Abstract
Single-stranded DNA or double-stranded DNA has the potential to adopt a wide variety of unusual duplex and hairpin motifs in the presence (trans) or absence (cis) of ligands. Several principles for the formation of those unusual structures have been established through the observation of a number of recurring structural motifs associated with different sequences. These include: (i) internal loops of consecutive mismatches can occur in a B-DNA duplex when sheared base pairs are adjacent to each other to confer extensive cross- and intra-strand base stacking; (ii) interdigitated (zipper-like) duplex structures form instead when sheared G·A base pairs are separated by one or two pairs of purine·purine mismatches; (iii) stacking is not restricted to base, deoxyribose also exhibits the potential to do so; (iv) canonical G·C or A·T base pairs are flexible enough to exhibit considerable changes from the regular H-bonded conformation. The paired bases become stacked when bracketed by sheared G·A base pairs, or become extruded out and perpendicular to their neighboring bases in the presence of interacting drugs; (v) the purine-rich and pyrimidine-rich loop structures are notably different in nature. The purine-rich loops form compact triloop structures closed by a sheared G·A, A·A, A·C or sheared-like Ganti·Csyn base pair that is stacked by a single residue. On the other hand, the pyrimidine-rich loops with a thymidine in the first position exhibit no base pairing but are characterized by the folding of the thymidine residue into the minor groove to form a compact loop structure. Identification of such diverse duplex or hairpin motifs greatly enlarges the repertoire for unusual DNA structural formation.
INTRODUCTION
In addition to the regular duplex structure with canonical G·C and A·T base pairs, many DNA sequences adopt a variety of unusual structures in either the single-stranded or double-stranded state. For example, in the double-stranded state, the dinucleotide (CG)2 repeats were found to adopt the left-handed Z-DNA structure (1,2). The existence of such structure in vivo has been verified recently by the discovery of proteins that can specifically recognize the left-handed duplex structure (3–5). Similarly, the single-stranded G-rich DNA sequences have long been found to adopt a stable G-tetrad quadruplex structure in vitro (6–8). Its presence in vivo has also been established recently by the discovery of enzymes that can specifically recognize such sequences to promote (9), cleave (10), or unwind (11–13) the G4-DNA structure. Since tandem repeat sequences are highly abundant in the eukaryotic genomes (14), there is thus great interest in trying to decipher the unusual structures of such repeat sequences (7,15–21), and hence their biological functions. In addition, single-stranded DNA sequences screened by the SELEX technology can form unique three-dimensional structures (aptamer) to match the target molecules (22). Moreover, single-stranded DNA virus (23) or plasmid (24) occasionally contains an unusual fold-back structure at its 3′ end to complete the replication process. Such unusual structural motifs may represent novel targets for pharmaceutical research.
In the present survey, some principles for the unusual DNA duplex and hairpin formation will be summarized through the observation of a number of recurring structural motifs associated with different sequences. Such motifs represent stable structural elements that may be of general importance. Comprehensive coverage of all such motifs may allow one to predict stable secondary structure from any specific nucleic acid sequences. The principles to be described in this article are: (i) cross-strand base/base stacking resulting from adjacent sheared base pairs; (ii) interdigitated base/base stacking resulting from separated sheared base pairs; (iii) base/deoxyribose stacking in single-residue GNA, ANA, ANC and GNC loops closed by a sheared base pair; (iv) minor groove interaction in pyrimidine-rich bi- or tri-loops; and (v) perpendicular base/base interaction.
Recently, a sheared RNA G·U pair was also observed in a conserved hairpin loop of the Escherichia coli 16S rRNA in solution (25) and in the large subunit ribosome structure in crystal (26). Other structural aspects of RNA motifs have been reviewed in several articles (27–32), and will not be further described here. Likewise, the multi-stranded DNA triplex and quadruplex structures have also been described in several recent review articles (7,33,34).
INTERNAL LOOP MOTIFS: SHEARED BASE PAIRING AND CROSS-STRAND STACKING
Recently, several three-dimensional structures of DNA duplexes containing a 2 × 2 (two consecutive mismatched base pairs) (35–39), or 4 × 4 (four consecutive mismatched base pairs) (40) internal loop have been determined. From the published UV melting data, it was shown that such unusual duplexes exhibit stability only slightly less than those containing canonical G·C or A·T base pairs. Examination of these internal loop structures shows that tandem sheared base pairs is the critical structural element responsible for their stability (41,42), which confers the duplexes with excellent cross-strand stacking between the sheared base pairs and excellent intra-strand base stacking between the sheared base pair and its flanking base pair.
Sheared G·A, A·A and A·C base pairs
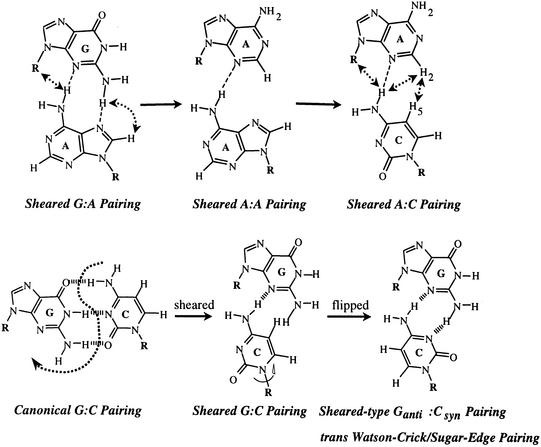
Incorporation of mismatch base pairs into DNA duplexes usually destabilizes the parent duplexes to varying degrees depending upon the nature of the incorporated mismatches. However, recent data have shown that tandem sheared purine·purine (36–38,42–44) or purine·pyrimidine (38) base pairs are the exceptions to this rule. Such tandem mismatches were found to be rather stable and are compatible with flanking Watson–Crick base pairs to form B-DNA like, although locally distorted, duplex structures. The schemes of the sheared G·A pairing and its extension to the A·A (44) and A·C (38) are shown in Figure 1. Such base pairing is special in that the H-bonding donor and acceptor in the Watson–Crick H-bonding sides are not employed. Instead, one purine base uses its functional groups in the minor groove edge to pair with the functional groups in the major groove edge of another purine or pyrimidine base (Fig. 2) to form the so-called sheared pairing. Such sheared pairing is well characterized by the extraordinary upfield shifting of the guanosine imino proton from the paired region (∼13 p.p.m.) to the unpaired region (∼10 p.p.m.), due to the fact that it does not participate in H-bonding (45,46). It therefore serves as a good marker for the NMR studies of nucleic acid molecules containing such motifs.
Figure 1.
Extension of the sheared G·A base pair (top left) to the sheared A·A base pair (top middle) and then to the sheared A·C base pair (top right). Two H-bonds (connected by dotted lines) are possible for a sheared G·A pair, while only one single H-bond is found for the sheared A·A and A·C base pairs. NOEs detected for such sheared base pairs are denoted by dotted arrows. The bottom figures show the formation of sheared-like (or trans Watson–Crick/Sugar-edge) Ganti·Csyn base pair (right) (32) from the canonical G·C base pair (left) via the intermediate sheared G·C base pair (middle). The flipping of cytosine base to the rare syn domain is necessary to prevent the steric hindrance between the G2NH2-CH5 protons (middle). It also facilitates the formation of a second G2NH2-C3N H-bond (right).
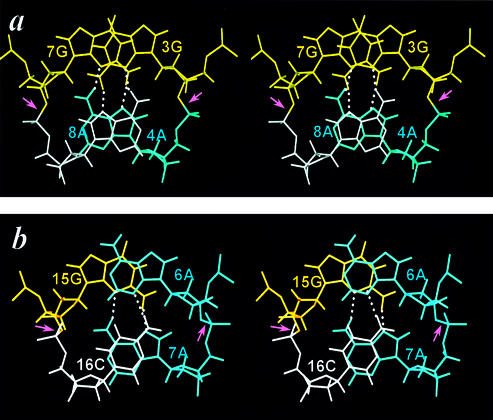
Figure 2.
The cross-eye stereo views of the cross-strand G/G and A/A stacks in the 5′-d(GA)/(AG)-5′ motif (a) and the cross-strand A/C and G/A stacks in the 5′-d(GC)/(AA)-5′ motif (b). The 8A and 16C nucleotides are drawn in white to illustrate the difference between the purine/purine (8A/4A) and purine/pyrimidine (7A/16C) stacks. The H-bonds are connected by white dotted lines and the unusual α(t) torsion angles connecting the tandem sheared base pairs marked by pink arrows. The transformation of the ζ torsion angles from the regular gauche– domain to the trans domain extends the phosphate backbone and allows the cross-strand stacking to occur.
The best evidence for a sheared G·A pair is the strong cross-strand A6NH2↔GH1′ and medium-strength A6NH2↔GH4′ NOEs (shown as dotted arrows in Fig. 1) (37) in H2O solution at low temperature (–5°C). Weak cross-strand G2NH2↔AH8 NOE could also be observed. Such NOEs unambiguously establish the sheared configuration of the G·A pairing in solution state. Similarly, a sheared A·C base pair configuration was also well characterized by the observation of cross-strand C4NH2↔AH1′, C4NH2↔AH2 and CH5↔AH2 NOEs (Fig. 1) in the H2O/NOESY spectrum at low temperature (47).
2 × 2 (GA)2 internal loops
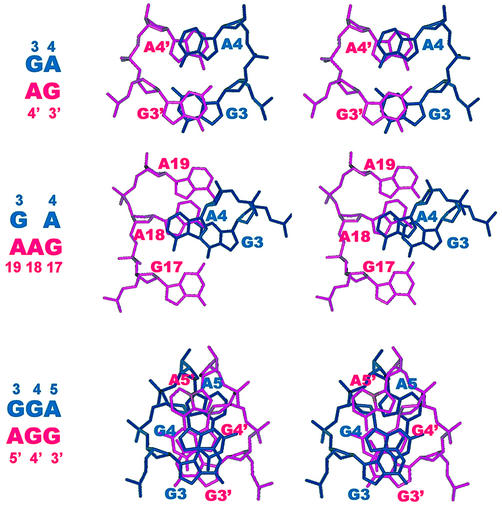
To date, no single-sheared G·A pair has ever been found to exist in a B-DNA duplex environment consisting of canonical G·C and A·T base pairs; only tandem sheared G·A paired motif can be embedded in a B-DNA (48,49) or A-RNA duplex (50). The reason for this is not yet clear, but may possibly be due to the fact that a single sheared base pairing causes considerable backbone torsion angle distortions and interrupts the regular intra-strand base stacking of a B-DNA duplex. Existence of a single sheared G·A pair is therefore not favored, except for serving as the closing base pair in a single-residue GNA loop (51–53). However, when two such sheared base pairs (a 2 × 2 internal loop) are present in tandem, a completely different story results. Excellent cross-strand G/G and A/A stacking (Fig. 2a) occur, with the original partial intra-strand stacking being completely replaced with the perfect inter-strand stacking. In addition, the sheared G·A pairs also exhibit excellent intra-strand stacking with their flanking canonical base pairs (43,54). This strong stacking may compensate for the backbone distortion from the ε(t)ζ(g–) domain to the ε(g–)ζ(t) domain necessary to accommodate such a dramatic change of base pairing. The transformation of backbone torsion angle from the ζ(g–) domain to the ζ(t) domain is correlated with the huge chemical shift changes (2 p.p.m. upfield shifting) for the phosphorus atoms connecting the adjacent sheared G·A base pairs (46,55).
2 × 2 (GC)/(AA) internal loops
As described above, strong cross-strand homo G/G and A/A stacking were observed for the classical tandem sheared G·A base pairs (Fig. 2a) (36,37,43,54). However, such cross-strand base stacking is not restricted to the homo purine bases. Hetero purine/purine (G/A) or purine/pyrimidine (A/C) cross-strand base stacking can also occur, as observed recently in the 5′-G15C16/A7A6-5′ motif, shown in Figure 2b. In this novel 2 × 2 internal loop of the human HIV-1 reverse transcriptase DNA inhibitor (56), the paired G15/A7 bases and A6/C16 bases were found to participate in sheared pairing, with the G15 base stacking upon the cross-stranded A6 base, and the A7 base upon the cross-stranded C16 base (Fig. 2b). It is also interesting to note that the stacking between the pyrimidine (C16) base and the purine (A7) base is quite good, with the six-membered ring of the C16 base perfectly stacking on top of the six-membered ring of the A7 base. Furthermore, the deoxyribose of the C16 residue also participates in stacking with the following 3′ end A17 base (another type of base/deoxyribose interaction, see below), with its H2′ proton situating almost on top of the A17 base (38). Such stacking is nicely correlated with the dramatic upfield shifting to –0.5 p.p.m. for the C16 H2′ proton (38). The participation of a smaller size pyrimidine residue (C16 in the present case) in the tandem sheared-pairing motif has thus drawn an interesting conclusion; while its base stacks well with the cross-stranded A7 base, its deoxyribose stacks instead with the following intra-stranded 3′ end A17 base. The entire pyrimidine moiety thus participates in stacking with the bases above and below to stabilize this unique motif.
4 × 4 internal loops
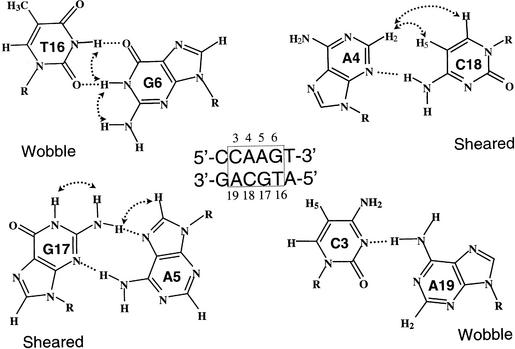
One of the major structural features of the 2 × 2 internal loop in the tandem sheared base pairs is the good intra-strand stacking between the sheared base pairs and their neighboring base pairs. Therefore it is possible to construct a stable 4 × 4 internal loop if the mismatches surrounding the tandem sheared base pairs can also provide good intra-strand stacking. Indeed it was recently found that the wobble T·G and wobble A·C base pairs serve as such mismatches to bracket the tandem sheared base pairs to form a stable duplex containing a series of four consecutive mismatches (40). As shown in Figure 3, the bases of the head-to-head H-bonded wobble G6·T16 and C3·A19 base pairs can align very well with the side-by-side H-bonded sheared G17·A5 and A4·C18 base pairs, respectively. These result in good T16/G17/A4/C3 stacking on one side and good G6/A5/C18/A19 stacking on the other side, with the cross-strand stacking occurring between the G17/A4 and A5/C18 bases. Such a 4 × 4 internal loop duplex is only slightly less stable than the canonically H-bonded duplex as judged from the UV-melting studies. Thus, when an oligonucleotide 21mer hairpin containing a canonically base-paired 5′-TAAA/ATTT-5′ segment is replaced by an all mismatched 5′-CAAG/ACGT-5′ segment, the melting temperature decreases only by ∼5°C (40). This is quite extraordinary, since in a regular DNA duplex, the melting temperature usually decreases by ∼10°C when a single mismatch is created (57), not to mention that four consecutive mismatches are simultaneously present in a 4 × 4 internal loop duplex as discussed here. Besides, all the mismatched internal-loop hairpin contains two H-bonds less (each AC mismatch has only one H-bond at a neutral pH condition, see Fig. 3) than the hairpin containing the canonical 5′-TAAA/ATTT-5′ segment. It is thus clear that next to H-bonding, the unique cross-strand and intra-strand base stacking also play significant roles in stabilizing double-helical nucleic acid structures of an unusual nature.
Figure 3.
The H-bonding scheme of the 4 × 4 5′-d(CAAG)/(ACGT)-5′ internal loop. The sheared A·C base pair and wobble A·C base pair are bonded through only one H-bond under a neutral pH condition. The critical NOEs used to identify such non-canonical base pairs are indicated by curved arrows.
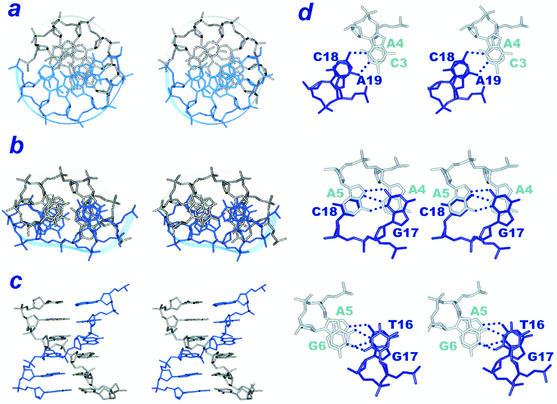
Such particular base stacks are clearly revealed in Figure 4, which shows the down-to-the-helical view and the major groove view of the base stacks of the entire 4 × 4 internal loop segment (Fig. 4b and c) and the nearest-neighbor stacks (Fig. 4d). It is clear from Figure 4b that the 4 × 4 internal loop segment does show an extraordinary base stacking arrangement. Unlike the regular 36° twisting angles adopted between the base pairs in a canonical duplex (Fig. 4a), the base pair twisting angles in the 4 × 4 internal loop segment fluctuate a lot, with those between the outer internal-loop base pair and the inner internal-loop base pair (C3·A19/A4·C18 and G6·T16/A5·G17) adopting rather large twisting angles of 66 ± 3° and 58 ± 4°, respectively, while that between the inner internal loop base pairs of A4·C18/A5·G17 adopting an even negative twisting angle of –11 ± 1° to accommodate the phenomenal cross-strand purine/purine (A4/G17) and purine/pyrimidine (A5/C18) stacking (Fig. 4c) (40). Another interesting feature of this 4 × 4 internal loop is the excellent stacking of all six-membered rings of every base in the internal loop, irrespective of the A, G, C or T residue (Fig. 4d). Such stacks, without doubt, contribute significantly to the stability of hairpins containing such 4 × 4 internal loop in the stem region.
Figure 4.
The cross-eye stereo views of the 4 × 4 5′-d(CAAG)/(ACGT)-5′ internal loop structure. (a) Down-to-the-helix view of the parent canonically paired 5′-d(CTAAAT)/(GATTTA)-5′ duplex, (b) down-to-the-helical view and (c) perpendicular-to-the-helical view of the duplex containing the 4 × 4 5′-d(CAAG)/(ACGT)-5′ internal loop. (d) The nearest-neighbor stacks of the 4 × 4 5′-d(CAAG)/(ACGT)-5′ internal loop. The perfect intra-strand (top and bottom) and cross-strand stacking (middle) of the base six-membered rings is clearly manifested in this figure.
Sequence dependence of the tandem G·A pairs
The sheared G·A pairing and cross-strand G/G and A/A base stacks were first discovered in the d(PyGAPu)2 motifs with a preceding 5′ end pyrimidine and a following 3′ end purine (48,54,58,59). Soon after it was discovered that the (GA)2 motif conformation changes dramatically when preceded and followed by alternative flanking base pairs (45). Thus, when the preceding 5′ end base of the d(GA)2 motif is switched from a pyrimidine to a purine and the following 3′ end base from a purine to a complementary pyrimidine, the resulting d(PuGAPy)2 sequence becomes less stable. Indeed, the G·A base pair in the d(AGAT)2 motif adopts a head-to-head G·A configuration instead with much poorer inter-strand purine– purine stacking (60), while the duplex containing the d(GGAC)2 motif reveals no stable structure at all as judged from the much broader NMR spectra (S.-H.Chou et al., unpublished data). Such a notion has, however, been somewhat expanded recently through the observation that the tandem G·A base pairs in the non-symmetrical 5′-d(PuGAPu)/d(PyAGPy)-5′ motifs also adopt the shearing modes and the characteristic cross-strand purine/purine stacking (37). It is also important to note that although tandem G·A base pairs appear frequently in the rRNA secondary structure (61), they exhibit quite different sequence dependent behavior (50). For example, the tandem G·A base pairs in the r(AGAU)2 motif were found to possibly adopt the sheared pairing modes, contrary to the head-to-head pairing mode mentioned above for a similar DNA sequence. Moreover, in the r(GGAC)2 sequence, the tandem G·A base pairs were found to adopt the head-to-head pairing mode, and is the most stable motif among all studied tandem G·A base pairs (62), although no stable species could be detected for the similar DNA sequences. Furthermore, the r(GAGC)2 motif, which has never been found to occur in DNA, was stably existent in RNA (50). DNA and RNA thus follow very different flanking sequence-dependent rules regarding the pairing in the (GA)2 motifs. The reasons for such subtle differences are still unclear.
Other sheared-pairing related DNA structures
Since shearing G·A pairing is such a unique structural element in DNA, it is natural to ask if it also happens in other multi-stranded DNA structures. Indeed, G·A pairing is found to be involved in some higher order DNA structures. For example, in the GGA triplet repeat sequence, the minor groove edge of the guanine base in the G-tetraplex structure can pair with the adenine base to form more complicated higher ordered motifs, like those of the A· (G·G·G·G·) pentad (63), A· (G·G·G·G·)·A hexad (64) or G(·A)·G(·A)·G(·A)·G heptad (65). Recently the structure of a DNA aptamer containing a l-argininamide-binding pocket has also been determined (66) and found to contain extensive base mismatches, including three sheared G·A pairs, to maximize the stacking interaction in an 18-residue loop. Such examples indicate that tandem sheared base-paired motifs are indeed important elements in forming unusual DNA structures.
INTERDIGITATED (ZIPPER-LIKE) STACKING
To date, many different types of DNA mismatch have been discovered (67). However, most of these mismatches remain paired co-planarly, irrespective of the configuration they adopt. Recently, however, an entirely novel interdigitated or zipper-like conformation for the mismatched G/G base pair (35,68–70), A/A base pair (71) or even the canonical G/C and A/T base pairs (72) was discovered, in which the base pairing is disrupted, and the unpaired bases are intercalated with each other, when such bases are bracketed by a pair of sheared G·A base pairs. Structural characteristic of such stacking is different from the cross-strand base/base stacking mentioned above, in that only a single residue is involved in stacking, with both of its base and deoxyribose involved in stacking.
Zipper-like 5′-GGA/AGG-5′ motif
The first zipper-like motif was discovered (35) in the purine-rich (TGGAA)n strand of the human centromeric tandem repeats (73,74), in which the interdigitated 5′-GGA/AGG-5′ motif is bracketed by a pair of canonical A·T base pairs. Although it is still not clear if the two complementary strands of the centromeric 5′-(TGGAA)n/(ACCTT)n-5′ tandem repeats actually separate in vivo, nuclear proteins recognizing the purine-rich (TGGAA)n strand with reasonably high affinity have been identified from the HeLa cell nucleus (74). Furthermore, the purine-rich strand alone exhibits the same thermal stability as the canonically paired duplex (74). There are thus reasons to believe that the two centromeric strands are separated in vivo, and that the zipper-like 5′-GGA/AGG-5′ motif of the purine-rich strand serves important biological functions.
The most remarkable structural feature of the 5′-GGA/AGG-5′motif is that the central two guanosine residues are not paired, but intercalate with each other (Fig. 5), with one unpaired guanine base stacked with another unpaired guanine base, and its deoxyribose stacked with the adenine base of the neighboring sheared G·A pair (35,68,69). Therefore, both the base and the deoxyribose of the unpaired guanosine residues participate in stacking with the bracketed G·A pairs in a way akin to the cytidine residue in the GCA single-residue loop structure (53,68), or to the guanosine residue in the AGC single-residue loop structure (47) (see Fig. 7b). This phenomenon is again responsible for the large upfield shifting of the H4′ proton of the unpaired guanosine residue (35), which is now under the strong ring-current shifting effect of the adenine base. Another interesting feature for the 5′-GGA/AGG-5′ motif is that since the central guanosines are unpaired, their 2NH2 groups are free to form H-bonds with the cross-strand phosphate backbone (35), which further stabilizes such interdigitated structural motifs. Therefore, such a zipper-like motif contains some unique structural features such as base/sugar stacking, cross-strand base-phosphate H-bonding, and cross-strand unpaired base stacking that are rarely observed in any other nucleic acid structures.
Figure 5.
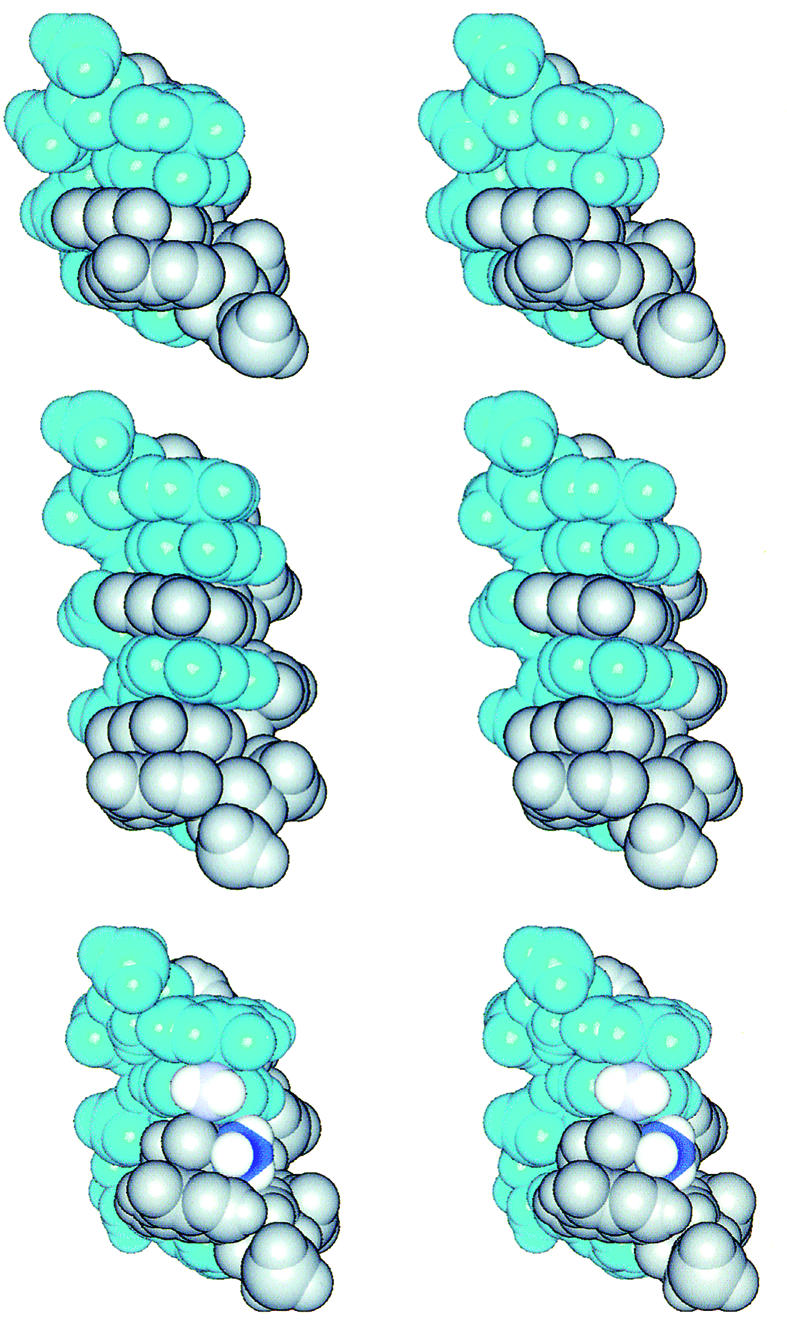
The cross-eye stereo pictures into the major groove of the interdigitated zipper-like d(GGA)2 (top), d(GGGA)2 (middle) and 5′-d(GTA)/(AAG)-5′ (bottom) motifs in space-filling. One strand is colored in blue, while another in gray to clearly illustrate the cross-strand stacking feature. In the 5′-d(GTA)/(AAG)-5′ (bottom) motif, the 6N atom of the intercalated adenine is colored in light blue and the methyl carbon of the intercalated thymine in deep blue.
Figure 7.
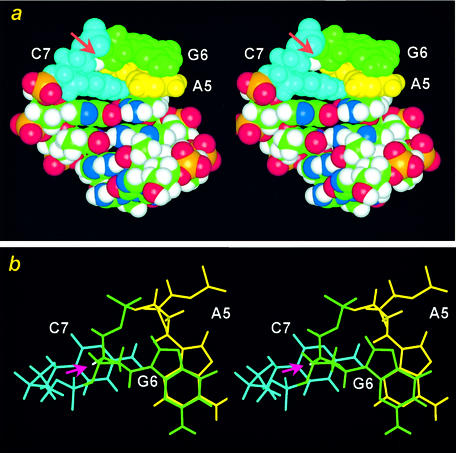
(a) The cross-eye stereo structure in space-filling mode of a DNA hairpin containing the AGC miniloop. The G6H4′ proton is situated directly above the cytidine base and is colored in white and marked with a pink arrow. (b) The cross-eye stereo structure of the AGC miniloop in stick mode viewed down to the helix. The simultaneous stacking of the base and deoxyribose of the loop G6 residue with the A5 and C7 bases of the closing sheared A·C pair is obvious in this view.
Zipper-like 5′-GGGA/AGGG-5′ and 5′-GAAA/AAAG-5′ motifs
Since most centromeric satellite sequences are asymmetric in the distribution of purine content and usually result in one strand being purine-rich vis-à-vis the other strand, it is natural to ask if the distinctive zipper-like 5′-GGA/AGG-5′ motif can be extended to the longer 5′-G(Pu)nA/A(Pu)nG-5′ sequences (n ≥ 2). Indeed, the 5′-G(Pu1Pu2)A/A(Pu1Pu2)G-5′ sequences were found to adopt the similar elongated zipper-like motifs, with the 5′-G(GG)A/A(GG)G-5′ sequence exhibiting the highest melting temperature, and the 5′-G(AA)A/A(AA)G-5′ sequence the lowest melting temperature (70). One of the reasons for this melting temperature difference is possibly due to the lack of cross-strand ANH2-phosphate H-bonds in the 5′-G(AA)A/A(AA)G-5′ motif, because ANH2 groups are not situated in a right position to form such a bond. Interestingly, in the 5′-G(G1G2)A/A(G2G1)G-5′ motif, the sugar conformations of the unpaired guanosines are not uniform, with the two outer zipper G1 deoxyriboses adopting the unusual C3′-endo conformation (as demonstrated by the very strong intra H8-H3′ NOE), while those of the inner zipper G2 deoxyriboses adopting an intermediate O4′-endo conformation (70). Such elongated zipper-like motifs exhibit a narrow minor groove, with the shortest inter-strand P–P distance being only 8.8 Å. This is also correlated with the short inter-strand sugar H1′–H1′ distances (as revealed by their medium-strength NOEs). Such a feature is rarely observed in any nucleic acid structure, except in the cytidine-rich ‘i’ motif, which also forms a four-stranded interdigitated structure and exhibits medium-strength inter-strand sugar H1′–H1′ NOEs (75). One stabilizing factor that deserves noting for such motifs is the anti-parallel stacking between the unpaired guanines in the zipper region (Fig. 5, bottom). Theoretical calculation has indicated that guanine base is much more polar than adenine base (76,77), and hence the stability of the G/G stacking would depend to a great extent upon whether they are engaged in anti-parallel (stabilizing) or parallel stacking (destabilizing). In the current 5′-G(GG)A/A(GG)G-5′ motif, the four central guanines are all engaged in anti-parallel stacking and therefore partially account for the extraordinary stability of such a highly distorted duplex structure.
The successful extension of zipper-like structures from the 5′-GGA/AGG-5′ motif to the 5′-GGGA/AGGG-5′ motif further implies the possibility of extending to even longer zipper structures, like that of the 5′-GGGGA/AGGGG-5′ motif. However, such sequences are prone to G-4 tetrad formation and are therefore difficult to study by either NMR or X-ray methods. Replacement of the zipper guanines by either inosine or adenine to interrupt the consecutive guanines may help observe such a long zipper structure.
Although the 5′-GAAA/AAAG-5′ motif is less stable than the 5′-GGGA/AGGG-5′ motif, a DNA duplex containing such a motif has recently been crystallized by the addition of Co++ ions in the solution and its structure determined by an X-ray diffraction method (71). Interestingly, the 5′-GGGA/AGGG-5′ motif in the solution state and the 5′-GAAA/AAAG-5′ motif in the solid state both form similar zipper-like structures with excellent overlap (70), although some difference in the base orientation and groove width was observed, due to the failure of the adenine zipper in the 5′-GAAA/AAAG-5′ motif to form cross-strand H-bonds.
A further point of interest about such motifs is that the isolated G-strand [(CTACGGGACCGA)n] of another evolutionarily conserved centromeric dodeca-satellite DNA sequence in Drosophila also forms highly stable intramolecular hairpin structures (Tm ∼75°C) that are stabilized by the formation of non-Watson–Crick G·A pairs (19). The homopurine tetranucleotide 5′-GGGA-3′ was found to be the major determinant in stabilizing this dodeca-satellite G-strand hairpin structure. Three possible pairing registers for this tetra-nucleotide have been proposed, including registers with a d(GA)2 motif, a d(GGA)2 motif, or even a d(GGGA)2 motif with two interacting G·G pairs between the G·A pairs (19). However, the exact nature of the G·G interactions and the non-Watson–Crick G·A pairing were not yet clear. But it is interesting to note that a single-stranded DNA binding protein that specifically binds the unstructured dodeca-satellite C-strand has recently been uncovered (21). This indicates that the purine-rich G-strand has the potential to separate from its complementary pyrimidine-rich C-strand to form altered DNA structures to organize the Drosophila centromere structure.
Zipper-like Watson–Crick base pair motifs
While studying the DNA analogs of the RNA E-like motif 5′-NGUAP/ZAAGQ-5′ (where N/Z and P/Q are complementary canonical base pairs) (78), it was accidentally discovered that the central T/A pair does not form a H-bonded base pair at all. Instead, the central T/A pair in the 5′-GTA/AAG-5′ motif exhibits considerable structural change, with their H-bonding completely disrupted and the two bases engaged in stacking with each other to form a zipper-like conformation bracketed by two sheared G·A base pairs (72) as shown in Figure 5 (bottom). Furthermore, such a zipper-like structure occurs not only in the 5′-GTA/AAG-5′ or 5′-GAA/ATG-5′ motif containing a potential T/A or A/T base pair, but also in the 5′-GGA/ACG-5′ or 5′-GCA/AGG-5′ motif containing a potential G/C or C/G base pair. Obviously, such special structures are stable enough to compensate for the energy loss of two or three H-bonds of the disrupted base pairs, since NMR spectra of the DNA hairpins containing such 5′-GXA/AYG-5′ motifs in the stem region reveal only one single set of resonances with no other species detected at all (72). While abundant and distinctive NOEs allowed the structural determination of such zippers with certainty, they can in fact be easily verified by the appearance of characteristic upfield-shifted signals of >3 p.p.m. for the T-H3 imino proton or >2 p.p.m. for the G-H1 imino proton of the central complementary X/Y pair (72). These indicate that the X/Y pair in the 5′-GXA/AYG-5′ motifs is not H-bonded either in canonical or Hoogsteen modes. The extraordinary upfield shifting of the X/Y H1′/H4′ proton signals by ∼1 and ∼2 p.p.m., respectively, is also consistent with the picture of a zipper-like structure, in which the unpaired X/Y deoxyriboses are stacked upon the adenine bases of the sheared G·A base pairs (72), akin to that shown in Figure 6 (bottom). Such extraordinary upfield shifting of deoxyribose protons usually implies the existence of an unusual deoxyribose/base stacking pattern not present in a canonical B-DNA environment, in which the deoxyriboses are located far away from the base stacking core and are not strongly affected by their ring-current shifting. The rather sharp proton signals of these zippers also indicate that they are very stable in the NMR time scale (72).
Figure 6.
Comparison of the stacking pattern between the 5′-GA/AG-5′ (top), 5′-GA/AAG-5′ (middle) and 5′-GGA/AGG-5′ motifs (bottom). One strand is colored in red and another strand in deep blue. While excellent cross-strand G/G and A/A stacking are obvious for the 5′-(GA)/(AG)-5′ motif (top), good anti-parallel cross-strand G/G stacking between the unpaired guanine bases (G4 and G4′), and between the deoxyriboses of the unpaired guanosines and their neighboring adenine bases [G4(deoxyribose)/A5′(base) and G4′(deoxyribose)/A5(base)] are observed for the 5′-(GGA)/(AGG)-5′ motifs (bottom). Intercalation of residue A18 into the sheared A4·G17 and G3·A19 base pairs of the 5′-GA/AAG-5′ motif causes considerable change in the stacking pattern, resulting in a novel cross-strand three-purine A4/A18/G3 stacking (middle).
The stereo space-filling pictures of the d(GGA)2 motif, d(GGGA)2 motif and 5′-d(GTA)/d(AAG)-5′ motif viewed into the major groove are shown in Figure 5 for comparison.
Non-symmetrical zipper-like 5′-GAA/AG-5′ motif: three-purine stacking
While base stacking in the above-mentioned symmetrical 2 × 2 and 4 × 4 internal loops and symmetrical zipper-like 3 × 3 and 4 × 4 interdigitated structures occur with an unusual two-sided stacking pattern (see Figs 4 and 6), that of the non-symmetrical 2 × 3 internal loop in the highly conserved 5′-G3A4/A19A18G17–5′ ‘bubble’ present at the 3′ end hairpin of the single-stranded DNA genome of parvoviruses (23) is completely different (Fig. 6, middle). This motif contains an unpaired adenosine stacked between two bracketed sheared G·A pairs. However, the extraordinary cross-strand G/G and A/A stacking in the tandem sheared G·A pairs has undergone considerable changes. A novel three-purine stacking pattern (Fig. 6) is observed instead; the inserted A18 base is completely un-stacked from its neighboring G17 and A19 bases, but stacked nicely with the cross-strand A4 and G3 bases to form a novel three-purine A4/A18/G3 stack that is different from the double G/G or A/A stack present in the 5′-(GA)/(AG)-5′ motifs (Fig. 6, top) or the (GGA)2 motif (Fig. 6, bottom). Also, unlike the case in a bulged duplex that usually exhibits an ∼20°C kink in the helical axis when the bulge is bracketed by canonical G·C or A·T base pairs (79), no significant kink is observed for the helix containing the bulged-adenine that is bracketed by sheared G·A pairs. These facts indicate that the structure of the non-symmetrical internal loop can be very different from that of the symmetrical internal loop. The well structured ‘5′-(GAA)/(AG)-5′’ internal loop in the parvovirus genomes can explain their resistance to the single-strand-specific endonuclease (23).
The above-mentioned zipper-like or interdigitated stacking is not restricted to the DNA only. It has also been observed in the crystal structure of a tRNA molecule to stabilize its tertiary folding (80) or in the solution structure of a theophylline-binding RNA aptamer (81).
Adjacent sheared base pairs versus separated sheared base pairs
From the description above of the unusual DNA conformations, basically two major types of unique motifs can be generated by the sheared base pairing: either a distorted duplex with well stacked adjacent sheared base pairs (36,37,40,43,44,46,48,82) or an interdigitated duplex motif bracketed by separated sheared base pairs (35,68–70,72). When two sheared base pairs occur in tandem (adjacent), they form a compact cross-strand stacking core that can be further extended in either direction by wobble base pairs to form a well stacked distorted duplex with four consecutive mismatches. The stable formation of such DNA duplex with four mismatches indicates that the energy loss from mismatch pairing can be compensated by the excellent cross-stand and intra-strand base stacking. On the other hand, when the tandem sheared base pairs are interrupted and separated by a G/G, or A/A mismatch, or by a T/A, or G/C canonical base pair, or even by two pairs of G/G mismatches, dramatically different DNA conformation are observed instead. The base pairs of the central residues, whether they are canonical or not, are broken. Instead, they are switched to the zipper-like or interdigitated conformation. The reason for such a dramatic switch is again possibly due to the need for cross-strand base stacking and cross-strand H-bonding to stabilize the consecutive mismatches.
HAIRPIN LOOP MOTIFS
UUCG (83), GNRA (84) and AGNN (85,86) tetraloops are important RNA structural motifs. The first and last residues in these tetraloops form a non-canonical base pair to close the loop. To date, two is considered the minimum number required for the unpaired residues in a stable RNA hairpin loop (85–88). On the other hand, DNA hairpin loops are more compact, and a single unpaired residue in the loops is usually sufficient when the loops are closed by a sheared G·A (51,52,89), A·A (90), A·C (24,47,91,92) or even a Ganti·Csyn base pair (93). Such compact DNA miniloops are occasionally found in the replication origins (94), promoter region (95,96), or enzyme (topoisomerase) binding site (91,92,97) that have potential to form cruciform structures. They also appear in the 3′ end fold-in structures of some linear DNA viruses (23,98) or linear plasmids (24). On the other hand, DNA loops with two or three unpaired residues are possible when the first loop residue is a thymidine, which can fold into the minor groove to exhibit specific interaction with the stem G·C base pairs (99–102). The purine-rich GGA sequence and its complementary pyrimidine-rich TCC sequence thus can each form different yet stable loop structures to facilitate the cruciform formation (100).
Recently, there have been systematic studies of thermodynamically stable DNA or RNA hairpin triloops or tetraloops using a temperature-gradient gel electrophoresis method (103–105). Some of the stable loop motifs identified from the combinatorial library are found in nature while others are not, which may serve as the new targets for NMR structural studies.
Single-residue GNA, ANA and ANC loops closed by a sheared mismatched base pair
The single-residue GNA (51–53,89), ANA (90) or ANC loops (47,97) are characterized by the simultaneous stacking of the base and deoxyribose of the unpaired N residue upon the closing guanine and adenine bases of the GNA loop (51,53,89), the closing adenine bases of the ANA loop (90) or the closing adenine and cytosine bases of the ANC loop (47,97) (Fig. 7b), respectively. Such sheared or side-by-side pairing nature of the closing base pairs makes it possible for a single residue in the loop to span two helical strands (Fig. 7b), which is unlikely when the closing base pair is a bonded by a head-to-head canonical G·C or A·T base pair. Such a base/deoxyribose stacking interaction is rarely observed in a regular right-handed B- or A-form DNA, although it occurs in the left-handed Z-form DNA (1), and in the above-mentioned zipper-like structures. The most distinguishing feature for the NMR spectra of such base/deoxyribose stacks is the observation of dramatically upfield shifting of the deoxyribose H4′ protons of the unpaired loop residues (53), which are situated directly upon the adenine or cytosine base (Fig. 7) that exhibit strong or medium ring-current shifting effect. This is revealed by the fact that the chemical shifts of the unpaired N-H4′ protons in the GNA and ANA loops shift upfield to the remarkable 1.9 p.p.m. region (more than 2 p.p.m.) (53,90), while those of the N-H4′s in the ANC or GNC loops (described in the next section) to the less dramatic 3.5 p.p.m. (∼0.5 p.p.m.) region (47), due to the fact that adenine is a stronger ring-current shifting base than the cytosine. Interestingly, the C-H4′ bond of the unpaired deoxyribose interacts with its stacked base in a perpendicular way. Such a perpendicular σ(C-H4′)/π(aromatic ring) interaction has also been occasionally observed in the three-dimensional structures of other nucleic acid and protein molecules (106,107).
Single-residue GNC loop closed by a sheared-like Ganti·Csyn base pair
The sheared-like Ganti·Csyn pairing is the most recent addition to this unusual family of compact DNA miniloops (Fig. 1) (93). It belongs to the trans Watson–Crick/Sugar-edge geometric family according to the nomenclature adopted for non-Watson–Crick base pairs by the Nucleic Acids Databank (32). It is special because, as described above, only sheared pairing of the mismatched types has been discovered in DNA to date. No canonical Watson–Crick G·C or A·T base pair has ever been found to assume a sheared-like pairing mode to close a compact DNA loop. Such an issue is not trivial, since the d(GXC) (X = G or C) tandem repeats, when expanded beyond a certain threshold, can lead to the fragile X syndrome, one of the disastrous human hereditary neurodegenerative diseases (108,109). Knowledge of the precise tertiary structure of these hairpins will help to understand their biological roles. In this respect, we have found that the 5′-(GCATCGXC GATGC)-3′ oligonucleotides fold into stable hairpins with compact GXC miniloops that are closed by a unique sheared Ganti·Csyn base pair. The key to the formation of stable sheared G·C base pairing is the rotation of the cytidine glycosidic angle from the classical anti-form to the unusual syn-form, after the translation of the complementary cytidine residue from the head-to-head position to the side-by-side position (Fig. 1, bottom). While the syn-conformation for pyrimidine is less favored due to the steric hindrance from the carbonyl O2 group of a pyrimidine to its H3′ as well as O4′ atoms (110), it is not unprecedented, and has been observed in the thymidines of out-of-alternation Z-DNA structure in both the solid (111,112) and solution (113) states, as well as in the cytidine of the L3 loop of the hepatitis delta virus ribozyme (114). However, it is important to note that these pyrimidines are not engaged in the sheared pairing mode, but in the canonical G·C or A·T pairing mode as happened in the out-of-alternation Z-DNA structure or is unpaired as in the L3 loop structure. The syn-cytidine nature is clearly revealed by the very strong intra-sugar CH6-CH1′ NOEs (93), which are either of comparable magnitudes to or stronger than those of the CH5-CH6 NOEs, indicating that the CH6–CH1′ distance is ∼2.5 Å or less, a value that is only possible when the glycosidic angle of cytidine is transformed into the syn domain. Interestingly, the sugar pucker of the syn-cytidine in such sheared Ganti·Csyn closing base pair remains in the C2′-endo domain, unlike those of the syn-cytidines in the out-of-alternation Z-DNA structure that are in the unusual C3′-endo domain (111,112).
Recently, a reversed closing sheared G·C pair was reported in the P loop structure of the peptidyl transferase center in the 50S ribosome to present three unpaired loop guanosine residues for RNA recognition (115). Such a special closing sheared G·C pair was also detected in the crystal structure of the 50S ribosome complex (26,32). However, it is interesting to note that such a reversed sheared RNA G·C pair is very different in nature from the sheared DNA G·C pair described in the present article in two important aspects. First, it achieves the unique G3N–C4NH2 and G2NH2–C3N H-bonding by locally reversing the backbone of the cytidine residue in the sheared G·C pair, resulting in considerable changes in the backbone torsion angles. Secondly, the glycosidic bond of the cytidine residue in the sheared G·C pair is conserved in the regular anti domain. Such a reversed sheared Ganti·Canti pairing is not possible for the d(GXC) triloops described here, because there is only one nucleotide in the loop to bridge the two opposite strands.
Minor groove interaction in pyrimidine-rich bi- or tri-loops
Contrary to the simultaneous base/deoxyribose stacking of the N residue upon the closing base pairs of the GNA, ANA, ANC or GNC loops, in which the first loop residue is a purine, the story is completely different for the pyrimidine-rich loops. When the first loop residue is a thymidine, it is looped into the minor groove instead (Fig. 8, top) to interact with the stem G·C base pairs through H-bonds (Fig. 8, bottom). No sheared closing pair is necessary to bridge the two strands for such pyrimidine-rich bi- or tri-loops; a head-to-head canonical or wobble T·G base pair is sufficient to close these loops. Several such stable pyrimidine-rich loops have been reported, including those containing the unpaired T1T2 (99,102), T1T2T3 (101) and T1C2C3 (100) residues. The best NMR evidence for the folding of the T1 residue of these loops into the minor groove is the observation of strong NOEs exhibited by the T1 residue to the H1′ protons at the stem residues several base pairs away (Fig. 8, bottom) (100,101). However, it is worth noting that when the first loop residue is a cytidine, no such well defined NOE could be observed (S.-H.Chou et al., unpublished results), possibly due to the lack of H-bonds between the folded cytidine and the stem base pairs.
Figure 8.
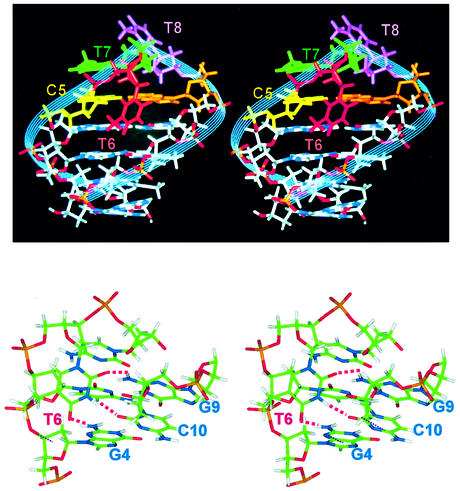
The cross-eye stereo pictures of the T6T7T8 triloop hairpin. The first loop thymidine residue (T6) folds into the minor groove, while the non-adjacent T7 and C5 bases exhibit excellent stacking (top). The H-bonding profile of the folded-in T6 residue with the surrounding stem base pairs is enlarged in the bottom figure, with the potential H-bonds being connected by dotted red lines.
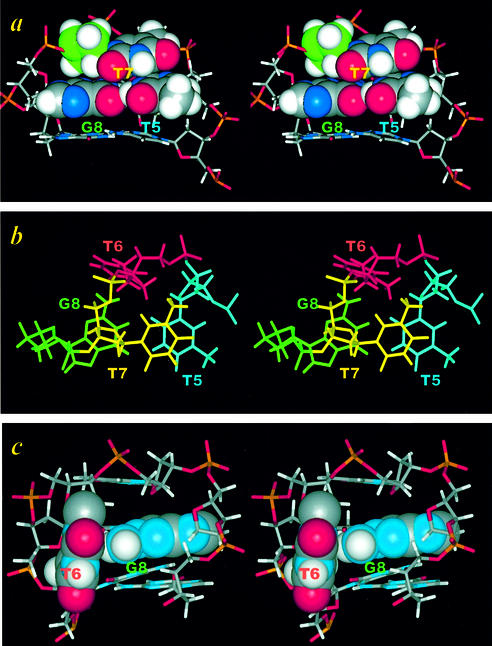
The loop interaction of the T5T6T7G8 loop is shown in Figure 9, which can be compared with those of A5G6C7 loop in Figure 7. Except for the folding of the T6 residue into the minor groove, the stacking feature of the T5T6T7G8 loop is very similar to that of the A5G6C7 or G5N6A7 loop, i.e. the base and deoxyribose of the T7 residue also stack nicely with the T5 and G8 bases of the closing base pair, respectively. Accordingly, the T7H4′ proton also experiences an upfield shifting of ∼1 p.p.m., due to the ring-current shifting effect of the guanine base. Moreover, a perpendicular interaction between the G8NH2 and the folded-in T6 base is observed in the T5T6T7G8 loop structure (Fig. 9c).
Figure 9.
The stereo cross-eye interaction pattern in the DNA TTTG loop. (a) The simultaneous stacking of the base and deoxyribose of the loop T7 residue upon the T5 and G8 bases of the closing wobble G·T pair. (b) The down-to-the-helix view of the folded-in T6 residue. The possible perpendicular G8NH2-T6 base interaction is plotted in space-filling mode in (c).
For an earlier account of the DNA loop structures, see the review article by Hilbers et al. (116).
PERPENDICULAR BASE–BASE INTERACTION MOTIF
The above description of zipper-like motifs designates a special interdigitated motif of disrupted canonical Watson– Crick base pairs when they are bracketed by sheared G·A base pairs in a cis (intrinsic) way. Likewise, a canonical Watson– Crick base pair can also be disrupted by a ligand in a trans (extrinsic) way. This is revealed recently by the determination of novel complex structures between the famous antitumor drug actinomycin D (ActD) with the specific DNA recognition sites 5′-GXC/CYG-5′ (where X/Y is a G·C or T·A Watson– Crick base pair) in the DNA hairpins closed by a mini ACC loop (117). In such complex structures (Fig. 10), the canonical G·C or T·A base pairs are disrupted and pushed out by the ActD chromophore. However, they are not disordered, but become perpendicular to the base plane and form specific H-bonds with the backbone phosphate group. Disruption of the canonical H-bonded base pair is clearly manifested by the dramatic upfield shifting of the TH3 or GH1 imino proton signals by >2 p.p.m. (117). The ordered structure of the displaced bases is also revealed by the similar non-exchangeable proton linewidths identical to those in the canonical H-bonded base pairs. Through some backbone adjustment, the ActD chromophore stacks nicely with the non-adjacent guanine bases to form an excellent guanine/chromophore/guanine hydrophobic stack in a way similar to that in the classical 5′-GC/CG-5′/ActD complex (118,119). The only difference is that the complementary cytosines of the guanine residues are now tilted away from, rather than toward, the hydrophobic core, due to the extra X/Y bases in the 5′-GXC/CYG-5′/DNA complexes (Fig. 10).
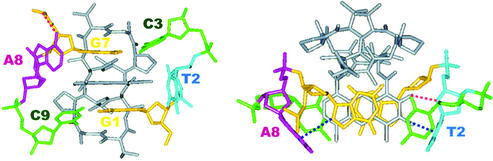
Figure 10.
The final ActD/TA complex structure in two different perspectives. The guanosine residues are drawn in yellow, cytosines in green, ActD in gray, the looped-out residues A8 in purple, and the looped-residues T2 in cyan. In the left figure, the complex structure was drawn viewed into the major groove. The planar chromophore of ActD passes through to the major groove to exhibit excellent hydrophobic stacking with the non-adjacent guanine bases. The perpendicular interactions between the G7H8-A8(π-ring) and the phenoxazone-3O-T2(π-ring) are obvious. In the right figure, the complex structure is drawn in the down-to-the-helix view.
Such a base looped-out structure is reminiscent of the ‘e’ motif in the CCG repeats (120), the cisplatin-induced interstrand cross-link structure (121), and the base-excised interdigitated structure (122). In the ‘e’ motif, the central cytidine residues in the (CCG)2 motif also do not form a C·C mismatch, but are ‘extrahelical’ and protrude into the minor groove without interfering with the conformation of the neighboring canonical G·C base pairs. Such a structure is unwound at the extrahelical sites and exhibits a wider minor groove to accommodate the extrahelical cytidines necessary for the hydrophobic interaction between the unpaired cytidines and the surrounding G·C base pairs. This phenomenon, along with the good cross-strand stacking between the non-adjacent guanine bases, may explain the stability of such an ‘e’ motif. Similarly, in the cisplatin-induced distorted structure, the cytidines in the 5′-GC/CG-5′ site were also determined to be extrahelical when the guanine bases were cross-linked by cisplatin, another widely used antitumor drug (121). In such a structure, the guanine bases are brought near by the N7-Pt-N7 linkage to exhibit good inter-stranded stacking. The extruded cytidines are perpendicular to the bases too. However, no specific interaction between these extruded cytidines with the duplex structure was described. Interestingly, base-excision of a self-complementary oligonucleotide containing two central G·T mismatches by the G·T/U-specific mismatch DNA glycosylase (MUG) has produced an unusual DNA structure that is very similar to the above-mentioned cisplatin-induced interstrand stacked structure (122). The abasic sugars are extruded from the double helix, and the two unpaired deoxyguanosines intercalate with each other instead and stack nicely with the adenines of the A·T base pairs above and below to form an interdigitated ‘A/G/G/A’ stack similar to that of the ‘G/G/G/G’ stack in the d(GGA)2 motif described above (35).
Recently, there are also reports about several unusual ActD/DNA binding motifs. For example, a crystallographic analysis of a complex structure between the ActD and DNA decamer d(CGATCGATCG) has resulted in an unexpected discovery of an ActD/5′-GATC-3′ binding motif that contains looped out AT dinucleotides (123). Another similar crystallographic study from the same group about the complex structure of ActD bound to the CTG triplet repeat has found that the ActD cyclic-pentapeptide lactone rings exhibit excellent contacts with the T·T mismatches to result with a stable Actd/(TGCT)2 motif (124). Furthermore, a solution NMR study of ActD binding with a seemingly ss-DNA d(CCGTTTTGTGG) oligomer has resulted in a stable ActD/(GT)2 mismatched motif with a TTT loop (125). These further examples of ActD binding with a variety of different DNA contexts demonstrate the great potential for ActD to interact with DNA in very flexible ways.
For more detailed reviews about the drug–DNA interactions with distorted canonical base pairs, see the review articles by Patel et al. (126), Yang and Wang (127) and Han and Gao (128).
CONCLUSION
As described in this review article, there are indeed several rather unusual DNA motifs that are fully compatible with the Watson–Crick base-paired duplex structure. Such motifs break the notion that DNA duplex is monotonous without much change in conformation, and offer novel recognition sites for ligands or proteins. These results also indicate that DNA duplex is flexible and is able to accommodate significant local distortions within a mainly canonically G·C/A·T base-paired duplex environment. However, deformation of DNA duplex structure can be even more dramatic in a global way. For example, barring the A-form DNA, or even the more drastic left-handed Z-form DNA (1,2), there have been reports that the two duplex strands can associate in a parallel way with reverse Watson–Crick base pairs (129–131) or with homopurine mismatches (132,133), or associate in an anti-parallel way, yet with pure Hoogsteen base pairing (134). Although it is still unclear whether such idiosyncratic duplex structures have biological functions or not, it is certainly an interesting area deserving much attention.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the National Science Council and the Chung-Zhen Agricultural Foundation Society of Taiwan, ROC for the instrumentation grants. S.-H.C. is the recipient of the outstanding research award of NSC and the outstanding research scholarship of Chung-Shan Foundation. This work was supported by the NSC grants 91-2113-M-005-014 to S.-H.C. and 91-2311-B-001-075 to A.H.-J.W.
REFERENCES
- 1.Wang A.H.-J., Quigley,G.J., Kolpak,F.J., van der Marel,G., van Boom,J.H. and Rich,A. (1981) Left-handed double helical DNA: variations in the backbone conformation. Science, 211, 171–176. [DOI] [PubMed] [Google Scholar]
- 2.Dickerson R.E., Drew,H.R., Conner,B.N., Wing,R.M., Fratini,A.V. and Kopka,M.L. (1982) The anatomy of A-, B- and Z-DNA. Science, 216, 475–485. [DOI] [PubMed] [Google Scholar]
- 3.Brown B.A. II, Lowenhaupt,K., Wilbert,C.M., Hanlon,E.B. and Rich,A. (2000) The Z alpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl Acad. Sci. USA, 97, 13532–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert A. and Rich,A. (2001) The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl Acad. Sci. USA, 98, 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz T., Behlke,J., Lowenhaupt,K., Heinemann,U. and Rich,A. (2001) Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA binding proteins. Nature Struct. Biol., 8, 761–765. [DOI] [PubMed] [Google Scholar]
- 6.Henderson E., Hardin,C.C., Walk,S.K., Tinoco,I.J. and Blackburn,E.H. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell, 51, 899–908. [DOI] [PubMed] [Google Scholar]
- 7.Patel D.J., Bouaziz,S., Kettani,A. and Wang,Y. (1999) Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric and triplet repeat disease DNA sequences. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, pp. 390–453.
- 8.Parkinson G.N., Lee,M.P.H. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 9.Muniyappa K., Anuradha,S. and Byers,B. (2000) Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol., 20, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H., Yabuki,A. and Maizels,N. (2001) A human nuclease specific for G4 DNA. Proc. Natl Acad. Sci. USA, 98, 12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- 12.Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res., 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y.-C., Shih,J.-W., Hsu,C.-L. and Lin,J.-J. (2001) Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem., 276, 47671–47674. [DOI] [PubMed] [Google Scholar]
- 14.Nowak R. (1994) Mining treasures from ‘Junk DNA’. Science, 263, 608–610. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T.A. (1993) Slippery DNA and diseases. Nature, 365, 207–208. [DOI] [PubMed] [Google Scholar]
- 16.Miwa S. (1994) Triplet repeats strike again. Nature Genet., 6, 3–4. [DOI] [PubMed] [Google Scholar]
- 17.Mitas M. (1997) Trinucleotide repeats associated with human disease. Nucleic Acids Res., 25, 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huertas D. and Azorin,F. (1996) Structural polymorphism of homopurine DNA sequences. d(GGA)n and d(GGGA)n repeats form intramolecular hairpins stabilized by different base-pairing interactions. Biochemistry, 35, 13125–13135. [DOI] [PubMed] [Google Scholar]
- 19.Ferrer N., Azorin,F., Villasante,A., Gutierrez,C. and Abad,J.P. (1995) Centromeric dedeca-satellite DNA sequences form fold-back structures. J. Mol. Biol., 245, 8–21. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz-Lombardia M., Cortes,A., Huertas,D., Eritia,R. and Azorin,F. (1998) Tandem 5′-GA:GA-3′ mismatches account for the high stability of the fold-back structures formed by the centromeric Drosophila dodeca-satellite. J. Mol. Biol., 277, 757–762. [DOI] [PubMed] [Google Scholar]
- 21.Cortes A., Huertas,D., Fanti,L., Pimpinelli,S., Marsellach,F.X., Pina,B. and Azorin,F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp 160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gold L., Polisky,B., Uhlenbeck,O. and Yarus,M. (1995) Diversity of oligonucleotide functions. Annu. Rev. Biochem., 64, 763–797. [DOI] [PubMed] [Google Scholar]
- 23.Astell C.R., Smith,M., Chow,M.B. and Ward,D.C. (1979) Structure of the 3′ hairpin termini of four rodent parvovirus genomes: nucleotide sequence homology at origin of DNA replication. Cell, 17, 691–703. [DOI] [PubMed] [Google Scholar]
- 24.Huang C.-H., Lin,Y.-S., Yang,Y.-L., Huang,S.-w. and Chen,C.W. (1998) The telomeres of streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol., 28, 905–916. [DOI] [PubMed] [Google Scholar]
- 25.Morosyuk S.V., Cunningham,P.R. and SantaLucia,J.,Jr (2001) Structure and function of the conserved 690 hairpin in E. coli 16S rRNA. II. NMR solution structure. J. Mol. Biol., 307, 197–211. [DOI] [PubMed] [Google Scholar]
- 26.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 27.Varani G. and Tinoco,I.,Jr (1991) RNA structure and NMR spectroscopy. Q. Rev. Biophys., 24, 479–532. [DOI] [PubMed] [Google Scholar]
- 28.Hermann T. and Westhof,E. (1999) Non-Watson–Crick base pairs in RNA–protein recognition. Chem. Biol., 6, R335–R343. [DOI] [PubMed] [Google Scholar]
- 29.Hermann T. and Westhof,E. (1999) Docking of cationic antibiotics to negatively charged pockets in RNA folds. J. Med. Chem., 42, 1250–1261. [DOI] [PubMed] [Google Scholar]
- 30.Moore P.B. (1999) Structural motifs in RNA. Annu. Rev. Biochem., 67, 287–300. [DOI] [PubMed] [Google Scholar]
- 31.Westhof E. and Fritsch,V. (2000) RNA folding: beyond Watson–Crick pairs. Structure, 8, R55–R65. [DOI] [PubMed] [Google Scholar]
- 32.Leontis N.B., Stoabugh,J. and Westhof,E. (2002) The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res., 30, 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang E. and Feigon,J. (1999) Structures of nucleic acid triplexes. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, pp. 355–388.
- 34.Radhakrishnan I. and Patel,D.J. (1994) DNA triplexes: solution structures, hydration sites, energetics, interactions and function. Biochemistry, 38, 11405–11416. [DOI] [PubMed] [Google Scholar]
- 35.Chou S.-H., Zhu,L. and Reid,B.R. (1994) The unusual structure of the human centromere (GGA)2 motif: unpaired guanosines stacked between sheared GA pairs. J. Mol. Biol., 244, 259–268. [DOI] [PubMed] [Google Scholar]
- 36.Green K.L., Jones,R.L., Li,Y., Robinson,H., Wang,A.H.-J., Zon,G. and Wilson,W.D. (1994) Solution structure of a GA mismatch DNA sequence, d(CCATGAATGG)2 determined by 2D NMR and structural refinement methods. Biochemistry, 33, 1053–1062. [DOI] [PubMed] [Google Scholar]
- 37.Chou S.-H., Tseng,Y.-Y., Chen,Y.-R. and Cheng,J.-W. (1999) Structural studies of symmetric DNA undecamers containing non-symmetrical sheared (PuGAPu):(PyGAPy) motifs. J. Biomol. NMR, 14, 157–167. [DOI] [PubMed] [Google Scholar]
- 38.Chou S.-H. and Tseng,Y.-Y. (1999) Cross-strand purine-pyrimidine stack and sheared purine:pyrimidine pairing in the human HIV-1 reverse transcriptase inhibitors. J. Mol. Biol., 285, 41–48. [DOI] [PubMed] [Google Scholar]
- 39.Gao Y.-G., Robinson,H., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (2000) Structure and recognition of sheared tandem GA base pairs associated with human centromere DNA sequence at atomic resolution. Biochemistry, 38, 16452–16460. [DOI] [PubMed] [Google Scholar]
- 40.Chou S.-H. and Chin,K.-H. (2001) Solution structure of a DNA double helix incorporating four consecutive non-canonical base pairs. J. Mol. Biol., 312, 769–781. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Zon,G. and Wilson,W.D. (1991) Thermodynamics of DNA duplexes with adjacent GA mismatches. Biochemistry, 30, 7566–7572. [DOI] [PubMed] [Google Scholar]
- 42.Chou S.-H., Zhu,L. and Reid,B.R. (1997) Sheared purine:purine pairing in biology. J. Mol. Biol., 267, 1055–1067. [DOI] [PubMed] [Google Scholar]
- 43.Chou S.-H., Cheng,J.-W., Fedoroff,O. and Reid,B.R. (1994) DNA sequence GCGAATGAGC containing the human centromere core sequence GAAT forms a self-complementary duplex with sheared G:A pairs in solution. J. Mol. Biol., 241, 467–479. [DOI] [PubMed] [Google Scholar]
- 44.Maskos K., Gunn,B.M., LeBlanc,D.A. and Morden,K.M. (1993) NMR study of G:A and A:A pairing in d(GCGAATAAGCG)2. Biochemistry, 32, 3583–3595. [DOI] [PubMed] [Google Scholar]
- 45.Cheng J.-W., Chou,S.-H. and Reid,B.R. (1992) Base pairing geometry in GA mismatches depends entirely on the neighboring sequence. J. Mol. Biol., 228, 1037–1041. [DOI] [PubMed] [Google Scholar]
- 46.Chou S.-H., Cheng,J.-W., Fedoroff,O.Y., Chuprina,V.P. and Reid,B.R. (1992) Adjacent G:A mismatch base pairs contain BII phosphodiesters in solution. J. Am. Chem. Soc., 114, 3114–3115. [Google Scholar]
- 47.Chou S.-H., Tseng,Y.-Y. and Wang,S.-W. (1999) Stable sheared A:C pair in DNA hairpins. J. Mol. Biol., 287, 301–313. [DOI] [PubMed] [Google Scholar]
- 48.Ebel S., Lane,A.N. and Brown,T. (1992) Very stable mismatch duplexes: structural and thermodynamic studies on tandem GA mismatches in DNA. Biochemistry, 31, 12083–12086. [DOI] [PubMed] [Google Scholar]
- 49.Ke S.-H. and Wartell,R.M. (1996) The thermal stability of DNA fragments with tandem mismatches at a d(CXYG):d(CY′X′G) site. Nucleic Acids Res., 24, 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter A.E., Wu,M. and Turner,D.H. (1994) The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry, 33, 11349–11354. [DOI] [PubMed] [Google Scholar]
- 51.Hirao I., Kawai,G., Yoshizawa,S., Nishimura,Y., Ishido,Y., Watanabe,K. and Miura,K.-i. (1994) Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res., 22, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshizawa S., Kawai,G., Watanabe,K., Miura,K.-i. and Hirao,I. (1997) GNA trinucleotide loop sequences producing extraordinarily stable DNA minihairpins. Biochemistry, 36, 4761–4767. [DOI] [PubMed] [Google Scholar]
- 53.Chou S.-H., Zhu,L. and Reid,R.R. (1996) On the relative ability of centromeric GNA triplets to form hairpins versus self-paired duplexes. J. Mol. Biol., 259, 445–457. [DOI] [PubMed] [Google Scholar]
- 54.Chou S.-H., Cheng,J.-W. and Reid,B. (1992) Solution structure of [d(ATGAGCGAATA)]2: adjacent GA mismatches stabilized by cross-strand base-stacking and BII phosphate groups. J. Mol. Biol., 228, 138–155. [DOI] [PubMed] [Google Scholar]
- 55.Chou S.-H. (2000) NMR studies of DNA structures containing sheared purine:purine and purine:pyrimidine base pairs. J. Biomol. Struct. Dyn., Conversation 11, 2, 303–316. [DOI] [PubMed] [Google Scholar]
- 56.Schneider D.J., Feigon,J., Hostomsky,Z. and Gold,L. (1995) High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry, 34, 9599–9610. [DOI] [PubMed] [Google Scholar]
- 57.Peyret N., Seneviratne,A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal AA, CC, GG and TT mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 58.Li Y., Zon,G. and Wilson,W.D. (1991) NMR and molecular modeling evidence for a GA mismatch base pair in a purine-rich DNA duplex. Proc. Natl Acad. Sci. USA, 88, 26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lane A., Martin,S.R., Ebel,S. and Brown,T. (1992) Solution conformation of a deoxynucleotide containing tandem G:A mismatched base pairs and 3′-overhanging ends in d(GTGAACTT)2. Biochemistry, 31, 12087–12095. [DOI] [PubMed] [Google Scholar]
- 60.Prive G.G., Heinemann,U., Chandrasegaran,S., Kan,L.S., Kopka,M.L. and Dickerson,R.E. (1987) Helix geometry, hydration and GA mismatch in a B-DNA decamer. Science, 238, 498–504. [DOI] [PubMed] [Google Scholar]
- 61.Gautheret D., Konings,D. and Gutell,R.R. (1994) A major family of motifs involving G:A mismatches in ribosomal RNA. J. Mol. Biol., 242, 1–8. [DOI] [PubMed] [Google Scholar]
- 62.Wu M. and Turner,D.H. (1996) Solution structure of r(GCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry, 35, 9677–9689. [DOI] [PubMed] [Google Scholar]
- 63.Zhang N., Gorin,A., Majumdar,A., Kettani,A., Chernichenko,N., Skripkin,E. and Patel,D.J. (2001) V-shaped scaffold: a new architectural motif identified in an A(GGGG) pentad-containing dimeric DNA quadruplex involving stacked G(anti)G(anti)G(anti)G(syn) tetrads. J. Mol. Biol., 311, 1063–1079. [DOI] [PubMed] [Google Scholar]
- 64.Kettani A., Gorin,A., Majumdar,A., Hermann,T., Skripkin,E., Zhao,H., Jones,R. and Patel,D.J. (2000) A dimeric DNA interface stabilized by stacked A(GGGG)A hexads and coordinated monovalent cations. J. Mol. Biol., 297, 627–644. [DOI] [PubMed] [Google Scholar]
- 65.Matsugami A., Ouhashi,K., Kanagawa,M., Liu,H., Kanagawa,S., Uesugi,S. and Katahira,M. (2001) An intramolecular quadruplex of (GGA)4 triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad and its dimeric interaction. J. Mol. Biol., 313, 255–269. [DOI] [PubMed] [Google Scholar]
- 66.Lin C.-H., Wang,W., Jones,R.A. and Patel,D.J. (1998) Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop. Chem. Biol., 5, 555–572. [DOI] [PubMed] [Google Scholar]
- 67.Saenger W. (1984) Principles of Nucleic Acid Structure. Springer, New York.
- 68.Zhu L., Chou,S.-H. and Reid,R.B. (1995) The structure of a novel DNA duplex formed by human centromere d(TGGAA) repeats with possible implications for chromosome attachment during mitosis. J. Mol. Biol., 254, 623–637. [DOI] [PubMed] [Google Scholar]
- 69.Zhu L., Chou,S.-H. and Reid,B.R. (1996) A single G-to-C change causes human centromere TGGAA repeats to fold back into hairpins. Proc. Natl Acad. Sci. USA, 93, 12159–12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chou S.-H. and Chin,K.-H. (2001) Quadruple-intercalated G-6 stacks: a possible motif in the fold-back structure of Drosophila centromeric dodeca-satellite? J. Mol. Biol., 314, 139–152. [DOI] [PubMed] [Google Scholar]
- 71.Shepard W., Cruse,W.B.T., Fourme,R., de la Fortelle,E. and Prange,T. (1998) A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 Å resolution. Structure, 6, 849–861. [DOI] [PubMed] [Google Scholar]
- 72.Chou S.-H. and Chin,K.-H. (2001) Zipper-like Watson–Crick base pairs. J. Mol. Biol., 312, 753–768. [DOI] [PubMed] [Google Scholar]
- 73.Prosser J., Frommer,M., Paul,C. and Vincent,P.C. (1986) Sequence relationships of three human satellite DNAs. J. Mol. Biol., 187, 145–155. [DOI] [PubMed] [Google Scholar]
- 74.Grady D.L., Ratliff,R.L., Robinson,D.L., McCanlies,E.C., Meyne,J. and Moyzis,R.K. (1992) Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl Acad. Sci. USA, 89, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gehring K., Leroy,J.-L. and Gueron,M. (1993) A tetrameric DNA structure with protonated cytosine:cytosine base pairs. Nature, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 76.Sponer J., Gabb,H.A., Leszczynski,J. and Hobza,P. (1997) Base-base and deoxyribose-base stacking interactions in B-DNA and Z- DNA: a quantum-chemical study. Biophys. J., 73, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spackova N., Berger,I. and Sponer,J. (2000) Nanosceond molecular dynamics of zipper-like DNA duplex structures containing sheared GA mismatch pairs. J. Am. Chem. Soc., 122, 7564–7572. [Google Scholar]
- 78.Gutell R.R., Cannone,J.J., Shang,Z., Du,Y. and Serra,M.J. (2000) A story: unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol., 304, 335–354. [DOI] [PubMed] [Google Scholar]
- 79.Rosen M.A., Live,D. and Patel,D.J. (1992) Comparative NMR study of An-bulge loops in DNA duplexes: intrahelical stacking of A, A-A and A-A-A bulge loops. Biochemistry, 31, 4004–4014. [DOI] [PubMed] [Google Scholar]
- 80.Rich A., Quigley,G.J. and Wang,A.H.-J. (1979) Conformational flexibility of the polypeptide chain. In Sarma,R.H. (ed.), Stereodynamics of Molecular Systems. Pergamon Press, New York, pp. 315–330.
- 81.Zimmermann G.R., Jenison,R.D., Wick,C.L., Simorre,J.-P. and Pardi,A. (1997) Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nature Struct. Biol., 4, 644–649. [DOI] [PubMed] [Google Scholar]
- 82.Gao X. and Patel,D.J. (1987) NMR studies of AC mismatches in DNA dodecanucleotides at acidic pH. J. Biol. Chem., 262, 16973–16984. [PubMed] [Google Scholar]
- 83.Cheong C., Varani,G. and Tinoco,I.,Jr (1990) Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature, 346, 680–682. [DOI] [PubMed] [Google Scholar]
- 84.Heus H.A. and Pardi,A. (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science, 253, 191–194. [DOI] [PubMed] [Google Scholar]
- 85.Wu H., Yang,P.K., Butcher,S.E., Kang,S., Chanfreau,G. and Feigon,J. (2001) A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J., 20, 7240–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lebars I., Lamontagne,B., Yoshizawa,S., Elela,S.A. and Fourmy,D. (2001) Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing. EMBO J., 20, 7250–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jucker F.M. and Pardi,A. (1995) Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry, 34, 14416–14427. [DOI] [PubMed] [Google Scholar]
- 88.Butcher S.E., Dieckmann,T. and Feigon,J. (1997) Solution structure of the converseved 16S-like ribosomal RNA UGAA tetraloop. J. Mol. Biol., 268, 348–358. [DOI] [PubMed] [Google Scholar]
- 89.Zhu L., Chou,S.-H. and Reid,B.R. (1995) Structure of a single-cytidine hairpin loop formed by the DNA triplet GCA. Nature Struct. Biol., 2, 1012–1017. [DOI] [PubMed] [Google Scholar]
- 90.Chou S.-H., Zhu,L., Gao,Z., Cheng,J.-W. and Reid,B.R. (1996) Hairpin loops consisting of single adenine residues closed by sheared A:A and G:G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin. J. Mol. Biol., 264, 981–1001. [DOI] [PubMed] [Google Scholar]
- 91.Amir-Aslani A., Mauffret,O., Bittoun,P., Sourgen,F., Monnot,M., Lescot,E. and Fermandjian,S. (1995) Hairpins in a DNA site for topoisomerase II studied by 1H- and 31P-NMR. Nucleic Acids Res., 23, 3850–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amir-Aslani A., Mauffret,O., Sourgen,F., Neplaz,S., Maroun,R.G., Lescot,E., Tevanian,G. and Fermandjian,S. (1996) The hairpin structure of a topoisomerase II site DNA strand analyzed by combined NMR and energy minimization methods. J. Mol. Biol., 263, 776–788. [DOI] [PubMed] [Google Scholar]
- 93.Chin K.-H. and Chou,S.-H. (2003) Sheared Ganti:Csyn base pair: a unique d(GXC) loop closure motif. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 94.Hirao I., Ishida,M., Watanabe,K. and Miura,K. (1990) Unique hairpin structures occurring at the replication origin of phage G4 DNA. Biochim. Biophys. Acta, 1087, 199–204. [DOI] [PubMed] [Google Scholar]
- 95.Glucksmann-Kuis M.A., Malone,C., Markiewicz,P. and Rothman-Denes,L.B. (1992) Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell, 70, 491–500. [DOI] [PubMed] [Google Scholar]
- 96.Glucksmann-Kuis M.A., Dai,X., Markiewicz,P. and Rothman-Denes,L.B. (1996) E. coli SSB activates N4 virion RNA polymerase promoters by stabilizing a DNA hairpin required for promoter recognition. Cell, 84, 147–154. [DOI] [PubMed] [Google Scholar]
- 97.Mauffret O., Amir-Aslani,A., Maroun,R.G., Monnot,M., Lescot,E. and Fermandjian,S. (1998) Comparative structural analysis by [1H,31P]-NMR and restrained molecular dynamics of two DNA hairpins from a strong DNA topoisomerase II cleavage site. J. Mol. Biol., 283, 643–655. [DOI] [PubMed] [Google Scholar]
- 98.Astell C.R., Chow,M.B. and Ward,D.C. (1985) Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J. Virol., 54, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ippel J.H., Lanzotti,V., Galeone,A., Mayol,L., van den Boogaart,J.E., Pikkemaat,J.A. and Altona,C. (1995) Conformation of the circular dumbbell d<pCGC-TT-GCG-TT>: structure determination and molecular dynamics. J. Biomol. NMR, 6, 403–422. [DOI] [PubMed] [Google Scholar]
- 100.Chou S.-H., Tseng,Y.-Y. and Chu,B.-Y. (1999) Stable formation of a pyrimidine-rich loop hairpin in a cruciform promoter. J. Mol. Biol., 292, 309–320. [DOI] [PubMed] [Google Scholar]
- 101.Chou S.-H., Tseng,Y.-Y. and Chu,B.-Y. (2000) Natural abundance heteronuclear NMR studies of the T3 mini-loop hairpin in the terminal repeat of the adenoassociated virus 2. J. Biomol. NMR, 17, 1–16. [DOI] [PubMed] [Google Scholar]
- 102.Chou S.-H., Chin,K.-H. and Chen,C.W. (2001) Enhanced DNA folding induced by thymine-CH3 and perpendicular guanine–thymine interaction. J. Biomol. NMR, 19, 33–48. [DOI] [PubMed] [Google Scholar]
- 103.Shu Z. and Bevilacqua,P.C. (1999) Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry, 38, 15369–15379. [DOI] [PubMed] [Google Scholar]
- 104.Nakano M., Moody,E.M., Liang,J. and Bevilacqua,P.C. (2002) Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg), d(cCNNGg) and d(gCNNGc). Biochemistry, 41, 14281–14292. [DOI] [PubMed] [Google Scholar]
- 105.Proctor D.J., Schaak,J.E. and Bevilacqua,J.M. (2002) Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry, 41, 12062–12075. [DOI] [PubMed] [Google Scholar]
- 106.Nishio M., Umezawa,Y., Hirota,M. and Takeuchi,Y. (1995) The CH/π interaction: significance in molecular recognition. Tetrahderon, 51, 8665–8701. [Google Scholar]
- 107.Umezawa Y. and Nishio,M. (2000) CH/π interaction in the crystal structure of TATA-box binding protein/DNA complexes. Bioorg. Med. Chem., 8, 2643–2650. [DOI] [PubMed] [Google Scholar]
- 108.Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- 109.Moore H., Greenwell,P.W., Liu,C.-P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haschemeyer A.E.V. and Rich,A. (1967) Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J. Mol. Biol., 27, 369–384. [DOI] [PubMed] [Google Scholar]
- 111.Wang A.H.-J., Gessner,R.V., van der Marel,G.A., van Boom,J.H. and Rich,A. (1985) Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc. Natl Acad. Sci. USA, 82, 3611–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schroth G.P., Kagawa,T.F. and Ho,P.S. (1993) Structure and thermodynamics of nonalternating CG base pairs in Z-DNA: the 1.3 Å crystal structure of the asymmetric hexanucleotide d(m5CGGGm5CG):d(m5CGCCm5CG). Biochemistry, 32, 13381–13392. [DOI] [PubMed] [Google Scholar]
- 113.Feigon J., Wang,A.H.-J., van der Marel,G.A., van Boom,J.H. and Rich,A. (1985) Z-DNA forms without an alternating purine–pyrimidine sequence in solution. Science, 230, 82–84. [DOI] [PubMed] [Google Scholar]
- 114.Lynch S.R. and Tinoco,I.,Jr (1998) The structure of the L3 loop from the hepatitis delta virus ribozyme: a syn cytidine. Nucleic Acids Res., 26, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Puglishi E.V., Green,R., Noller,H.F. and Puglisi,J.D. (1997) Structure of a conserved RNA component of the peptidyl transferase centre. Nature Struct. Biol., 10, 775–778. [DOI] [PubMed] [Google Scholar]
- 116.Hilbers C.W., Heus,H.A., van Dongen,M.J.P. and Wijmenga,S.S. (1994) The hairpin elements of nucleic acid structure: DNA and RNA folding. In Eckstein,F. and Lilley,D.M.J. (eds.), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, Heidelberg, Vol. 8, pp. 56–103.
- 117.Chou S.-H., Chin,K.-H. and Chen,F.-M. (2002) Looped out and perpendicular: Deformation of Watson–Crick base pair associated with actinomycin D binding. Proc. Natl Acad. Sci. USA, 99, 6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu C. and Chen,F.-M. (1996) Actinomycin D binds strongly and dissociates slowly at the dGpdC site with flanking T/T mismatches. Biochemistry, 35, 16346–16353. [DOI] [PubMed] [Google Scholar]
- 119.Chen H., Liu,X. and Patel,D.J. (1996) DNA bending and unwinding associated with actinoymcin D antibiotics bound to partially overlapping sites on DNA. J. Mol. Biol., 258, 457–479. [DOI] [PubMed] [Google Scholar]
- 120.Gao X., Huang,X., Smith,G.K., Zheng,M. and Liu,H. (1995) New antiparallel duplex motif of DNA CCG repeats that is stabilized by extrahelical bases symmetrically located in the minor groove. J. Am. Chem. Soc., 117, 8883–8884. [Google Scholar]
- 121.Huang H., Zhu,L., Reid,B.R., Drobny,G.P. and Hopkins,P.B. (1995) Solution structure of a cisplatin-induced DNA interstrand cross-link. Science, 270, 1842–1845. [DOI] [PubMed] [Google Scholar]
- 122.Barrett T.E., Savva,R., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct. Nature Struct. Biol., 5, 697–701. [DOI] [PubMed] [Google Scholar]
- 123.Robinson H., Gao,Y.-G., Yang,X.-L., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (2001) Crystallographic analysis of a novel complex of actinomycin D bound to the DNA decamer CGATCGATCG. Biochemistry, 40, 5587–5592. [DOI] [PubMed] [Google Scholar]
- 124.Hou M.-H., Robinson,H., Gao,Y.-G. and Wang,A.H.-J. (2002) Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res., 30, 4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chin K.-H., Chen,F.-M. and Chou,S.-H. (2003) Solution structure of ActD/5′-CCGTTTTGTGG-3′ complex: drug interaction with tandem GT mismatches and hairpin loop backbone. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patel D.J., Mao,B., Gu,Z., Hingerty,B.E., Gorin,A., Basu,A.K. and Broyde,S. (1998) Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol., 11, 391–407. [DOI] [PubMed] [Google Scholar]
- 127.Yang X.-L. and Wang,A.H.-J. (1999) Structural studies of atom-specific anticancer drugs acting on DNA. Pharmacol. Ther., 83, 181–215. [DOI] [PubMed] [Google Scholar]
- 128.Han X. and Gao,X. (2001) Sequence specific recognition of ligand–DNA complexes studied by NMR. Curr. Med. Chem., 8, 551–581. [DOI] [PubMed] [Google Scholar]
- 129.van de Sande J.H., Ramsing,N.B., Germann,M.W., Elhorst,W., Kalisch,B.W., Kitzing,E.V., Pon,R.T., Clegg,R.C. and Jovin,T.M. (1988) Parallel stranded DNA. Science, 241, 551–557. [DOI] [PubMed] [Google Scholar]
- 130.Otto C., Rippe,K., Thomas,K., Ramsing,N.B. and Jovin,T.M. (1991) The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson–Crick. Biochemistry, 30, 3062–3069. [DOI] [PubMed] [Google Scholar]
- 131.Parvathy V.R., Bhaumik,S.R., Chary,K.V.R., Govil,G., Liu,K., Howard,F.B. and Miles,H.T. (2002) NMR structure of a parallel-stranded DNA duplex at atomic resolution. Nucleic Acids Res., 30, 1500–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robinson H., van Boom,J.H. and Wang,A.H.-J. (1994) 5′-CGA motif induces other sequences to form homo base-paired parallel-stranded DNA duplex: the structure of (G-A)n derived from four DNA oligomers containing (G-A)3 sequence. J. Am. Chem. Soc., 116, 1565–1566. [Google Scholar]
- 133.Wang Y. and Patel,D.J. (1994) Solution structure of the d(TCGA) duplex at acidic pH. A parallel-stranded helix containing C+C, G:G and A:A pairs. J. Mol. Biol., 242, 508–526. [PubMed] [Google Scholar]
- 134.Abrescia N.G.A., Thompson,A., Huynh-Dinh,T. and Subirana,J.A. (2002) Crystal structure of an antiparallel DNA fragment with Hoogsteen base pairing. Proc. Natl Acad. Sci. USA, 99, 2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]