting their viability or polarity (Yamamoto et al. 1999; Miki et al. 2002) has limited the study of dynein in the mitotic cell cycle. We now find that, like Peg1, dynein promoted the end-on capture of microtubule tips by the cortex at cell ends and the instability of these microtubules once they had been captured. Thus, when dynein was absent, microtubules spent longer at cell tips and frequently continued to elongate and curl around these tips.
A second distinction from wild-type cells that was shared between dynein and peg1 mutants was the excessive mobility of the bright zones within individual interphase microtubule bundles. These zones are assumed to represent regions of greater microtubule density within bundles (Streiblova and Girbardt 1980; Busch and Brunner 2004; Carazo-Salas et al. 2005). They are generally found near the nucleus, where they undergo modest movements back and forth within the bundles, but rarely stray to the ends of these bundles. Recent studies localized γ-tubulin microtubule nucleation complexes to discrete sites within these bundles, and provided evidence for microtubule growth within the bundles (Sawin et al. 2004; Zimmerman and Chang 2005). Thus, these brighter zones may be generated by microtubule nucleating complexes generating microtubules to maintain the integrity of individual bundles. The excessive movement of these zones from one end of the cell to another within bundles when either dynein or Peg1 is missing indicates that these molecules restrain the movement of these zones within bundles. Perhaps they anchor the minus ends of one microtubule to the lattice of neighboring microtubules to prevent the kinesin Klp2 from inducing excessive sliding within bundles (Carazo-Salas et al. 2005). Consistently, the functional Peg1-GFP fusion protein Peg1.N2GFP accumulated in discrete foci within interphase microtubule bundles near the nuclei of starving cells; however, Peg1 does not appear to play a role in microtubule nucleation. A second GFP fusion protein, Peg1.C4GFP, that did not fully support Peg1’s interphase microtubule-destabilizing function and led to excessive migration of overlap zones within microtubule bundles was seen further away from the nucleus than Peg1.N2GFP. Thus, although Peg1.C4GFP was recruited to sites of action, its inability to work properly there meant that Peg1 complexes were no longer restrained to regions near the nucleus and the restraint on the overlap zone was relieved.
The curling of many microtubules around cell ends suggested that these microtubule ends were unable to detect when they encountered the cell cortex when Peg1 or Dhc1 was missing. Given the association of counterparts of both molecules with each other and microtubule ends in other systems (Han et al. 2001; Li et al. 2005; Mimori-Kiyosue et al. 2005), we expected to see association of the two molecules and find both molecules at microtubule tips in S. pombe. The two proteins did co-precipitate and Peg1 was found at microtubule tips; however, we were unable to detect a Dhc1.C4GFP fusion protein in vegetative cells (although we were able to see these Dhc1.C4GFP molecules expressed from this native locus lining the cortex and associating with the SPB during meiotic prophase) (Yamamoto et al. 1999; data not shown). The inability to see Dhc1.C4GFP could be due to camera threshold sensitivity as we could only detect Peg1.N2GFP at microtubule tips with cameras containing highly sensitive electron amplification chips, and the scarcity of Dhc1 protein and mRNA in whole-cell extracts suggests that Dhc1 levels are very low, indeed (Yamamoto et al. 1999). However, an alternative explanation could be that dynein association is a highly transient interaction that has the immediate impact of destabilizing the microtubules to which they are recruited. In this scenario, Dhc1.C4GFP would not accumulate above detection thresholds because the docking site would be rapidly lost. This would contrast with the +TIPs Tea2, Mal3, and Tip1 that need to stay on micro-tubule ends in order to transport significant levels of Tea1 to cell ends. A final possibility is that dynein could be required to load Peg1 or additional factors at the minus ends for transportation to influence dynamics at plus ends.
Peg1 reduces the rate of microtubule polymerization at cell tips
The rate of polymerization of interphase microtubules slows after they establish end-on associations with the cortex at cell tips (Tran et al. 2001). Compromising Peg1 and, to a lesser extent, Dhc1 function reduced the degree to which contact with the tip slowed microtubule polymerization, suggesting that the deceleration exploited some aspect of Peg1/Dhc1 function. Somewhat paradoxically, slowing was seen once more when both molecules were absent. A similar effect was seen on polymerization pausing intervals; peg1.1 and dhc1.Δ reduced pausing, yet pausing was seen once more in dhc1.Δ peg1.1 backgrounds. Perhaps similar mechanisms underlie pausing and slowing of microtubule polymerization.
The stochastic nature of microtubule capture in peg1.1 and dhc1.Δ
Loss of Peg1 and Dhc1 function did not stop all of the microtubules that encounter the cell cortex from establishing end-on associations. Furthermore, different imaging conditions generate diversity in microtubule behavior at cell tips (Tran et al. 2001). What could account for this variability? The impact of these molecules might be due to the angle at which they contact the cortex. Alternatively, different interphase microtubule bundles may differ in their requirements for Dhc1 and Peg1. In this case, Dhc1 and Peg1 could represent one of several means by which end-on association can be regulated, so that the stochastic recruitment of a different spectrum of MAPs before cell ends are encountered may render some microtubules competent to dock via a Peg1/Dhc1-independent mechanism, while others that fail to pick up these additional factors are reliant on Peg1/Dhc1. In higher eukaryotes, different +TIPs do, indeed, promote distinct interactions with the cortex. EB1 and APC bind the formin Diaphanous to stabilize microtubules in response to Rho signaling, and yet Rac and Cdc42 signaling capture microtubule ends via IQGAP1 and CLIP170 (Fukata et al. 2002; Watanabe et al. 2004; Wen et al. 2004).
Peg1 and dynein
Peg1 localization relies on dynein: The two molecules associate, and compromising the function of either gene generates similar phenotypes. Furthermore, many of the phenotypes arising from loss of Peg1 function are only seen when dynein is present. However, the relationship between these two molecules is more complex than a simple issue of CLASP and dynein acting in concert to some common goal. Thus, although the ability of Peg1 to restrain microtubule numbers in interphase bundles requires an active dynein complex, depletion of dynein itself does not similarly enhance the intensity of these bundles. This means that Peg1 function must modify dynein function to decrease microtubule polymerization within interphase bundles. Similarly, while Peg1 requires dynein to destabilize microtubules in tip1.Δ mal3.Δ cells, loss of dynein itself does not increase their stability, so once more the consequences of loss of Peg1 and loss of Dhc1 are not equivalent, despite the fact that both are recessive mutations and peg1.1 is epistatic to dhc1.Δ.
Materials and methods
Strains, cell culture, and molecular biology
Strains are listed in Supplementary Table 1. Standard fission yeast and molecular biology approaches were used throughout (Moreno et al. 1991). peg1.1-suppressing plasmids were cloned from a genomic library (Barbet et al. 1992). The peg1 coding sequences of peg1.1 were amplified with Pfu DNA polymerase in five independent PCR reactions that were pooled for sequencing. Repeating this procedure identified the same mutation. C-terminal tagging of peg1 +, dhc1 +, and tip1 + with 4GFP, 6HA, 9myc, and 6Pk was done by the PCR approach of Bähler et al. (1998) using pSM1023, pYM14, pYM18, and pFA6a-6Pk as templates (Maekawa et al. 2003; Janke et al. 2004). peg1 + was deleted by the same PCR approach using pFA6a-hphMX6 (Hentges et al. 2005). The marker switch approach of MacIver et al. (2003a), in which the ura4 + marker was inserted 936 base pairs (bp) upstream of the peg1 + ATG, was used to generate fusions in which the tag was located at the N terminus of Peg1. For two-hybrid analysis, peg1 +, tip1 +, tea1 +, and mal3 + cDNAs were cloned into pMM5, pMM6 (Pereira et al. 1999) as BamHI fragments and processed according to Pereira et al. (1999).
Cell biology
Standard fluorescence procedures that used calcofluor or AP9.2 (anti-Sad1), TAT1 (anti-α-tubulin) antibodies were as described previously (Woods et al. 1989; Moreno et al. 1991; Hagan and Yanagida 1995). Images were obtained using a Quantix camera (Photometrics) with Metamorph software (Universal Imaging). For live cell imaging to measure MT dynamics, a Perkin Elmer spinning disk confocal or Deltavision Spectris system was used. Spinning disk images were captured as sets of 10 12 × 0.6-μm slices with 5-sec delay. Each 10-slice stack took a maximum of 150 msec to capture. For Figures 1 (C, E, and G), 2, 3, 5B, 6 (B [main panel], C, D [top panel], and F), and Supplementary Figures S1, S4, and S5, the Z series was compressed to a maximal projection. For Figure 6 (A, D [bottom panel], E, and G) three sections were compressed to generate the images, while for Figure 7, one plane was imaged and for Supplementary Figure S2, one plane is shown. For live imaging on the Deltavision Spectris system 20 0.3-μm consecutive slices were continuously captured with a single passage through a set of 20 slices taking 6.5 sec. Cells were grown in filter-sterilized EMM2 supplemented media. The procedures of Brunner and Nurse were replicated for pnmt1GFPatb2 strains (Brunner and Nurse 2000). All image analysis used Imaris (Bitplane) software. For each genotype, 50 microtubules were imaged for growth and shrinkage rates and >100 for other parameters. For all live cell imaging, cells were mounted in a Bioptechs FC2 chamber with lectin (Sigma, L2380; 0.5 mg/mL). TRITC-labeled lectin was from Sigma (L5264).
Biochemistry
Standard Western blotting approaches following TCA precipitation and immunoprecipitation of cell extracts were as described previously (MacIver et al. 2003b), with the exception that immunocomplexes were washed in 25 mM HEPES (pH 7.5), 50 mM K-acetate, 1 mM MgCl2, 1 mM EDTA, 1% Nonidet p40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL aprotinin, 1 μg/mL pepstatin, 1 μg/mL leupeptin, 15 mM β-glycero-phosphate, 0.1 mM Na3VO4, and 50 mM NaF. For Western blotting and immunoprecipitation, anti-Pk (336) (Serotec), anti-myc (9E10), and anti-HA (12CA5) (CRUK) monoclonal antibodies were used. For Dhc1.HA immunoprecipitation and related control reactions, extracts were prepared by lysing cells with a Grentsch liquid-nitrogen pestle and mortar.
Acknowledgments
We thank Takashi Toda, Dick McIntosh, Kayoko Tanaka, Isabel Alvarez, Dan Mulvihill, Paul Nurse, Tony Carr, Elmar Schiebel, Corina Vietmeier-Decker, Inga Karig, Anne Kerres, Fred Chang, Olaf Nielsen, Frederique Bidard-Michelot, Roger Tsien, and Keith Gull for reagents. This work was supported by Cancer Research UK (CRUK) and Deutsche Forschungsgemeinschaft (SFB 590).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.381306.
References
- Akhmanova A., Hoogenraad C.C. Microtubule plus-end-tracking proteins: Mechanisms and functions. Curr. Opin. Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., et al. CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W.J., Carr A.M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe . Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Beinhauer J.D., Hagan I.M., Hegemann J.H., Fleig U. Mal3, the fission yeast homolog of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Hackney D.D. The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J. Biol. Chem. 2005;280:12299–12304. doi: 10.1074/jbc.M413620200. [DOI] [PubMed] [Google Scholar]
- Browning H., Hayles J., Aveline L., Nurse P., McIntosh J.R. A kinesin-like protein required for proper cellular morphology in Schizosaccharomyces pombe . Mol. Biol. Cell. 1998;9 [Google Scholar]
- Browning H., Hayles J., Mata J., Aveline L., Nurse P., McIntosh J.R. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 2000;151:15–28. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Hackney D.D., Nurse P. Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 2003;5:812–818. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- Brunner D., Nurse P. CLIP170-like tip1 spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Busch K.E., Brunner D. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.R., Antony C., Nurse P. The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science. 2005;309:297–300. doi: 10.1126/science.1113465. [DOI] [PubMed] [Google Scholar]
- Coquelle F.M., Caspi M., Cordelieres F.P., Dompierre J.P., Dujardin D.L., Koifman C., Martin P., Hoogenraad C.C., Akhmanova A., Galjart N., et al. LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol. Cell. Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.-Q., Chikashige Y., Haraguchi T., Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Drummond D.R., Cross R.A. Dynamics of interphase microtubules in Schizosaccharomyces pombe . Curr. Biol. 2000;10:766–775. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- Dujardin D., Wacker U.I., Moreau A., Schroer T.A., Rickard J.E., De Mey J.R. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 1998;141:849–862. doi: 10.1083/jcb.141.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Galjart N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- Garcia M.A., Vardy L., Koonrugsa N., Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt M.K., Savoian M.S., Riparbelli M.G., Massarelli C., Callaini G., Glover D.M. Klp67A destabilises preanaphase microtubules but subsequently is required to stabilise the central spindle. J. Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Lustig R.J., Berlin A., Chang F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 2001;11:836–845. doi: 10.1016/s0960-9822(01)00235-4. [DOI] [PubMed] [Google Scholar]
- Goodson H.V., Skube S.B., Stalder R., Valetti C., Kreis T.E., Morrison E.E., Schroer T.A. CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil. Cytoskeleton. 2003;55:156–173. doi: 10.1002/cm.10114. [DOI] [PubMed] [Google Scholar]
- Hagan I.M. Fission yeast microtubules. J. Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan I.M., Hyams J.S. The use of cell-division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe . J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. The product of the spindle formation gene sad1 +associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Liu B., Zhang J., Zuo W., Morris N.R., Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., Carr A.M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe . Yeast. 2005;15:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Inoue Y.H., do Carmo Avides M., Shiraki M., Deak P., Yamaguchi M., Nishimoto Y., Matsukage A., Glover D.M. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster . J. Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y.H., Savoian M.S., Suzuki T., Mathe E., Yamamoto M.T., Glover D.M. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 2004;166:49–60. doi: 10.1083/jcb.200402052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. Beyond self-assembly: From microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G.G., Akhmanova A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell. 2005;16:5334–5345. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Komarova Y., Modesti M., Wyman C., Hoogenraad C.C., Goodson H.V., Lemaitre R.P., Drechsel D.N., van Munster E., Gadella T.W., Jr, et al. Conformational changes in CLIP-170 regulate its binding to micro-tubules and dynactin localization. J. Cell Biol. 2004;166:1003–1014. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laycock J.E., Savoian M.S., Glover D.M. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J. Cell Sci. 2006;119:2354–2361. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K., Van Vactor D. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 2004;42:913–926. doi: 10.1016/j.neuron.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Li J., Lee W.-L., Cooper J.A. NudEL targets dynein to microtubule ends through Lis1. Nat. Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver F.H., Glover D.M., Hagan I.M. A ‘marker switch’ approach for targeted mutagenesis of genes in Schizosaccharomyces pombe . Yeast. 2003a;20:587–594. doi: 10.1002/yea.983. [DOI] [PubMed] [Google Scholar]
- MacIver F.H., Tanaka K., Robertson A.M., Hagan I.M. Physical and functional interactions between polo kinase and the spindle pole component Cut12 regulate mitotic commitment in S. pombe . Genes & Dev. 2003b;17:1507–1523. doi: 10.1101/gad.256003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M., Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., Earnshaw W.C. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., Rieder C.L. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Nurse P. Tea1 and the microtubular cyto-skeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Mathe E., Inoue Y.H., Palframan W., Brown G., Glover D.M. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oo-genesis. Development. 2003;130:901–915. doi: 10.1242/dev.00315. [DOI] [PubMed] [Google Scholar]
- Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic pro-phase in fission yeast. Mol. Biol. Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Pasqualone D., Huffaker T.C. Stu1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Grueneberg U., Knop M., Schiebel E. Interaction of the yeast γ-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;18:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Lourenco P.C., Snaith H.A. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Streiblova E., Girbardt M. Microfilaments and cytoplasmic microtubules in cell division cycle mutants of Schizosaccharomyces pombe . Can. J. Microbiol. 1980;26:250–254. doi: 10.1139/m80-038. [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., Canman J.C., Salmon E.D., Mitchison T.J. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T., Marsh L., Doye V., Inoue S., Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Take-fuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Wen Y., Eng C.H., Schmoranzer J., Cabrera-Poch N., Morris E.J., Chen M., Wallar B.J., Alberts A.S., Gundersen G.G. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. J. Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M.J., Blake-Hodek K., Kosco K., Hwang E., You L., Huffaker T.C. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., Macrae T.H., Baines A.J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal-antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., West R.R., McIntosh J.R., Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S., Chang F. Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]