Abstract
DXplain is an expert system designed to suggest a list of diseases that are associated with a set of clinical findings entered by a health profession student or practitioner. It has been widely used for almost 20 years, during which time many new functions and capabilities have been added. We discuss the ways in which different classes of user interact with the system and which functions are most commonly used.
1. Introduction
Development of DXplain began in 1984 with explicit design criteria. The system was to be “user-friendly,” usable by a physician with limited computer skills and without the need for training in its operation. We thus planned from the outset to use standard medical terminology with extensive use of synonyms and spelling correction. The program was intended to have comprehensive content, covering the broad domain of general internal medicine as well as related specialties and many pediatric diseases. The system was designed to explain its reasoning rather than acting as a “black box” oracle. We also planned for the system to grow, evolve and improve based on experience and user input. (1,2)
National distribution of DXplain began in 1987 over the dial-up AMANET. After AMANET ceased operation in 1990, DXplain continued to be distributed over dial-up networks until 1995. Between 1991 and 1996 we also distributed DXplain as a stand-alone version that could be loaded on an individual PC. This approach was abandoned as the availability of Internet access provided a better alternative to dealing with the problems of installation support, knowledge base updating and communication between the users and the developers. (3)
The current DXplain knowledge base (KB) includes 2241 diseases and over 4800 clinical findings (symptoms, signs, epidemiologic data and laboratory and radiologic findings). The average disease description includes 52.8 findings (with a range from 10 to over 100). Each disease/finding dyad has two numbers describing the relationship, one representing the frequency with which the finding occurs in the disease and the other the degree to which the presence of the finding suggests consideration of the disease. There are thus 2 x 52.8 x 2241, or 236,650 individual data points in the KB representing disease/finding relationships. In addition, each finding has an associated disease-independent Term Importance from 1 to 5 indicating how important it is to explain the presence of the finding. Each disease also has two associated values: one that is a crude approximation of its prevalence (very common, common, rare or very rare) and the other of its importance, ranked between 1 and 5, intended to reflect the impact of not considering the disease if it is present.
The most common mode of use of DXplain allows the user to input information about a patient case and see a list of diseases that would explain some or all of the findings entered. The list is displayed in two separate tables, one of common and very common diseases and one of rare and very rare diseases. Within each of these groups, the diseases are listed in rough order of degree of support by the entered findings, with flags for diseases that are well or very well supported. The user can click on any disease to see why it is on the list and which of the entered findings are not in the disease description, as well as which additional findings would be commonly found in the disease. In addition, each disease has up to ten selected literature references, chosen to be good clinical descriptors rather than the most recent research papers. Other commonly used functions of DXplain are Disease Information, which provides a description of any disease in the KB, and Differential Diagnosis for a finding, which lists the diseases associated with any finding in the KB.
During a case entry, the user is initially requested to enter three “demographic terms:” age, gender and duration of symptoms. Age and gender must be entered. While duration is not forced, the large majority of users provide all three terms. Thereafter, they are presented with a blank text entry box into which they type clinical findings using narrative text. After submitting the demographic entries and their initial findings, they are presented with an ordered list of diseases that could be associated with one or more of the findings. On the same page, they have a text-entry box for addition of further findings and also four system-selected questions about the case. These “Findings Present?” findings are selected because each has the potential to reinforce, or at times eliminate, the diseases that are high on the DXplain list. Beside each “Finding Present?” is a radio box for selection of Yes, No or Unknown, and also a “?” that when selected will explain why DXplain considers this finding useful. If the user selects Yes, the finding is added to the case description, and if No, the finding is included as not present. Choosing Unknown prevents the same finding being offered again in later iterations of this function.
2. Major Changes with the evolving system
Transition to the Web
Transition to distribution over the Internet was a logical evolution. Because DXplain is constantly evolving and changing, knowledge base updates are made on a frequent basis. No other method of distribution so easily permits us to be sure that users will have the latest version available as soon as possible. This is important to both ensure that any KB changes are immediately available to all users and to allow addition of new or newly prominent diseases (such as SARS) to the version seen by all users. A number of ancillary benefits are available with Internet distribution. One of these is making our disease references hot-links to Medline and also allowing the use of appropriate URL’s as additional references. An important characteristic of DXplain is that we encourage user input, with two-way communication about the system expedited. There is a Feedback button on every DXplain page, so that users can forward comments to the developers. We also interrogate users periodically, as discussed below under User Survey analysis.
Expansion of database
New diseases emerge and old diseases reappear in new formats. We have added these changes to DXplain on a regular basis. Since new laboratory tests are continually developed and become widely used, we add these and place them into appropriate disease descriptions. From January 1, 2000 until March 14, 2005, we have added 160 new findings and 66 new diseases to the knowledge base. The medical editors scan the literature on a regular basis, and use new clinical case series and review articles to update the database. Since textbooks tend to use such vague terms as “often” or “generally,” we focus on reports that provide detailed quantitative information about clinical findings in a series of cases.
Newer features: Focus, Disease Compare and Finding Information
Based largely on user input, we have added a number of functions to DXplain. In some cases, a user may consider one or more clinical findings to be particularly important. DXplain allows the user to “focus” on one or more of the findings entered, and DXplain will display only those diseases in which the selected findings occur. “Disease Compare” allows the user to select two or more diseases and see a side-by-side table comparing the findings in each disease. Because eponymic signs are not always familiar to all users, and other findings are best illustrated with an image, we added the capability of having a short text description or graphic associated with any of the Findings, and have added text or images to 426 Findings.
3. Experience with use
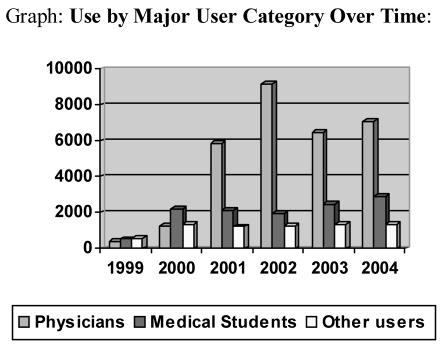
From March 1996, when DXplain distribution was switched entirely to Internet use, until December 2004, 46,319 users logged 122,324 sessions. Use over the last five years is summarized in the Graph and detailed in the Table. Use has grown from 1459 users in 1999 to 11,411 in 2004. The large increase from 2000 to 2001 reflected the addition of DXplain to the Merck Medicus web site aimed at individual physicians. The Table also shows the breakdown by class of user. Of total users, 54% were practicing physicians, 25% medical students and 6% resident physicians. The remaining 15% were non-practicing physicians, nurses, nurse practitioners, physician assistants, other health professionals and non-medical personnel (notably medical librarians and informaticians). This proportion has been relatively steady over the past three years. The increased use by first and second year medical students and by nurses reflects in part increased use of DXplain as a formal class assignment in courses on medical decision-making at several hospitals and medical schools.
Graph.
Use by Major User Category Over Time:
Table.
Yearly DXplain users by background category
| User Background Category | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | Total | |
|---|---|---|---|---|---|---|---|---|
| Attending Physician | 241 | 589 | 5117 | 8544 | 5884 | 6517 | 26894 | (54.3%) |
| House Staff | 148 | 640 | 717 | 580 | 509 | 508 | 3102 | ( 6.3%) |
| Physician NP | 0 | 0 | 37 | 62 | 59 | 62 | 220 | ( 0.4%) |
| Med Student (First 2 yrs) | 214 | 862 | 1130 | 1236 | 1499 | 2028 | 6969 | (14.1%) |
| Med Student (Second 2 yrs) | 310 | 1342 | 972 | 755 | 951 | 913 | 5243 | (10.6%) |
| Physician Assistant | 0 | 0 | 81 | 167 | 120 | 99 | 467 | ( 0.9%) |
| Nurse | 73 | 309 | 289 | 276 | 422 | 445 | 1814 | ( 3.6%) |
| Nurse Practitioner | 0 | 0 | 41 | 97 | 86 | 129 | 353 | ( 0.7%) |
| Admin or Research Staff | 105 | 223 | 158 | 128 | 108 | 98 | 820 | ( 1.6%) |
| Other Health Professional | 239 | 548 | 453 | 383 | 459 | 429 | 2511 | ( 5.1%) |
| Non-Medical Personnel | 129 | 257 | 197 | 215 | 193 | 183 | 1174 | ( 2.4%) |
| Total | 1459 | 4772 | 9192 | 12443 | 10290 | 11411 | 49567 |
(These data are based on unique users each year; therefore a user may be counted in more than one year.)
How do DXplain users use the system?
Of total use from 1999 through 2004, the average user logged 2.3 sessions, but this average included a wide range. Analysis of 52,626 sessions during a two-year block, January 2003–December 2004, showed that 11,184 of 18,952 (59%) individual users used the system only once. Of more frequent users, 5453 (29%) used DXplain 2 to 4 times; 1377 users (7%) logged 5 to 9 sessions and there were 938“ heavy users” (5%) who each used the system 10 or more times. Use did not vary greatly by class of user, with the breakdown of the average number of sessions per user almost the same for all groups, with slightly more sessions/user for first/second year medical students (2.6) and slightly fewer for nurses (1.6) than the overall average of 2.3.
Most DXplain users enter only a small number of findings per case, with an average of 4 non-demographic findings. Perhaps not surprisingly, medical students tended to be more thorough in entering findings. When looking only at terms entered directly by the user, and not from the “Findings Present?” prompts, over 25% of medical students entered more than 5 non-demographic findings, compared to 11–12% of house staff and attending physicians.
Which added DXplain features are most used?
Software developers frequently have ideas about what features will be desirable, but these features are not always seen as useful by the end-user. As DXplain developers we have added many new features that we found useful and exciting that have not had widespread use. The most typical use of DXplain is entry of a set of findings to see a list of associated diseases with one or more requests for information about a specific disease or finding. This is done by 59% of first and second year medical students, 51% of third and fourth year students and 45% of house staff and attending physicians. The second most common pattern is where the user enters a set of findings to see a disease list with no request for finding or disease information. Between 10 and 20% of use involves only a request for information about one or more diseases or findings with no entry of a set of findings.
The “Findings Present?” function is by far the most widely used added feature. Fully 42% of users select one or more findings off this selection box. We see many instances in which the user enters only one or two non-demographic terms directly and then selects dozens from the “Findings Present?” list. Of interest, this function is used less by first and second year medical students and “other” (largely librarians and informaticians) than by users with more clinical experience.
The “Focus” feature is used by between 6 and 9% of all users.
The ability to save and later retrieve a set of case findings was added at user request, but is rarely used. Only 3-4% of users actually use this feature. The “other” category has the highest use, at 9%, perhaps because this group frequently demonstrates DXplain to others rather than entering actual cases.
“Disease compare” is a relatively new feature, added in 2002. This allows the user to select two or more diseases and see associated findings compared in a graphic table in order to allow the user to easily review what findings the diseases have in common and which findings most readily distinguish a disease from another. In 2004, this function was used by 3–4% of users except for a much higher use by nurses and nurse practitioners, possibly because use of this function was emphasized in several formal courses for nurses that incorporated use of DXplain. This, along with the limited use of Focus and Save/Retrieve case, again suggests that nurses are using DXplain more as a textbook or part of a formal training program in diagnosis and less to help in reaching a diagnosis in a specific patient.
4. User Survey analysis
After every 3rd session, if at least one disease is considered supported, users are asked to answer three questions about the case they have just entered: what disease they believe the patient to have, what disease(s) were not on the DXplain list that should have been, and what disease(s) they had not previously considered were now under consideration because of DXplain.
We use the second question, about diseases “missed” by DXplain as an important source of quality assurance for the system. Every “missing disease” submitted under this question is reviewed by one or more of the physician editors. During the two year period 11/14/2001 to 10/23/2003, 1666 total questionnaires were answered. Of these, 417 (25%) noted a “missing disease” in question 2. A random sample of the “missing diseases” submitted during a one-month period was reviewed in detail. 45% of the reported “missing diseases” were felt after review to not be supported by the findings entered. One example: the findings entered were “child, prolonged, localized rash feet, conjunctivitis, headache” and the disease listed as missed was “leukemia.” We can only postulate that this user knew more about the patient (in this instance, perhaps an elevated white blood cell count) in this case (and similar ones) than was entered in the system. In 24% of cases, the “missing” disease was actually included on the DXplain list. In 6% the “missing disease” was not a disease (eg “use of estrogen products”). In 6% the disease was not part of the DXplain KB; most of these were felt to be trivial diseases (eg, ganglion) not appropriate for addition. In 20% of the entries reviewed, the disease should have been on the list and edits were made to the KB; re-entering the findings from the original case then resulted in the disease showing up on the list. Ongoing review of these user surveys is second only to literature review in providing KB updates and improvements.
Lessons learned
While user input is valuable in improving a system, requested features may be of limited value to the vast majority of users, and it would be a mistake to expend large amounts of resources based on a few requests. Not surprisingly, easy to use features that require minimal user effort are much more heavily used that are those that demand more thinking or typing. Thus the very heavy use of the “Findings Present?” function, which requires only responding with a click to system prompts.
Conclusion
Just as medical knowledge is continually expanding, an expert system must expand and evolve. We feel that it is the evolutionary nature of DXplain that has contributed to its continued and growing use over the past two decades. Users at all levels of medical and nursing training and practice can benefit from use of expert systems, though patterns of use vary.
References
- 1.Barnett GO, Cimino JJ, Hupp JA, Hoffer EP. Dxplain - an evolving diagnostic decision-support system. JAMA. 1987;258:67–74. doi: 10.1001/jama.258.1.67. [DOI] [PubMed] [Google Scholar]
- 2.Barnett GO, Hoffer EP, Packer MS, et al. DXplain - a demonstration and discussion of a diagnostic decision support system. Proc. of 16th SCAMC, Baltimore, MD, November, 1992, p. 822 [PMC free article] [PubMed]
- 3.Barnett GO, Hoffer E, Kim RJ, Famiglietti KT. Dxplain on the World Wide Web. Proc of the 1996 AMIA Annual Fall Symposium, October 1996, p. 988.