Abstract
Several biomedical vocabularies are often used by clinical applications due to their different domain(s) of coverage, intended use, etc. Mapping them to a reference terminology is essential for inter-systems interoperability. Manual vocabulary mapping is labor-intensive and allows room for inconsistencies. It requires manual searching for synonyms, abbreviation expansions, variations, etc., placing additional burden on the mappers. Furthermore, local vocabularies may use nonstandard words and abbreviations, posing additional problems. However, much of this process can be automated to provide decision-support, allowing mappers to focus on steps that absolutely need their expertise. We developed hybrid algorithms comprising of rules, permutations, sequence alignment and cost algorithms that utilize the UMLS SPECIALIST Lexicon, a custom knowledgebase and a search engine to automatically find probable matches, allowing mappers to select the best match from this list. We discuss the techniques, results from assisting to map a local codeset, and scope for generalizability.
INTRODUCTION
A combination of biomedical vocabularies (both standard vocabularies such as SNOMED, LOINC, etc. and local vocabularies for lab, billing, etc.) is often required to meet the requirements of a complex clinical information system. It is practically impossible for one vocabulary to cover all these applicable domains. For a clinical information system to effectively use them, these vocabularies need to be mapped to a single vocabulary, often called a Reference Terminology.
The Reference Terminology serves as the superset of all vocabularies used by the organization, allowing translation between various terminologies, and seamless integration between various clinical applications that use multiple vocabularies[1]. In the absence of a Reference Terminology, a single clinical application will need to implement many of the aforementioned vocabularies natively, leading to difficulties in relating and translating between the concepts in multiple terminologies. The mapping process becomes more challenging when an institution uses non-standard terminologies such as local charge codes, lab codes, etc., which do not adhere to well-accepted standards[2].
In this article, we discuss a hybrid approach we developed to map a local codeset named CodeSet-1 (CS-1) to the 3M Healthcare Data Dictionary (3M HDD, our Reference Terminology). We present our results from applying this technique for mapping CS-1 to 3M HDD, and initial attempts at generalizing these algorithms to other medical vocabularies.
BACKGROUND
The hybrid techniques described in this paper were developed to map CS-1 to the 3M HDD. CS-1 is an internally developed charge codeset used by a large hospital to capture all billable items. We needed to map CS-1 to the 3M HDD to facilitate integration with other applications that use the 3M HDD as the underlying vocabulary. The goal is to translate between CS-1 and CPT-4 (Current Procedural Terminology) or HCPCS (Healthcare Common Procedure Coding System), and the 3M HDD already has CPT-4 and HCPCS mapped to it.
CS-1 was developed over a period of time by multiple users without adhering to vocabulary standards[2], and has more than 10,000 concepts. The surface forms in CS-1 were highly abbreviated. CS-1 lacked formal definitions, concept uniqueness, and used nonstandard abbreviations. The representations (surface forms) were abbreviated in an inconsistent fashion, and often the abbreviations were clumped together as a single word. CS-1 often used different acronyms to denote the same word in different concepts. It also used the same acronym for different words in different concepts.
CS-1 often used a single word such as the name of a chemical substance to denote a lab test done to detect that substance using a specific method. It also used the just name of prosthesis or a catheter to denote a specific procedure for its implantation. In addition, CS-1 had several misspellings. CS-1 also had variability in terms of punctuations and space within and after acronyms, space between individual words, etc. CS-1 lacked expanded surface forms (“long names”) for the concepts. CS-1 ID and Representations (shown in Table 1) were the only data available to perform the mapping.
Table 1.
Examples of non-standard surface forms in CS-1 and their equivalent standard forms obtained from their 3M HDD Mappings.
| CS-1 ID | CS-1 REPRESENTATION | EQUIVALENT STANDARD FORM |
|---|---|---|
| 4523 | ACETONEQUNT | Acetone, serum; quantitative |
| 6534 | ALBUMINUR | Urine Albumin |
| 5634 | BMW GWIRE | Guide Wire |
| 7354 | BONEMAR ASP | Bone marrow; aspiration only |
| 8676 | EMERGENCY ROOM | Emergency Department Visit |
| 4356 | EMINASE 30 UNITS | Injection, anistreplase, per 30 units |
| 8567 | LYMPATIC SCAN | Lymphatic Scan |
| 1321 | NASOPHARNYX | Radiologic examination; neck, soft tissue |
| 6566 | LONG ARM ADULT CA | Application, cast; shoulder to hand (long arm) |
Due to this limitation, automapping using traditional automated keyword searches, and other hybrid methods[3] including Natural Language Processing were ineffective. Human expertise was required to understand these non-standard terms. But manually translating non-standard abbreviations into standard forms (shown in Table 1), and then finding their synonyms for the thousands of concepts in CS-1 was inefficient.
These limitations necessitated the development of a new method to automatically translate these terms into a standard form, and find their possible synonyms. We developed a translation algorithm to generate possible standard forms for each CS-1 concept, and a synonym generator to find possible synonyms for these multiple standard forms. We then integrated this algorithm with a search engine to automatically search the HDD for matches, and present them to a vocabulary mapper for human review and a final mapping decision. These algorithms were deployed as a single web application named ‘HyperSearch’. An important goal was to ensure that the algorithms being built were generalizable to other biomedical vocabularies.
METHODS
The HyperSearch application consists of two closely related components – the translation and synonym generation (TranSyn) component, and the search engine component.
TranSyn component
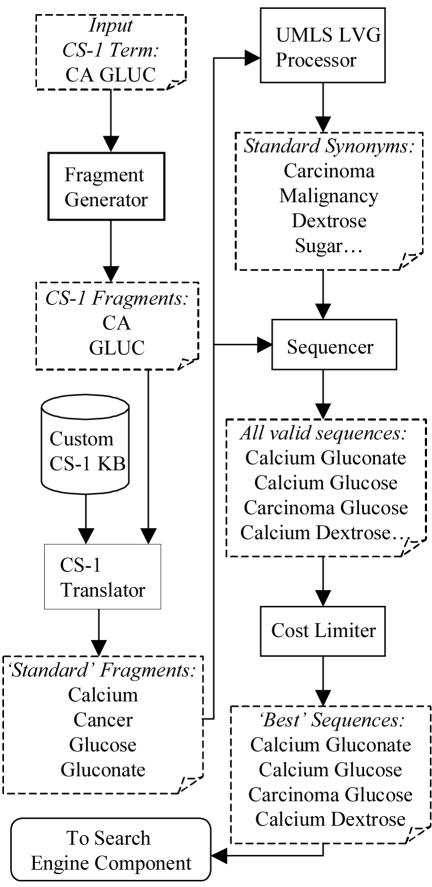
Due to the use of non-standard abbreviations in CS-1, the non-standard representations are first translated into their standard forms by the translation algorithm. Next, the synonym generation algorithm generates synonyms, abbreviation expansions, etc. The translation and synonym generation algorithms are closely integrated and work in tandem, and hence are called together as the ‘TranSyn’ component. An overview of the algorithm with an example is shown in Figure 1. The TranSyn component uses many algorithms similar to those used in DNA Sequencing[4], and are frequently referred to in this paper for better understanding. The individual steps in this algorithm are as follows.
Figure 1.
‘TranSyn’ Component Flow Diagram
Fragment Generator
The first step in translating the term is the generation of synonyms. Due to the highly variable nature of the abbreviations in CS-1, it was not possible to use semantic or linguistic methods. We were required to using syntactic parsing methods.
The Fragment Generator takes the input term and splits it into individual tokens, and tags each token with its position in the input string. It uses spaces, punctuation and special characters as delimiters. This causes splitting valid terms such as ‘W/O’ (meaning ‘without’) into two separate tokens, but this is essential to handle inconsistencies in the input data. Such splitting of valid multi-token terms into individual tokens is compensated for by the custom CS-1 Translator.
CS-1 Translator and Knowledgebase
The CS-1 Translator is a rule-based translation and disambiguation engine that finds the correct meaning of tokens in the input term from all possible alternatives. For instance, the token ‘I’ may mean ‘incision’, ‘iodine’, ‘intra’, etc. But in the context of ‘I&D’, it denotes ‘incision’. The tokenizer splits the input term at each occurrence of punctuations, special characters and spaces. So, we developed a context sensitive disambiguation engine to accommodate for special characters. This engine compared the current token to the unmodified input string to determine the context for accurate translation. This two-step process was used due to inconsistent use of special characters and punctuations in CS-1.
Furthermore, the substitution engine tries to find the match that involves the largest number of tokens. For example, ‘A.V.’ had several meanings such as ‘arteriovenous’ or ‘atrioventricular’. But ‘A.V.R.’ meant ‘aortic valve replacement’. So, the CS-1 Translator used a greedy algorithm to find substitutions for the largest number of tokens as a single string, starting at any given position. In this cause, the Translator would substitute ‘A.V.R.’ (three tokens) in one go as ‘aortic valve replacement’, before processing ‘A.V.’ (two tokens). Specific directives were defined for each specific word, telling the Translator whether or not to continue replacing shorter matches, if a longer match is already found.
After translation, we had a collection of meaningful substrings instead of the arbitrary words present in CS-1. Each of these substrings was tagged with its positional information in the unmodified input string. This information is required for the Sequencer later in the process, as described below.
The word meanings used for substitution, rules for context-sensitive disambiguation, and directives to process shorter matches were all defined in the CS-1 Knowledgebase, which was created by one of the authors (SKN) by manual review of CS-1 over a period of one week of full-time effort.
UMLS LVG Processor
The translated tokens obtained from the previous step were further queried against the UMLS SPECIALIST Lexicon using the UMLS LVG API to obtain all possible synonyms and abbreviation expansions[5]. This was done in order to match a CS-1 concept against all its possible synonyms in the 3M HDD.
At the end of this stage, we have several substrings of the original input string and several synonyms of these substrings. This is comparable to DNA Sequencing where a given DNA sample is amplified and digested to produce several fragments, each starting and ending at different positions in the original DNA strand. However, we gathered unique synonyms rather than multiple copies as is done in DNA Sequencing.
The synonym fragments, though meaningful, are not useful by themselves to find matches in the target vocabulary – the 3M HDD. These synonyms of the substrings need to be aligned together to obtain meaningful synonyms of the whole input term. This is done by the Sequencer.
Sequence and Cost Limiter
The Sequencer and the Cost Limiter together implement a shortest-path algorithm, a classical algorithm used in Artificial Intelligence. The Sequencer strings various synonyms of substrings together in the correct order to recreate entire synonyms of the original input string. A popular example of path finding algorithms is a DNA sequence alignment algorithm[4] that strings together the amplified DNA fragments. Another popular application is in rail and air reservation programs[6].
The Sequencer starts with a substring beginning at the first token, and tries to find another substring that begins at the position where the current one ends. Once it finds such a substring, the Sequencer appends it to the former. This process is iterated to find all possible paths from the start of the input term to its end, using all possible substrings that start and end sequentially. Since ‘connecting’ substrings alone are used, the nonsensical ‘dangling’ synonyms are eliminated. We are finally left with only the meaningful synonyms of the entire input term (‘successful paths’).
For each ‘successful path’ that is created, a ‘cost’ is computed. The cost depends on the number of substrings utilized and the length of each substring. Large number of substrings increases the cost, whereas longer substrings decrease it. When the cost is calculated, the synonyms with the least cost are often the most accurate[7]. The cost threshold was set at the 80th percentile after pilot tests, on the basis of ‘noise’ in the generated synonyms. The synonyms whose costs exceeded the 80th percentile were discarded.
The output contains the most meaningful synonyms of the entire input term. These are then passed to the search engine to obtain their respective matches. The search engine supports Boolean queries with multiple items. So, we create a large search string that combines all the synonyms with an ‘OR’ query. Even when one of the synonyms is an exact (or near-exact) match for a 3M HDD concept, it generates a very good match rank and score to the correct target concept. The search engine interface and implementation is described below.
Search Engine Component
We used Lucene[8] – an open source search engine written in Java and released by the Apache Software Foundation, as a component of HyperSearch. This search engine supports several useful features that help in obtaining matches even when the search string and the target string do not match exactly. This provides significant advantage over trying to match using SQL queries, or developing a string comparison algorithm from scratch. A notable feature of Lucene is fuzzy search, which helps to search terms that are misspelled or amalgamated together (e.g. ‘ACETONEQUNT’, ‘CATHTER’).
Lucene supports various ‘analyzers’ to preprocess the input terms, to allow for imprecise matches. We used built-in analyzers that ignored the word order, case, punctuations, articles and some selected prepositions that did not add much meaning (e.g. ‘for’, ‘to’, etc). We also used a stemming algorithm (‘Porter Stemmer’) that normalized word inflections (tenses, singular or plural, adverbs, etc). Thus, ‘run’ will match ‘ran’, ‘runs’, ‘running’, etc. This helped to obtain matches even when there are minor insignificant variations between the search string and the target string.
The output is provided to the vocabulary mapper as a list consisting of the rank, score, the unique concept identifier and the surface form in the 3M HDD. The mapper can then select the correct match from the list displayed. The whole process is repeated for each term in the CS-1 codeset. Thus, HyperSearch provides valuable support to mappers to make decisions, and frees them from performing mundane tasks, leading to an increase in consistency, efficiency and accuracy.
Practical Considerations and Implementation
We decided to use a mix of homegrown and third party components due to various practical considerations. We used the UMLS SPECIALIST Lexicon and Lucene due to their extensive features, coverage and support. However, we developed our own specialized algorithms for translation and synonym generation to achieve high performance. HyperSearch was written in Java and deployed on a Tomcat server running on Linux (kernel 2.4) on a Pentium 4 processor with 1 GB RAM. With this architecture, we were able to achieve sub-second response time for most input terms.
RESULTS
To evaluate the accuracy of our hybrid algorithms (‘TranSyn’), we compared the percentage of terms matched before and after implementing the TranSyn component. The Search Engine component remained the same. The test was done by sending one CS-1 term (its representation) at a time to HyperSearch, and storing the results into a database. The results consisted of a ranked list of possible matches from the 3M HDD. These results were compared with the manual mapping done for the same terms by a human mapper at the client’s institution and were provided to us.
To be considered a positive match, the correct match as previously determined by the human mapper should be within the top 10 concepts returned by HyperSearch. We limited this to the top 10 results to have a very strict measurement, though more matches may be allowed in the real world, as described below.
The results are summarized in the following table. The ‘Before’ (Version 1) results are from a version with only the search engine and without any TranSyn component. The ‘After’ (Version 2) results denote those from a version with the search engine and all TranSyn components described in this paper.
In addition to the top 10, the top 30 results were also analyzed separately for Version 2, since a mapper could see at least 30 rows of results at a glance, without scrolling the page, which is an acceptable limit for usability[9]. Several incremental developmental versions were built, with accuracy metrics falling between Versions 1 and 2.
About 60% terms that were not mapped by the latest version (Version 2) were drug brand names, which were not included in the CS-1 Knowledgebase or the UMLS Specialist Lexicon. Other terms were misspellings, or ‘not elsewhere classified’ terms, which were not captured by the CS-1 Knowledgebase. These accounted for more than 15% of the unmatched terms.
Overall, HyperSearch in its current form shows 62.8% accuracy in returning a correct match among the top 10 results, and 70% among the top 30%.
DISCUSSION
The combination of algorithms described in this paper performs well to interpret and disambiguate terms in a highly variable and inconsistent vocabulary, find the synonyms and their matches in the target vocabulary. The accuracy of this tool can be improved by enriching the knowledgebase further.
The modules in HyperSearch are also readily generalizable to other vocabularies. The CS-1 translator and knowledgebase are the only vocabulary specific components. We deactivated the CS-1 Translator component and built a version that is currently used for mapping SNOMED CT to the 3M HDD. This version shows much higher accuracy than our experience with CS-1, due to the standardized representations in SNOMED CT. Accuracy measurements for this version are not available at the time of this writing.
Thus, in addition to mapping a non-standardized local vocabulary, the study also shows the generalizability of these methods to map any given biomedical vocabulary. Furthermore, this project also emphasizes need for good vocabulary design, including the need for standardized representations.
Table 2.
Comparison of HyperSearch versions with and without TranSyn hybrid algorithms
| Before | After | |||
|---|---|---|---|---|
| Version Number | Version 1 | Version 2 | ||
| Technique | Search Engine Alone | Search Engine + TranSyn | ||
| Number of CS-1 concepts tested | 4102 | 4102 | ||
| Matched in top 10 | 870 | 21.2% | 2576 | 62.8% |
| Matched in top 30 | Not measured | 2875 | 70.1% | |
| Matches grouped by result rank: | ||||
| Rank 1 | 369 | 9% | 1521 | 37% |
| Rank 2 | 138 | 3.4% | 325 | 7.9% |
| Rank 3 | 91 | 2.2% | 211 | 5.1% |
| Rank 4 | 51 | 1.2% | 139 | 3.4% |
| Rank 5 | 48 | 1.2% | 99 | 2.4% |
| Ranks 6 to 10 | 173 | 4.2% | 281 | 6.9% |
| Ranks 11 to 15 | Not measured | 125 | 3% | |
| Ranks 16 to 20 | Not measured | 65 | 1.6% | |
| Ranks 21 to 25 | Not measured | 66 | 1.6% | |
| Ranks 26 to 30 | Not measured | 43 | 1% | |
Acknowledgements
The authors thank Chengjian Che, MD at the University of Utah Department of Medical Informatics for the batch search module used to run batch queries to measure accuracy, and Jacqueline Roe at 3M Health Information Systems for compiling the list of abbreviations used in CS-1. We also thank various Open Source developers for their invaluable contributions that were used in this project.
References
- 1.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc AMIA Annu Fall Symp. 1997;640:4. [PMC free article] [PubMed] [Google Scholar]
- 2.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 3.Solbrig HR, Elkin PL, Ogren PV, Chute CG. A formal approach to integrating synonyms with a reference terminology. Proc AMIA Symp. 2000:814–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho Junior SA. Sequence Alignment Algorithms. London: King's College; 2003.
- 5.Browne AC, Divita G, Aronson AR, McCray AT. UMLS language and vocabulary tools. AMIA Annu Symp Proc. 2003:798. [PMC free article] [PubMed] [Google Scholar]
- 6.Pallottino S, Scutella MG. Shortest Path Algorithms in Transportation models: classical and innovative aspects: Department of Informatics, University of Pisa; 1998. Report No.: TR-97.
- 7.Carberry S. Understanding Pragmatically Ill-formed Input. In: The 10th International Conference on Computational Linguistics; 1984; 1984. p. 200–206.
- 8.Apache Lucene. [cited March 1, 2005]; Available from: http://lucene.apache.org
- 9.Nielsen J. Designing Web Usability: New Riders Publishing, USA; 2000.