Abstract
Curved DNA fragments are often found near functionally important sites such as promoters and origins of replication, and hence sequence-dependent DNA curvature prediction is of great utility in genomics and bioinformatics. In light of this, an assessment of three different dinucleotide step parameters (based on gel retardation as well as crystal structure data) is carried out. These parameters (BMHT, LB and CS) are evaluated quantitatively for their ability to predict correctly the experimental results of a large set of nucleic acid sequences containing A-tracts as well as GC-rich motifs. This set contained around 40 synthetic as well as natural sequences whose solution properties have been well characterized experimentally. All three models could account reasonably well for curvature in the various DNA sequences. The CS model, where dinucleotide parameters are calculated from crystal structure data, consistently shows slightly better correlation with experimental data. Our simple analysis also indicates that presently available trinucleotide parameters fail to predict curvature in some of the well-characterized sequences. The study shows that the dinucleotide parameters with some further refinement can be used to predict sequence-dependent curvature correctly in genomic sequences.
INTRODUCTION
The role of intrinsic curvature of DNA in biologically important functions such as transcription, replication, recombination and chromatin organization has been well documented (1–10) over the last two decades. It has also been shown that elements of curved DNA are often found near functionally important sites such as promoters and origins of replication (1–10). Due to the functional importance of DNA curvature, biologists have long sought a biophysical explanation of this property (11–22). Initial observation of curvature in the sequences having periodic A-tract repeats led to various theories which tried to explain DNA curvature based on unique properties of oligo(A) tracts (23–24). However, observation of many more curved DNA fragments with sequence elements other than A-tracts necessitated formulation of a more general model. This led to a series of models based on sequence-dependent dinucleotide and trinucleotide parameters (25–37). These parameter sets were based on various experimental techniques such as X-ray crystallography, NMR, gel electrophoresis, cyclization kinetics, etc. In the light of the potential utility of DNA structure prediction in genomics and bioinformatics, it is essential to have an evaluation of various local parameters to arrive at an accurate estimate of the macroscopic curvature of nucleic acids. Though it has been thought that trinucleotide models (35–37) are necessary to explain DNA curvature in some of the sequences, dinucleotide models remain the simplest forms of nearest neighbor descriptions. Also, Liu and Beveridge (27) recently have re-examined the possibility that a dinucleotide parameter set, based on a refined method, can explain the curvature of A-tract as well as non-A-tract sequences. In view of this, we have carried out an assessment of different dinucleotide parameter sets.
Due to lack of enough data, some of the initially proposed dinucleotide parameters were based on a low number of observations. Some of the parameters are calculated using similar methods and give similar results. Considering this, we have selected three dinucleotide parameter sets, which are based on large data sets and different methodologies, i.e. BMHT (based on gel mobility data), CS (based on crystal structure data) and LB (based on gel mobility as well as crystal structure data).
The BMHT model, proposed by Bolshoy et al. (25), is based on DNA gel retardation values. They estimated 16 roll and tilt wedge angles based on independent gel mobility experiments performed on a training set of 54 different sequences. In this calculation, the twist values were fixed at those proposed earlier by Kabsch et al. (38). When choosing the starting values, previously determined roll and tilt values of the AA step (23) were taken into consideration. The resulting set of parameters have appreciable wedge angles in the case of dinucleotide steps AG/CT, CG/CG, GA/TC, GC/GC in addition to AA/TT. The BMHT model proves satisfactory for the sequences in the training set. However, due to its bias towards the AA/TT step, it fails to account for unusual cases such as unconventional helical phasing sequences proposed by Dlakic and Harrington (39) and the GGGCCC type of motif (36).
The CS model is based on mean values of dinucleotide step parameters obtained from B-DNA crystal structure data (26,40). Unlike the BMHT parameter set, the CS model is not based on any fitting procedure and hence can be upgraded as more data on novel structures become available. The striking feature of the model is the absence of large roll and tilt for the AA/TT dinucleotide step. Another unique feature of this parameter set is that it includes two distinctly different arrangements of CA/TG steps corresponding to the B-I and B-II conformations. The roll and tilt values corresponding to the B-II conformation are used when the CA/TG step is either preceded by a pyrimidine or followed by a purine, e.g. in CCA, TCA, CAA or CAG and their self-complementary sequences involving TG. Though the CS parameter set underestimates curvature of A-tract sequences, it gives a reasonably good agreement with the experimentally observed trends. The CS model was also shown to predict the curvature correctly in several naturally occurring sequences (26,40).
Recently, Liu and Beveridge (27) calculated dinucleotide step parameters based on a refined prediction method (the LB model). They carried out an optimization procedure using initial parameter values based on oligonucleotide crystal structure data. Further parameter optimization was carried out against the same experimental values used by Bolshoy et al. (25), using simulated annealing. However, in contrast to the BMHT model, all the 30 parameters (three base step parameters: roll, tilt and twist for the 10 unique base steps) were taken into consideration. During the optimization, the parameter variation is restricted within one standard deviation of crystal structure values. Due to the protocol followed, the LB model does not digress much from the crystal structure AA roll values, unlike the BMHT model. The LB model was tested (27) on a number of sequences including the GGGCCC motif (41) and helical phasing sequences (39) mentioned above. It is claimed (27) that it can account satisfactorily for the bending data for a wide variety of sequences.
The three different dinucleotide step parameters have been examined here for their ability to predict DNA structure correctly. The parameter sets are tested on a data set of nucleic acid fragments for which gel mobility data are available. Our data set is much larger and more diverse (encompassing A-tract-containing as well as non A-tract sequences) than previous studies. We find that the dinucleotide model, based on careful analysis of crystal structure data, consistently gives a more satisfactory correlation with experimental results when unrelated sequences are considered. The analysis, in addition, shows that trinucleotide parameters, based on DNase I sensitivity and nucleosomal positioning preferences, can also fail to predict curvature in some of the most extensively studied sequences.
METHODS
DNA structure prediction
The DNA duplex structures of the sequences, for which the gel mobility data are available (Table 1), were studied with the help of in-house software NUCGEN (42). For this purpose, three sets of dinucleotide parameters, i.e. BMHT (25), CS (26) and LB (27), were used to generate 96mer or longer fragments of DNA. Since it is not clear which is the best among the different measures used for quantifying varying degrees of DNA curvature, the predicted structures were analyzed for all of the following measures.
Table 1. The sequences of the synthetic DNA oligomers analyzed in this study.
| No. | Sequence | Name | Repeat unit (bp) | Experimental RL values (96mer) |
|---|---|---|---|---|
| ‘A-tract’ sequences (49) | ||||
| 1 | (GTGTAAAAAG GTGTAAAAAG GTGTAAAAAA G)n | Ab2R31 | 31 | 1.27 |
| 2 | (TCGGAAAAAA GTCGGAAAAA G)n | Abad21 | 21 | 1.28 |
| 3 | (TCGGAAAAAG TCGGAAAAAG TCGGAAAAAA G)n | Abad31 | 31 | 1.29 |
| 4 | (TGAGAAAAAC TGAGAAAAAC TGAGAAAAAA C)n | Ag2R31 | 31 | 1.30 |
| 5 | (GACTAAAAAA TGACTAAAAA T)n | Agood21 | 21 | 1.42 |
| 6 | (GACTAAAAAT GACTAAAAAT GACTAAAAAA T)n | Agood31 | 31 | 1.42 |
| 7 | (TCGGAAAAAA TGACTAAAAA G)n | Amix21 | 21 | 1.34 |
| 8 | (GGGCAAAAAA CGGGCAAAAA C)n | Ast21 | 21 | 1.39 |
| 9 | (GGGCAAAAAC GGGCAAAAAC GGGCAAAAAAC)n | Ast31 | 31 | 1.40 |
| Unconventional helical phasing DNA motifs (39)a | ||||
| 10 | (GGGCCCGAGA GAAAAAAGAG AGGGCCCAAA AA)n | AA-out | 32 | 1.00 |
| 11 | (GGGCCCAAAA AGAGAGGGCC CGAGAGAAAA AA)n | GC-out | 32 | 1.16 |
| 12 | (GGGCCCGAGA GGGCCCAAAA AAGAGAGAAA AA)n | GA-out | 32 | 1.29 |
| Sequences with A4T4 and T4A4 motifs (50,51) | ||||
| 13 | (CAAAATTTTG)n | (CA4T4G)n | 10 | 1.43 |
| 14 | (CTTTTAAAAG)n | (CT4A4G)n | 10 | 1.00 |
| 15 | (GAAAATTTTC)n | (GA4T4C)n | 10 | 1.40 |
| 16 | (GTTTTAAAAC)n | (GA4T4C)n | 10 | 1.00 |
| GGGCCC-type motif (41,52,53)b | ||||
| 17 | (GAGGGCCCTA)n | – | 10 | 1.27 |
| Mixed set of sequences (54) | ||||
| 18 | (GGCAAAAACG)n | A5N5 | 10 | 1.93 |
| 19 | (CCGGCAAAAA CGGGC)n | A5N10 | 15 | 1.02 |
| 20 | (TCTCTAAAAA ATATATAAAA A)n | A5N6 | 21 | 1.40 |
| 21 | (GAAAATTTTC)n | H-A4T4 | 10 | 1.70 |
| 22 | (GTTTTAAAAC)n | H-T4A4 | 10 | 1.08 |
| 23 | (GGCAAAATTT TCGAAAATTT T)n | QH-A4T4 | 21 | 1.50 |
| 24 | (GGCTTTTAAA ACGTTTTAAA A)n | QH-T4A4 | 21 | 1.26 |
| 25 | (CGCATGTCAC CGACGCATGA G)n | NA-CATG | 21 | 1.10 |
| 26 | (CTCCGGATAG GCTCCGGATA G)n | NA-CCGG | 21 | 1.10 |
| 27 | (TCATGCGACT ATATGCGACT A)n | NA-TATA | 21 | 1.04 |
| 28 | (GCATGAGATC GGCATGAGAT C)n | NA-GAGA | 21 | 1.00 |
aThe RL values correspond to the 200 bp sequence.
bThe RL value correspond to the experiment conducted in the presence of Mg2+ ions.
Curvature calculation using least square circle (LSC) fit
A circle can be fitted to the atomic coordinates of a DNA molecule curved in a plane. The radius of this circle can be taken as a measure of the curvature of the DNA fragment. The smaller the radius, the more curved is the DNA fragment and vice versa. We used a least square fitting method to trace a circle through the base pair centers that define the three-dimensional path of the DNA. For all the generated DNA molecules, the calculated values of the r.m.s.d., from the best-fit plane, were found to be very small, indicating that the molecules are near planar. To avoid erroneous fitting, we also examined the r.m.s.ds from the fitted line (rmsL) as well as the circle (rmsC). The trajectory with the smaller r.m.s.d. was assigned as being the correct one. The DNA curvature is measured in terms of curvature units (43), where one curvature unit corresponds to the mean DNA curvature in the crystallized nucleosome (radius = 42.8 Å).
Curvature (LSC) = 42.8 Å/radius of the best fit circle
Koo and Crothers (24) have reported that relative gel mobility (RL) is a linear function of curvature squared. Hence for carrying out a quantitative analysis, we have plotted (LSC)2 against RL.
Ratio of end to end distance to the contour length (d/lmax)
Given the atomic coordinates of a DNA fragment, the end to end distance ‘d’ is the shortest distance between centers of the first and the last base pairs. The ratio of ‘d’ to total length traced by the DNA path, ‘lmax’ is used as one of the measures of curvature (26,39,44–46). For an ideally straight molecule, this ratio will be unity, while for a completely closed circle the ratio will be zero. Hence, the d/lmax ratio varies between limits of zero and one. The more curved the DNA, the smaller this ratio is.
Ratio of moments of inertia (Imax/Imin)
To describe the global nature of DNA structure, an ellipsoid can be fitted to the DNA construct. The magnitudes of the moments of inertia of the ellipsoid can be used to define the shape of the molecule. One of the three components of moments of inertia for a linear rod-like molecule will be very large compared with the others, while all the three components will be of similar magnitude for a molecule with a small radius of curvature and folded into a globular shape. The ratio of the maximum component (Imax) to the minimum component (Imin) can serve as a measure of relative elongation of the object (26,39,44–48). The Imax/Imin ratio will be unity for a perfect circle. For a particular structure, the Imax/Imin ratio varies inversely with respect to the LSC.
Cumulative and successive bending angles
Vectors connecting every nth base pair center can define the DNA trajectory. The successive bending angle can be defined as the angle between consecutive vectors, while the cumulative bending angle at a particular point is the angle between the vector at that point and the first vector. The magnitude of the bending angle and the position of maximum bending can provide important information about the varying geometry along the DNA molecule (27,36), particularly for non-repetitive DNA sequences. For bending angle calculations, vectors joining every 10th base pair, i.e. the nth and (n + 10)th base pair, have been used. The cumulative bending angle at the nth position corresponds to the angle made by the vector joining the nth and (n + 10)th base pair centers with the vector joining the first base pair center to the 10th.
For each generated DNA duplex model, all the above measures were calculated and were used for the comparison with experimental data. We validated the values obtained for each measure by visual inspection of three views of the DNA structures, after orienting the molecule in the mutually perpendicular x–y, y–z and z–x planes.
Calculation of curvature using the ‘BEND’ program
The curvature values have also been calculated using the BEND program (36). In this analysis, two trinucleotide parameters based on nucleosomal positioning preferences (36,37) and DNase I sensitivity (35) are included in addition to the three dinucleotide parameter sets. The curvature, in this algorithm, is defined as the angle between averaged normal vectors 31 bp apart.
RESULTS AND DISCUSSION
As mentioned above, considering the need of sequence-dependent DNA structure prediction, we have carried out a comparison between three different dinucleotide parameter sets. The evaluation of parameters is based on quantitative comparison as far as possible. We have carried out the comparison on a much larger set of sequences, which cover a wider spectrum of dinucleotide types, than used in previous studies.
Comparison of predicted curvature with the experimental data derived from gel mobility experiments
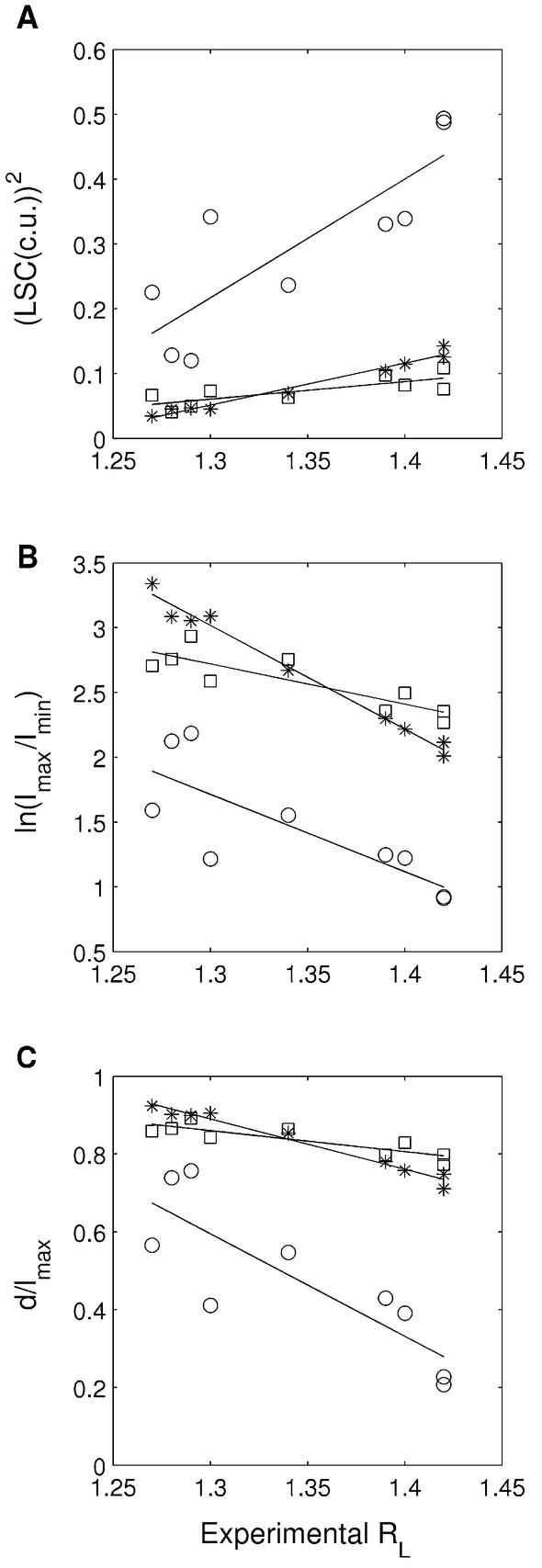
‘A-tract’ sequences. Polymeric sequences with A-tracts have been studied extensively and their gel mobility has been correlated with the extent of bending. A systematic study of 21mer and 31mer repeat-containing phased A6 and A5 tracts interspersed with non-A-tracts has been carried out by Haran et al. (49). They reported a set of gel retardation values and the corresponding curvature values, obtained using the formula mentioned earlier (24). The DNA models of these sequences generated using BMHT and CS parameter sets have already been tested (26,49) by comparing the predicted curvature with the experimental one. However, since there was no such analysis using the LB model, for the sake of completeness, we have carried out the same kind of comparison for the 96 bp long DNA fragments generated using all the three parameter sets (Table 1: sequences 1–9). While correlating with experimental data, we have included Imax/Imin and d/lmax values in addition to LSC. From Figure 1, it is obvious that all the three parameter sets predict the curvature well. The CS model predicts the experimental data significantly better than the other two. Both the parameters based on crystal structure data (CS and LB models) predict considerably less curvature than the BMHT model, which is itself based on similar gel mobility data but still predicts higher curvature values than the experimental ones. The scaling factors, for BMHT, CS and LB parameters, required to bring the average theoretical curvature into agreement with experimental curvature values are 0.69, 1.33 and 1.33, respectively. The slight difference in BMHT scaling factor, from the value reported (0.63) by Haran et al. (49), may be due to the slightly different methods followed for DNA structure generation and for calculating curvature.
Figure 1.
Comparison between the experimentally determined gel retardation values (RL) and the predicted curvature calculated using dinucelotide parameters, i.e. BMHT (open circles), CS (stars) and LB (open squares), for repetitive ‘A-tract’ sequences (Table 1: sequences 1–9). The experimental RL values (Table 1) constitute the x-axis, while predicted curvature in terms of LSC, Imax/Imin and d/lmax is on the y-axis. In the case of Imax/Imin, the ln(Imax/Imin) values are plotted on the y-axis and, in the case of LSC, the (LSC)2 values are plotted. The predicted and the experimental values correspond to the 96mer DNA sequences. (A) The correlation between RL values and (LSC)2 calculated using BMHT parameters (R = 0.84), CS parameters (R = 0.99) and LB parameters (R = 0.78). (B) The correlation between RL values and ln(Imax/Imin) calculated using BMHT parameters (R = –0.80), CS parameters (R = –0.99) and LB parameters (R = –0.86). (C) The correlation between RL values and d/lmax calculated using BMHT parameters (R = –0.84), CS parameters (R = –0.99) and LB parameters (R = –0.86).
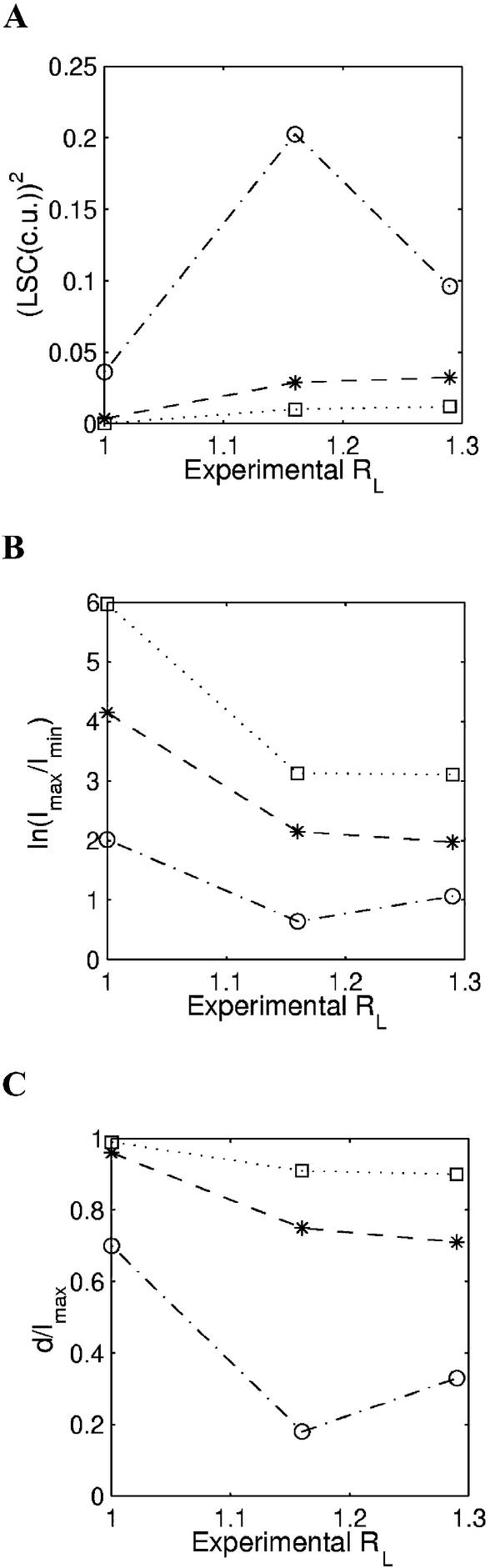
Unconventional helical phasing of DNA motifs. Dlakic and Harrington (39) analyzed polynucleotides with identical composition but different combinations of AAAAA(A), GGGCCC and GAGAG motifs. The sequences were synthesized such that one of the above motifs is out of phase, while the other two motifs are in phase with the DNA helix (see Table 1: sequences 10–12) and are referred to as AA-out, GC-out and GA-out. According to the gel mobility data, GA-out shows maximum curvature, while AA-out seems to be straight and GC-out shows intermediate curvature. It was suggested that trinucleotide models are more successful than dinucleotide models in predicting the curvature of these sequences (39). In a separate study, the dinucleotide model proposed by Liu and Beveridge (27) was shown to predict the correct trend qualitatively. We analyzed the structures of these sequences using three different geometrical measures (LSC, d/lmax and Imax/Imin described above). These DNA structures were generated for 200 bp long fragments using the three dinucleotide parameter sets. As in previous studies, we could also carry out only a qualitative comparison with experimental trends, since the number of sequences in this set is too small for a quantitative analysis (Fig. 2). In agreement with the earlier report (39), DNA structures generated using the BMHT model do not show experimentally observed trends for these three sequences. The BMHT parameter set predicts all the three sequences, including AA-out, to be curved, and GC-out is predicted to be more bent than GA-out, both these predictions being contrary to the observed data. On the other hand, for all three sequences, the CS and LB models do slightly better, in that AA-out is predicted to be straight and the other two to be curved, in agreement with the observed experimental trend (Fig. 2). However, the LB model predicts very low curvature for the moderately bent GC-out and GA-out sequences. The CS model predicts the gel mobility trends marginally better, though the predicted curvature of GC-out is still less than the expected value.
Figure 2.
Comparison between the experimentally determined gel retardation values (RL) and the predicted curvature for repetitive DNA sequences having unconventional helical phasing motifs (Table 1: sequences 10–12), using dinucelotide parameters, i.e. BMHT (open circles and dashed and dotted line), CS (stars and dashed line) and LB (open squares and dotted line). The experimental RL values (Table 1) constitute the x-axis, while predicted curvature in terms of (A) LSC, (B) Imax/Imin and (C) d/lmax is on the y-axis. In the case of Imax/Imin, the ln(Imax/Imin) values are plotted on the y-axis and in the case of LSC, the (LSC)2 values are plotted.
Sequences with an A4T4 motif are bent while those with T4A4 motif are straight. Hagerman (50,51), using a set of elegant experiments, showed that DNA polymers with different polarity of oligo(dA)–oligo(dT) tracts behave very differently with respect to their gel mobilities. The four representative sequences are listed in Table 1 (sequences 13–16). It can be seen (Table 2) that all three parameter sets are able to reproduce the observed trend qualitatively. It should be noted that the training set of sequences used for optimization of the LB and BMHT parameters includes the sequences CT4A4G and GT4A4C as well as a number of sequences similar to CA4T4G and GA4T4C. Hence it is not surprising that these two models predict the trend correctly. It is also clear that the BMHT model is far superior in predicting the difference between the highly curved A4T4 and non-curved T4A4 sequences than the other two models. It is surprising that in spite of using the same training set, the discrimination between these sequences is far less in the case of the LB model, which predicts very low curvature for the highly curved A4T4 sequences. It is also quite interesting that the CS model, which is not biased in a similar manner, is in agreement with experimental data for three of the sequences. The only exception is GA4T4C, which is correctly predicted to be bent, but the curvature is slightly less than that expected from gel mobility data. It should be noted that all three models show some context dependence in the case of the highly curved A4T4 motif, while experimentally they show nearly identical gel retardation.
Table 2. Comparison between the experimental and predicted values for the 96 bp repetitive sequences with an A4T4 motif and T4A4 motif (Table 1: sequences 13–16).
| Sequence | Experimental RL | BMHT | CS | LB | |
|---|---|---|---|---|---|
| LSC | (CA4T4G)n | 1.43 | 0.55 | 0.62 | 0.12 |
| (CT4A4G)n | 1.01 | 0.03 | 0.01 | 0.04 | |
| (GA4T4C)n | 1.40 | 0.43 | 0.26 | 0.14 | |
| (GT4A4C)n | 1.01 | 0.03 | 0.09 | 0.08 | |
| Imax/Imin | (CA4T4G)n | 1.43 | 3.8 | 3.0 | 53.9 |
| (CT4A4G)n | 1.01 | 242.9 | 332.9 | 202.8 | |
| (GA4T4C)n | 1.40 | 5.9 | 15.4 | 40.9 | |
| (GT4A4C)n | 1.01 | 273.9 | 96.1 | 90.7 | |
| d/lmax | (CA4T4G)n | 1.43 | 0.46 | 0.34 | 0.97 |
| (CT4A4G)n | 1.01 | 1.00 | 1.00 | 1.00 | |
| (GA4T4C)n | 1.40 | 0.65 | 0.86 | 0.95 | |
| (GT4A4C)n | 1.01 | 1.00 | 0.98 | 0.98 |
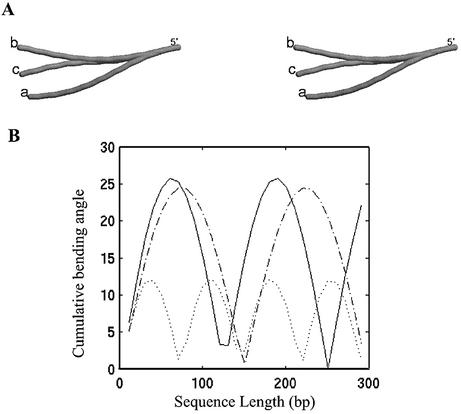
GGGCCC type of motif. It has been observed experimentally that a non-A-tract sequence, containing the GGGCCC motif, in phase with helix periodicity (Table 1: sequence 17), in the presence of Mg2+, also shows curvature with magnitude similar to phased A-tracts (41,52,53). In a previous analysis by Goodsell and Dickerson (36), it was noted that only trinucleotide parameters could account for curvature due to the GGGCCC motif, while the dinucleotide models fail to do the same. This study included only the BMHT model out of the three dinucleotide models examined here. It has been reported recently that the structure of a polymer with the repeat sequence, d(GAGGGCCCTA)n, as predicted by the LB model, shows a bending profile indicating significant curvature, though not as pronounced as for A-tracts (27). We therefore evaluated the three parameter sets in the context of this sequence. Since there are no related sequences to compare in this case, we visually examined the bending profile for the structures generated using the three parameters (Fig. 3). The largest cumulative bending angles corresponding to the BMHT, CS and LB parameters are 26°, 25° and 12°, respectively (as shown in Fig. 3B). The corresponding bending angle values for the structure of an A-tract sequence (Table 1: sequence 1), which has a similar gel mobility, are 162° (BMHT), 141° (CS) and 83° (LB). The other measures also indicate that the magnitude of this bending is very small compared with the A-tract sequence and cannot be considered very substantial. An inspection of the bending angle profile over a longer stretch also gives a clear indication of the serpentine nature of the structures (Fig. 3B). Though the bending angle value corresponding to the LB model is similar to that reported by Liu and Beveridge (27) and smaller than the values obtained for the BMHT and CS models, we tend to conclude that the magnitude of predicted bending is insignificant over a longer fragment.
Figure 3.
(A) The stereo pairs of the DNA structures of a 96mer fragment with repetitive (GAGGGCCCTA)n sequence (Table 1: sequence 17) generated using (a) BMHT parameters, (b) CS parameters and (c) LB parameters. All the structures are represented as cylinders passing through base pair centers. The structures are superimposed for the first 10 pairs of C1′ atoms at the 5′ end. (B) The cumulative bending angle profile for the GC-rich sequence over a length of 300 bp. The cumulative bending angle values (y-axis), calculated using BMHT parameters (continuous line), CS parameters (dashed and dotted line) and LB parameters (dotted line) are plotted along the sequence length (bp).
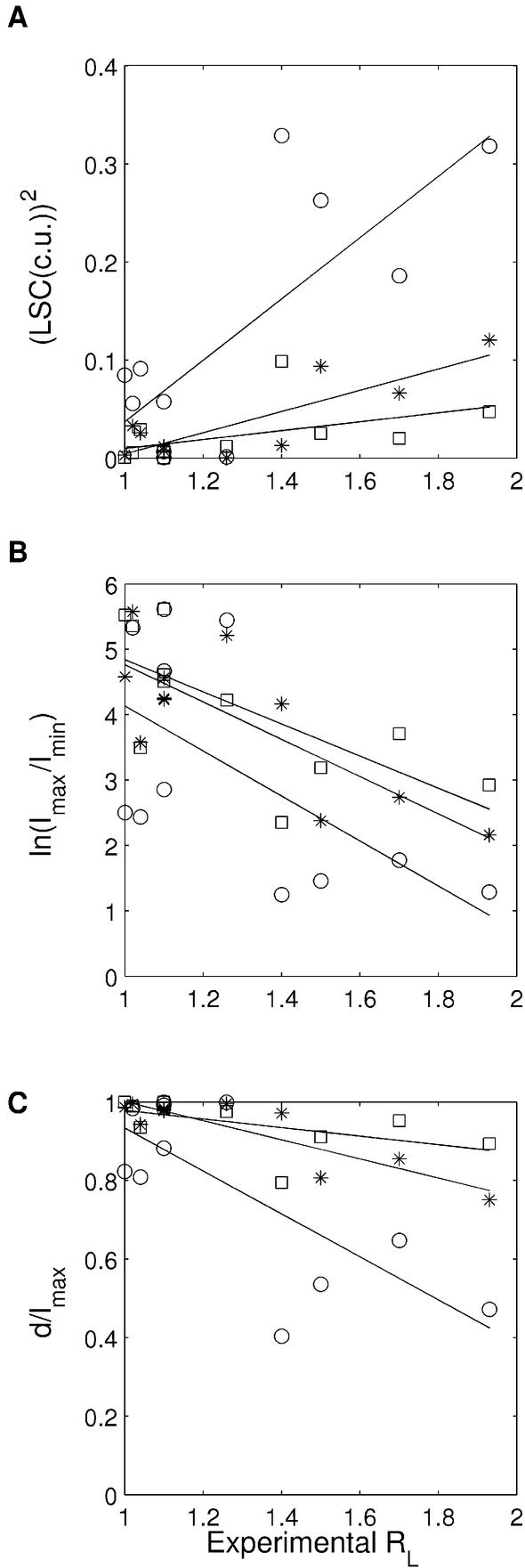
Mixed group of sequences studied in a single set of experiments. Some of the sets of sequences discussed above have only a small number of related sequences and hence are insufficient for quantitative comparison with the trends shown by experimental data. Another problem is that each of them is designed to study a particular type of sequence motif and hence each set has very similar sequences. At the same time, different experimental conditions under which the various sets are studied do not allow pooling of all the experimental results together. This confines the comparison of the prediction with experimental data to a qualitative level. However, recently, gel retardation data have become available for a diverse set of sequences which have been studied under identical experimental conditions by Ussery et al. (54). This set of sequences has A-tract-containing as well as GC-rich sequences (Table 1: sequences 18–28). They also re-examined the two sequences, (GA4T4C)n and (GT4A4C)n, studied earlier by Hagerman (50,51), and reported slightly different retardation values. We have also retained them, so that we can carry out quantitative comparison between the experimentally determined gel retardation values (RL) and the predicted curvature calculated as LSC, Imax/Imin and d/lmax (Fig. 4). The correlation coefficients, corresponding to the three dinucleotide parameters (given in the legend to Fig. 4), clearly show that the CS model follows the experimental trend better than the other two parameter sets. However, as in other cases, the range of curvature predicted by the CS and LB parameters is smaller than for the BMHT set.
Figure 4.
Comparison between the experimentally determined gel retardation values (RL) and the predicted curvature for the mixed set of sequences studied by Ussery et al. (54) (Table 1: sequences 18–28), using dinucleotide parameters, i.e. BMHT (open circles), CS (stars) and LB (open squares). The experimental RL values (Table 1) constitute the x-axis, while predicted curvature in terms of LSC, Imax/Imin and d/lmax is on the y-axis. The experimental and predicted values correspond to the 96mer DNA sequences. In the case of Imax/Imin, the ln(Imax/Imin) values are plotted on the y-axis and, in the case of LSC, the (LSC)2 values are plotted. (A) The correlation between RL values and (LSC)2 calculated using BMHT parameters (R = 0.77), CS parameters (R = 0.83) and LB parameters (R = 0.48). (B) The correlation between RL values and ln(Imax/Imin) calculated using BMHT parameters (R = –0.60), CS parameters (R = –0.79) and LB parameters (R = –0.69). (C) The correlation between RL values and d/lmax calculated using BMHT parameters (R = –0.75), CS parameters (R= –0.88) and LB parameters (R = –0.55).
Other natural sequences from prokaryotes and eukaryotes. Regions of curved DNA are known to be present in many naturally occurring DNA sequences from prokaryotes and eukaryotes (10,20,55). Since most of the sequences have phased A-tracts, it is not surprising that all the models generated using any of the three parameter sets predict them to be bent.
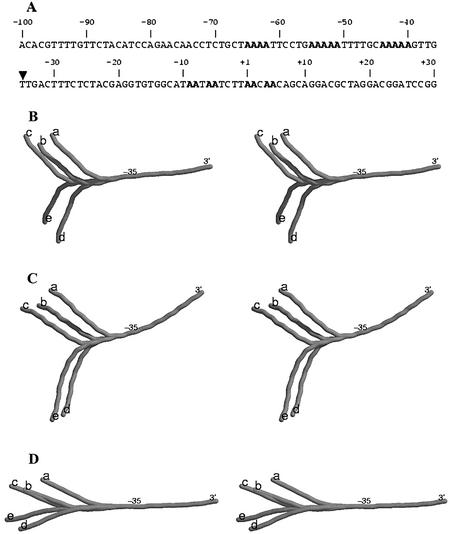
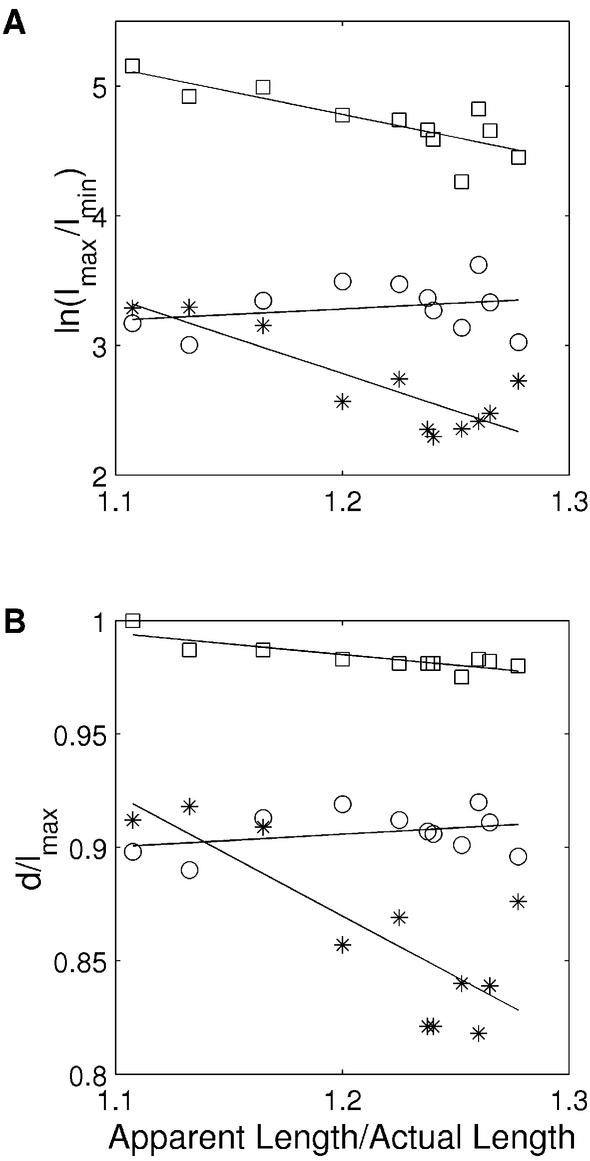
One of the most interesting and systematic studies on curvature of a natural sequence is that of the Alu156 promoter. The Alu156 promoter (Fig. 5A) isolated from the Bacillus subtilis bacteriophage SP82 has two curved elements, one immediately upstream and another downstream of the –35 position (56,57). The curvature in the upstream region is attributed to the presence of phased A-tracts, while the downstream region has two pairs of adenines at positions –5 and +5. Due to its curved nature, the wild-type Alu156 promoter migrates with retarded gel mobility, a rate consistent with a molecule 22% larger than its actual size. McAllister and Achberger (57) altered the rotational orientation of the upstream curved DNA using short DNA insertions of 6, 9, 11, 13, 15, 17, 19, 21, 25 and 29 bp between this curved DNA and the promoter region downstream of the –35 position. When these mutant promoters were analyzed, it was found that changes in rotational orientation of the two curved DNA fragments correlated with changes in promoter function. The most efficient mutant promoters contained insertions of 11 and 21 bp, and insertions of 15 and 25 bp resulted in the least efficient mutant promoters. Since 11 and 21 bp insertion mutants maintain the helical phase of DNA, it is expected that the two curved elements will be in the same rotational orientation as that in the wild type, albeit further apart. For the same reason, in 15 and 25 bp insertion mutants, the direction of the upstream curve element will be opposite with respect to wild type. All three dinucleotide models are successful in predicting curvature in the A-tract-rich upstream region (Fig. 5). The superposition of the mutant structures by aligning the 3′ ends (corresponding to the base pairs between positions –35 and +30) of mutants with the wild-type structure clearly shows that the three dinucleotide models can predict correct rotational orientation in the 11, 21, 15 and 25 bp insertion mutants (Fig. 5). This difference in orientation is most pronounced in the structures predicted by the CS model and least for the LB parameters. The CS model is also superior to other models in predicting curvature in the region downstream of the –35 position. Here it is important to stress that the sequence downstream of the –35 region, in contrast to the upstream region, does not have any A-tracts except for pairs of adenines at positions –5 and +5. The quantitative comparison of the ratio of apparent to actual length, based on the gel mobilities of these sequences, with Imax/Imin and d/lmax values (Fig. 6), also confirms that the CS model predicts the experimental trend very well. [These structures showed large deviations from the circle as well as line fit (see Methods), and hence the curvature calculated in terms of LSC is not considered.]
Figure 5.
(A) The nucleotide sequence of the wild-type Alu156 promoter spanning the region –100 to +30. The site of insertions (–35 position) is indicated by a filled triangle. The runs of adenines in the upstream region and the two pairs of adenines at positions +5 and –5 are shown in bold. (B–D) The stereo pairs of the DNA structures predicted for (a) the 130mer wild-type Alu156 promoter sequence shown in (A) and for its mutants with (b) 11 bp, (c) 21 bp, (d) 15 bp and (e) 25 bp inserts. All the structures are represented as cylinders passing through the base pair centers The structures have been generated using different dinucleotide parameters, i.e. BMHT (B), CS (C) and LB (D). The 3′ end and the –35 position (the site of DNA insertions) are indicated. Identical regions of the various structures for insertion mutants and the wild type, corresponding to the base pairs between positions –35 and +30, have been superimposed. The different orientations of the upstream region in the various length insertion mutants are clearly seen.
Figure 6.
Comparison between the experimentally determined ratio of the apparent length to the actual length and the predicted curvature for the 130mer Alu156 wild-type promoter sequence and for its mutants with insertions of 6, 9, 11, 13, 15, 17, 19, 21, 25 and 29 bp. The dinucleotide parameters, i.e. BMHT (open circles), CS (stars) and LB (open squares), were used to predict curvature of these sequences. The ratio (apparent length/actual length) constitutes the x-axis, while predicted curvature in terms of Imax/Imin and d/lmax is on the y-axis. In the case of Imax/Imin, the ln(Imax/Imin) values are plotted on the y-axis. (A) The correlation between the ratios of the apparent length to the actual length and ln(Imax/Imin) values calculated using BMHT parameters (R = 0.26), CS parameters (R = –0.86) and LB parameters (R = –0.81). (B) The correlation between the ratios of apparent length to actual length and d/lmax calculated using BMHT parameters (R = 0.33), CS parameters (R = –0.80) and LB parameters (R = –0.84).
Comparison with structures determined using NMR
An accurate determination of nucleic acid structures in solution is now possible due to the recent developments in NMR techniques based on residual dipolar couplings. The solution structure of an oligonucleotide containing an A-tract has been reported recently and helps explain the anomalously slow gel mobility of polymers of this sequence (19). The solution structure of the dodecamer d(GGCAAAAAACGG) (hereafter referred as A6) has an overall helix bend of 19°, while the structure of the control sequence d(GGCAAG AAACGG) with an AT to GC transition in the center of the A-tract has been reported to be bent by 9° (henceforth the control sequence will be referred to as A2G1A3). In addition, a polymer with the A6 sequence repeat showed anomalously retarded gel mobility which was considerably reduced in the case of the A2G1A3 polymer. The NMR structures provided a structural basis for this change in mobility. Based on the NMR structures, a bend of ∼190° was estimated for the d(A6N4)10 sequence and a bend of 90° for the control sequence d(A2G1A3N4)10 (19). These estimated bending angles were found to be in agreement with the gel mobility data.
We generated structures of these two sequences using the three different dinucleotide parameter sets. The bending angles for the d(A6N4)10 sequence are 254° (BMHT), 145° (CS) and 44° (LB), while those for the d(A2G1A3N4)10 sequence are 65° (BMHT), 87° (CS) and 19° (LB). The values of radius of curvature (in Å), calculated from the corresponding bending angles, are 103 (NMR), 77 (BMHT), 134 (CS) and 448 (LB) for the A6 sequence. The bending angle values clearly show that the BMHT and CS parameters correctly predict the relative curvature of the two sequences.
Relationship to previous comparative studies
To date, there have been only a few systematic attempts to compare the various parameter sets in terms of their predictive power using well-defined geometric measures. Tung et al. (45) were the first to compare the Tung–Harvey model (TH model) and an AA-wedge model. The TH model is based on conformational energy calculations (34), while the AA-wedge model attributes DNA curvature to the wedge angle in an AA dinucleotide (23). The analysis was carried out on the seven sequences described by Hagerman (50,51). The comparison was qualitative and based on different geometrical measures. Tung et al. concluded that both parameter sets require improvements to predict the DNA structure correctly. The TH model was better than the AA model in predicting the curvature as a function of fragment length. However, the TH model was shown to be qualitatively incorrect in predicting the observed difference in ANTN and TNAN loci, which are accounted for successfully by the AA model.
In a more detailed study, Goodsell and Dickerson (36) compared six bending models including AA-wedge, TH and BMHT dinucleotide models and a trinucleotide model (24,25,30–32,36,37) on four test sequences, comprising phased and out-of-phase A-tracts, a phased GGGCCC motif in the repeat sequence (GAGGGCCCTA)n (Table 1: sequence 17) and the kinetoplast sequence from Leishmania tarentolae. This comparison was carried out based on the successive bending angle profile over the sequence. They observed that, even though all the models could correlate the extent of local bending with the macroscopic curvature behavior for the three sequences having phased A-tracts, the five dinucleotide models, including the BMHT model, fail to predict any curvature for the sequence with a GGGCCC motif. The only trinucleotide model included in this study was that based on nucleosome positioning preferences (36,37). Based on this analysis, it was concluded that only trinucleotide models could reproduce the curvature in this sequence. However, in a recent study (27), it was shown that dinucleotide parameters such as in the LB model could also predict curvature for this sequence. Though our analysis, discussed above, indicates that all the three models predict some amount of curvature for the (GAGGGCCCTA)n sequence, we support the previous view that the magnitude of the predicted curvature cannot be considered to be very significant. At the same time, it may be pertinent to note that this sequence is only observed to be curved in the presence of divalent cations (41).
The contribution of the GGGCCC motif to DNA curvature was also studied by Dlakic and Harrington (39), with different flanking sequences, containing two other motifs, i.e. AAAAA(A) and GAGAG (Table 1: sequences 10–12). They also stated that trinucleotide models are apparently required for explaining the curvature in these DNA sequence motifs. The analysis included four dinucleotide parameter sets (including the BMHT model) (25,28,31–33) along with two trinucleotide sets (35–37) and three geometrical measures, i.e. the radius of gyration, EED (same as d/lmax) and RMI (similar to Imax/Imin). The evaluation of models was based on qualitative trends. Our analysis, as discussed in the section on ‘Unconventional helical phasing of DNA motifs’, confirms that the BMHT model does not predict the gel mobility trend correctly for these sequences. However, the CS and LB models, which were not included in the earlier analysis, both predict the correct trends. Thus, the CS and LB models, which did not predict significant curvature for the (GAGGGCC CTA)n sequence, could, however, predict the correct trends for sequences which contain the GGGCCC motif in a different sequence context.
Analysis using trinucleotide parameters
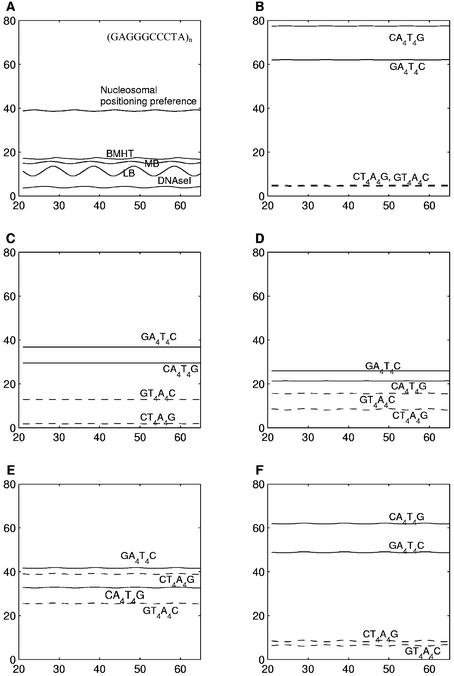
In this context, the sequence (GAGGGCCCTA)n with a GGGCCC motif (Table 1: sequence 17) was re-examined, using the trinucleotide parameters. We used the ‘BEND’ program (36), which allowed us to include the two sets of trinucleotide parameters, based on DNase I sensitivity (35) and nucleosomal positioning preferences (36,37), in addition to the three dinucleotide parameters (BMHT, CS and LB). It is interesting to see that while the trinucleotide model based on nucleosomal positioning preferences is able to predict the high curvature reported for this sequence correctly, the DNase I sensitivity-based trinucleotide parameters totally fail to account for it (Fig. 7A). The structures predicted using dinucleotide parameters, i.e. BMHT, CS and LB, have small curvature values that are intermediate between those predicted by the two trinucleotide parameters.
Figure 7.
(A) The curvature profiles generated by the ‘BEND’ program (36) for the repetitive sequence (GAGGGCCCTA)n. The profiles corresponding to the two trinucleotide parameters (nucleosomal positioning preferences and DNase I sensitivity) are shown along with those corresponding to BMHT, CS and LB parameters. The curvature profiles are plotted on the y-axis against the sequence length (in bp) on the x-axis. (B–F) The curvature profiles, generated using the ‘BEND’ program (36), for the repetitive sequences (CA4T4G)n, (CT4A4G)n, (GA4T4C)n and (GT4A4C)n. The profiles correspond to (B) BMHT parameters, (C) CS parameters, (D) LB parameters and the two trinucleotide parameters, (E) nucleosomal positioning preference and (F) DNase I sensitivity. The curvature profiles are plotted on the y-axis against the sequence length (in bp) on the x-axis.
We also carried out a comparison of predicted curvature for the A4T4 and T4A4 sequences (50,51) using trinucleotide as well as dinucleotide models vis-à-vis the experimental data. As discussed above, the (CA4T4G)n and (GA4T4C)n sequences are bent, while the (CT4A4G)n and (GT4A4C)n sequences are straight (Table 1: sequences 13–16). The nucleosomal positioning preference model could not predict this trend (Fig. 7E). In contrast the DNase I sensitivity-based trinucleotide parameters (Fig. 7F) and dinucleotide param eters (Fig. 7B, C and D) can correctly account for the experimentally observed trends.
In the case of the A-tract sequence set (Table 1: sequences 1–9), the average curvature predicted using both the trinucleotide sets showed poor correlation with experimental RL. The calculated correlation coefficients between the average predicted curvature and the experimental RL values for the nucleosomal positioning preference model and DNase I sensitivity-based model are 0.32 and 0.13, respectively. On the other hand, in the case of the mixed sequence set studied by Ussery et al. (54) (Table 1: sequences 18–28), the trinucleotide parameter sets showed quite good correlation with experimental data (correlation coefficient: 0.80 for the nucleosomal positioning preference model and 0.68 for the DNase I model).
It has often been thought that trinucleotide parameters necessarily represent an improvement over the dinucleotide-based descriptions (36,39,58). However, our simple yet comprehensive analysis shows that the currently available trinucleotide parameters fail to predict curvature in some of the most extensively studied sequences. This may be because the gel mobility data essentially reflect static DNA curvature, while trinucleotide parameters, especially the DNase I sensitivity model, reflect the dynamic ability of DNA to adopt a bent conformation. Correct assessment of the ability to predict the DNA flexibility/bendability requires a comparison with experimental data other than those based on gel mobility. It should also be mentioned that the present trinucleotide parameters are based on relative scales and not on actual structural information such as roll, tilt and twist angles. A trinucleotide model based on such three-dimensional structural studies requires a much larger data set than presently available, and hence a rigorous assessment of trinucleotide parameters to predict curvature is not possible.
CONCLUSION
Various dinucleotide models (BMHT, CS and LB) can account reasonably well for intrinsic bending observed in different sets of sequences, especially those containing A-tracts. Such bending has been reported experimentally from gel mobility studies and corresponds to static curvature. Interestingly, a recent NMR study confirms the occurrence of such bending for an A-tract-containing sequence. The dinucleotide models can also predict very well the curvature for other sets of sequences, such as those studied by Dlakic and Harrington (39) and by Ussery et al. (54), which have GC-rich motifs located in different sequence contexts. The only exception is the sequence with a phased GGGCCC motif, where all three dinucleotide models predict only slight curvature. More experimental studies corresponding to novel sequences are needed to refine these parameters and properly account for the curvature in GC-rich sequences. Our analysis also indicates that presently available flexibility-based trinucleotide parameters fail to predict curvature in some of the well-characterized sequences. At present, the paucity of structural data for arriving at reliable tri- or tetranucleotide parameters precludes a quantitative assessment of their ability to predict DNA curvature.
It is interesting that the CS model, which is not based on any kind of fitting procedures, shows better quantitative agreement with the experimentally observed trends than the other two dinucleotide parameter sets. It has often been stated that the crystal structures of oligonucleotides are subject to crystal packing forces and hence may not represent the solution structure. In this context, the success of the CS model, which is based on crystal structure data, in predicting the observed bending of a DNA oligomer in solution is quite noteworthy and indicates that the local geometry of dinucleotide steps may be quite similar in the two environments.
Sequence-dependent bending of DNA is observed at a number of biologically important sites such as promoters, and hence correct understanding of the structure of DNA in genomic sequences is of immense importance. Hence, the success of the dinucleotide parameters, especially that of the CS model, in predicting curvature in natural sequences such as the Alu156 promoter and its insertion mutants, is quite significant. It suggests that the dinucleotide parameters with some further refinements are quite capable of correctly predicting DNA curvature in genomic sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor N. V. Joshi for valuable discussions. A.K. is a recipient of a senior research fellowship from the University Grants Commission, Government of India. This work was supported by a grant from DBT, India.
REFERENCES
- 1.Hagerman P.J. (1990) Sequence-directed curvature of DNA. Annu. Rev. Biochem., 59, 755–781. [DOI] [PubMed] [Google Scholar]
- 2.McFall S.M., Chugani,S.A. and Chakrabarty,M.A. (1998) Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene, 223, 257–267. [DOI] [PubMed] [Google Scholar]
- 3.McGill G. and Fisher,D.E. (1998) DNA bending and the curious case of Fos/Jun. Chem. Biol., 5, R29–R38. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Martin J., Rojo,F. and de Lorenzo,V. (1994) Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev., 58, 268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polaczek P., Kwan,K., Liberies,D.A. and Campbell,J.L. (1997) Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol. Microbiol., 26, 261–275. [DOI] [PubMed] [Google Scholar]
- 6.Henriquez V., Milisavljevic,V., Kahn,J.D. and Gennaro,M.L. (1993) Sequence and structure of cmp, the replication enhancer of the Staphylococcus aureus plasmid pT181. Gene, 134, 93–98. [DOI] [PubMed] [Google Scholar]
- 7.Widlund H.R., Kuduvalli,P.N., Bengtsson,M., Cao,H., Tullius,T.D. and Kubista,M. (1999) Nucleosome structural features and intrinsic properties of the TATAAACGCC repeat sequence. J. Biol. Chem., 274, 31847–31852. [DOI] [PubMed] [Google Scholar]
- 8.Muller H.P. and Varmus,H.E. (1994) DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J., 13, 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald D.J., Dryden,G.L., Bronson,E.C., Williams,J.S. and Anderson,J.N. (1994) Conserved patterns of bending in satellite and nucleosome positioning DNA. J. Biol. Chem., 269, 21303–21314. [PubMed] [Google Scholar]
- 10.Zahn K. and Blattner,F.R. (1985) Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature, 317, 451–453. [DOI] [PubMed] [Google Scholar]
- 11.Nelson H.C., Finch,J.T., Luisi,B.F. and Klug,A. (1987) The structure of an oligo(dA)·oligo(dT) tract and its biological implications. Nature, 330, 221–226. [DOI] [PubMed] [Google Scholar]
- 12.Goodsell D.S., Kaczor-Grzeskowiak,M. and Dickerson,R.E. (1994) The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T–A steps. J. Mol. Biol., 239, 79–96. [DOI] [PubMed] [Google Scholar]
- 13.Goodsell D.S., Kopka,M.L., Cascio,D. and Dickerson,R.E. (1993) Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc. Natl Acad. Sci. USA, 90, 2930–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGabriele A.D. and Steitz,T.A. (1993) A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J. Mol. Biol., 231, 1024–1039. [DOI] [PubMed] [Google Scholar]
- 15.DiGabriele A.D., Sanderson,M.R. and Steitz,T.A. (1989) Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc. Natl Acad. Sci. USA, 86, 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatzky-Schwartz M., Arbuckle,N.D., Eisenstein,M., Rabinovich,D., Bareket-Samish,A., Haran,T.E., Luisi,B.F. and Shakked,Z. (1997) X-ray and solution studies of DNA oligomers and implications for the structural basis of A-tract-dependent curvature. J. Mol. Biol., 267, 595–623. [DOI] [PubMed] [Google Scholar]
- 17.Umemoto K., Sarma,M.H., Gupta,G. and Sarma,R.H. (1990) Effect of the methyl group on DNA bending and curvature: structure of d(GA4U4C)2 in solution. Biochemistry, 29, 4714–4722. [DOI] [PubMed] [Google Scholar]
- 18.Sarma M.H., Gupta,G. and Sarma,R.H. (1988) Structure of a bent DNA: two-dimensional NMR studies on d(GAAAATTTTC)2. Biochemistry, 27, 3423–3432. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald D., Herbert,K., Zhang,X., Pologruto,T., Lu,P. and Polgruto,T. (2001) Solution structure of an A-tract DNA bend. J. Mol. Biol., 306, 1081–1098. [DOI] [PubMed] [Google Scholar]
- 20.Marini J.C., Effron,P.N., Goodman,T.C., Singleton,C.K., Wells,R.D., Wartell,R.M. and Englund,P.T. (1984) Physical characterization of a kinetoplast DNA fragment with unusual properties. J. Biol. Chem., 259, 8974–8979. [PubMed] [Google Scholar]
- 21.Wu H.M. and Crothers,D.M. (1984) The locus of sequence-directed and protein-induced DNA bending. Nature, 308, 509–513. [DOI] [PubMed] [Google Scholar]
- 22.Kahn J.D. and Crothers,D.M. (1992) Protein-induced bending and DNA cyclization. Proc. Natl Acad. Sci. USA, 89, 6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulanovsky L.E. and Trifonov,E.N. (1987) Estimation of wedge components in curved DNA. Nature, 326, 720–722. [DOI] [PubMed] [Google Scholar]
- 24.Koo H.S. and Crothers,D.M. (1988) Calibration of DNA curvature and a unified description of sequence-directed bending. Proc. Natl Acad. Sci. USA, 85, 1763–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolshoy A., McNamara,P., Harrington,R.E. and Trifonov,E.N. (1991) Curved DNA without A–A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bansal M. (1996) Structural variations observed in DNA crystal structures and their implications for protein–DNA interaction. In Sarma,R.H. and Sarma,M.H. (eds), Biological Structure and Dynamics, Proceedings of the Ninth Convention. Adenine Press, New York, NY, Vol. 1, pp. 121–134.
- 27.Liu Y. and Beveridge,D.L. (2001) A refined prediction method for gel retardation of DNA oligonucleotides from dinucleotide step parameters: reconciliation of DNA bending models with crystal structure data. J. Biomol. Struct. Dyn., 18, 505–526. [DOI] [PubMed] [Google Scholar]
- 28.Ulyanov N.B. and James,T.L. (1995) Statistical analysis of DNA duplex structural features. Methods Enzymol., 261, 90–120. [DOI] [PubMed] [Google Scholar]
- 29.Jernigan R.L., Sarai,A., Ting,K.L. and Nussinov,R. (1986) Hydrophobic interactions in the major groove can influence DNA local structure. J. Biomol. Struct. Dyn., 4, 41–48. [DOI] [PubMed] [Google Scholar]
- 30.Calladine C.R., Drew,H.R. and McCall,M.J. (1988) The intrinsic curvature of DNA in solution. J. Mol. Biol., 201, 127–137. [DOI] [PubMed] [Google Scholar]
- 31.Cacchione S., De Santis,P., Foti,D., Palleschi,A. and Savino,M. (1989) Periodical polydeoxynucleotides and DNA curvature. Biochemistry, 28, 8706–8713. [DOI] [PubMed] [Google Scholar]
- 32.De Santis P., Palleschi,A., Savino,M. and Scipioni,A. (1990) Validity of the nearest-neighbor approximation in the evaluation of the electrophoretic manifestations of DNA curvature. Biochemistry, 29, 9269–9273. [DOI] [PubMed] [Google Scholar]
- 33.Gorin A.A., Zhurkin,V.B. and Olson,W.K. (1995) B-DNA twisting correlates with base-pair morphology. J. Mol. Biol., 247, 34–48. [DOI] [PubMed] [Google Scholar]
- 34.Tung C.S. and Harvey,S.C. (1986) Base sequence, local helix structure and macroscopic curvature of A-DNA and B-DNA. J. Biol. Chem., 261, 3700–3709. [PubMed] [Google Scholar]
- 35.Brukner I., Sanchez,R., Suck,D. and Pongor,S. (1995) Sequence dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J., 14, 1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodsell D.S. and Dickerson,R.E. (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res., 22, 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satchwell S.C., Drew,H.R. and Travers,A.A. (1986) Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol., 191, 659–675. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W., Sander,C. and Trifonov,E.N. (1982) The ten helical twist angles of B-DNA. Nucleic Acids Res., 10, 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dlakic M. and Harrington,R.E. (1998) Unconventional helical phasing of repetitive DNA motifs reveals their relative bending contributions, Nucleic Acids Res., 26, 4274–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaich A.K., Bhattacharyya,D., Brahmachari,S.K. and Bansal,M. (1994) CA/TG sequence at the 5′ end of oligo(A)-tracts strongly modulates DNA curvature. J. Biol. Chem., 269, 7824–7833. [PubMed] [Google Scholar]
- 41.Brukner I., Susic,S., Dlakic,M., Savic,A. and Pongor,S. (1994) Physiological concentration of magnesium ions induces a strong macroscopic curvature in GGGCCC-containing DNA. J. Mol. Biol., 236, 26–32. [DOI] [PubMed] [Google Scholar]
- 42.Bansal M., Bhattacharyya,D. and Ravi,B. (1995) NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. CABIOS, 11, 281–287. [DOI] [PubMed] [Google Scholar]
- 43.Trifonov E.N. and Ulanovasky,L.E. (1987) Inherently curved DNA and its structural elements. In Wells,R.D. and Harvey,S.C. (eds) Unusual DNA Structures. Springer-Verlag, Berlin, Germany, pp. 173–187.
- 44.Srinivasan A.R., Torres,R., Clark,W. and Olson,W.K. (1987) Base sequence effects in double helical DNA. I. Potential energy estimates of local base morphology. J. Biomol. Struct. Dyn., 5, 459–496. [DOI] [PubMed] [Google Scholar]
- 45.Tung C.S., Garcia,A.E., Marr,T.G., Mitra,S.K. and Burks,C. (1988) Measures of DNA curvature. In Olson,W.K., Sarma,M.H., Sarma,R.H. and Sundaralingam,M. (eds), DNA Bending and Curvature. Adenine Press, New York, NY, Vol. 3, pp. 265–278.
- 46.Bansal M., Bhattacharya,D. and Mohanty,D. (1991) DNA bending: a natural consequence of base sequence dependent variability. In Balaram,P. and Ramaseshan,S. (eds), Molecular Conformation and Biological Interactions. Indian Academy of Sciences, Bangalore, India, pp. 347–362.
- 47.Jernigan R.L., Sarai,A., Shapiro,B. and Nussinov,R. (1987) Relationship between curved DNA conformations and slow gel migration. J. Biomol. Struct. Dyn., 4, 561–567. [DOI] [PubMed] [Google Scholar]
- 48.Olson W.K., Marky,N.L., Jernigan,R.L. and Zhurkin,V.B. (1993) Influence of fluctuations on DNA curvature. A comparison of flexible and static wedge models of intrinsically bent DNA. J. Mol. Biol., 232, 530–554. [DOI] [PubMed] [Google Scholar]
- 49.Haran T.E., Kahn,J.D. and Crothers,D.M. (1994) Sequence elements responsible for DNA curvature. J. Mol. Biol., 244, 135–143. [DOI] [PubMed] [Google Scholar]
- 50.Hagerman P.J. (1986) Sequence-directed curvature of DNA. Nature, 321, 449–450. [DOI] [PubMed] [Google Scholar]
- 51.Hagerman P.J. (1985) Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry, 24, 7033–7037. [DOI] [PubMed] [Google Scholar]
- 52.Brukner I., Jurukovski,V., Konstantinovic,M. and Savic,A. (1991) Curved DNA without AA/TT dinucleotide step. Nucleic Acids Res., 19, 3549–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brukner I., Dlakic,M., Savic,A., Susic,S., Pongor,S. and Suck,D. (1993) Evidence for opposite groove-directed curvature of GGGCCC and AAAAA sequence elements. Nucleic Acids Res., 21, 1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ussery D.W., Higgins,C.F. and Bolshoy,A. (1999) Environmental influences on DNA curvature. J. Biomol. Struct. Dyn., 16, 811–823. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka K., Muramatsu,S., Yamada,H. and Mizuno,T. (1991) Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol. Gen. Genet., 226, 367–376. [DOI] [PubMed] [Google Scholar]
- 56.McAllister C.F. and Achberger,E.C. (1988) Effect of polyadenine-containing curved DNA on promoter utilization in Bacillus subtilis. J. Biol. Chem., 263, 11743–11749. [PubMed] [Google Scholar]
- 57.McAllister C.F. and Achberger,E.C. (1989) Rotational orientation of upstream curved DNA affects promoter function in Bacillus subtilis. J. Biol. Chem., 264, 10451–10456. [PubMed] [Google Scholar]
- 58.Brukner I., Sanchez,R., Suck,D. and Pongor,S. (1995) Trinucleotide models for DNA bending propensity: comparison of models based on DNase I digestion and nucleosome packaging data. J. Biomol. Struct. Dyn., 13, 309–317. [DOI] [PubMed] [Google Scholar]