Abstract
A number of important applications in medicine and biomedical research, including quality of care surveillance and identification of prospective study subjects, require identification of large cohorts of patients with a specific diagnosis. Currently used methods are either labor-intensive or imprecise. We have therefore designed DITTO – a tool for identification of patients with a documented specific diagnosis through analysis of the text of physician notes in the electronic medical record.
Evaluation of DITTO on the example of diabetes mellitus, hypertension and overweight has shown it to be rapid and highly accurate. DITTO processed 1.7×105 notes/hr with sensitivity ranging from 74 to 96%, and specificity from 86 to 100%. Its accuracy substantially exceeded the performance of currently used techniques for each of the three diseases. DITTO can be adapted for use in another healthcare facility or to detect a different diagnosis. DITTO is an important advancement in the field, and we plan to continue to work to enhance its functionality and performance.
Introduction
A number of important applications in medicine and biomedical research require identification of large patient cohorts with a particular diagnosis. These include, among others, quality of care surveillance1, identification of prospective subjects for a research study2 and clinical decision support.
Several approaches have been used to identify patients with specific conditions, including death certificates3, billing data4–6, and surveys7. Each of these methods has its own shortcomings and sensitivity remains relatively low. Manual chart review remains the gold standard for identification of individuals diagnosed with a particular disease. This is a labor-intensive process that is not scalable to the level needed in a medium to large-size healthcare facility. Consequently, billing data is currently most commonly used for large-scale applications despite the above deficiencies.
As most elements of the medical record are increasingly computerized, more data becomes available for computer-assisted analysis. In particular, physician notes are a very rich source of clinical information8, and are now commonly available in digital format. However, the information in physician notes is unstructured and its analysis presents a technical challenge.
There have been a number of attempts to identify diagnoses from the text of physician notes. Most of the early reports were characterized by low sensitivity and specificity9–12. More recently, both academic13 and commercial14 tools were reported to have attained high accuracy in identifying clinical concepts from free text. However, these tools require extensive manual training on the data set and are slow (take c. 1 second or more per report)14, 15. Additionally, commercially available software is expensive and most academic systems are not freely available to the public.
We therefore have designed a software tool DITTO (Diagnosis Identification Through Textual element Occurrences) that accurately and rapidly identifies patient cohorts with a particular diagnosis through analysis of the text of physician notes, and can be employed in healthcare enterprises. We have implemented DITTO to identify cohorts of patients with documented diagnoses of diabetes mellitus, hypertension, and overweight – common diseases, which in turn place patients at risk for multiple complications, including heart, liver, and kidney disease.
Materials and Methods
Software and Hardware
DITTO has been implemented in Perl 5.6.1. It was tested under Linux RedHat 9.1 OS on a Pentium IV 3.06 GHz system with 2 GB of RAM.
Design
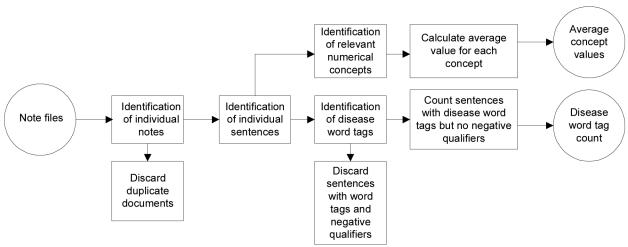
The algorithm of the text analysis is schematically represented in Figure 1. DITTO takes as input one or more files that contain the text of all physician notes for the patient population being studied. It subsequently performs the following steps:
Figure 1.
Schematic representation of the DITTO algorithm
Identification of individual notes
Identification and removal of duplicate documents
Identification of individual sentences
Identification of disease word tags in the sentences. For each of the three conditions we have compiled a list of word tags - morphemes, words or phrases that are specific for the disease (Table 1). These included the diagnosis itself, related adjectives and acronyms, and medications and procedures used exclusively to treat the disease.
Identification of negative qualifiers in the sentences. For each of the three conditions we have compiled a list of negative qualifiers – words or phrases which, when encountered in the same sentence as a disease word tag, made it unlikely that the sentence asserted that the patient had the disease. There were four main categories of negative qualifiers:
references to another disease (e.g. diabetes insipidus)
family history
non-confirmations (e.g. rule out diabetes)
negations The first category of negative qualifiers was disease-specific (Table 1) while the other three were common for all diseases (Table 2).
Identification of relevant numerical concepts Diagnosis of a number of medical conditions is based entirely or in part on numerical thresholds of various physiological or biochemical measurements (e.g. the diagnosis of overweight is based on body mass index ≥ 25). These measurements are frequently reported in physician notes and can be very valuable for ascertainment of diagnosis. We therefore designed DITTO to extract blood pressure values when identifying patients with hypertension and body weight, height, and body mass index (ratio of body weight to height squared) when identifying overweight patients.
Count of sentences with disease word tags but without negative qualifiers.
Calculation of average values of relevant numerical concepts. The number of sentences with disease word tags but without negative qualifiers and average values of relevant concepts were subsequently used to establish whether or not the patient had the disease (the thresholds were disease specific as discussed below).
Table 1.
Disease-Specific Word Tags and Negative Qualifiers.
| Disease | Word Tags | Disease-specific negative qualifiers |
|---|---|---|
| Diabetes mellitus | diabet* *IDDM medications (33 generic and brand names) |
insipidus gestational pregnancy pregnant |
| Hypertension | hypertens* HTN high [BP] elevated [BP] |
pregnancy pregnant pulmonary [HTN] portal [HTN] |
| Overweight | obes* overweight high BMI elevated BMI increased BMI gastric bypass gastric banding sibutramine |
wildcard expansion allowed in this direction
[HTN] can be either HTN or hypertension
[BP] can be either BP or blood pressure
Table 2.
Negative Qualifiers Common for All Diseases
| Family history | Non-confirmation | Negation |
|---|---|---|
| FH* family parents daughter son children child sibling brother mother father sister nephew niece |
work up for check for at risk for screen rule out |
no not doesn’t didn’t denies unknown N/A negative unlikely |
Data
In order to be able to assess sensitivity and specificity of DITTO, a patient population at high risk for diabetes, hypertension and obesity was selected. All outpatient physician notes and billing data were obtained for all adult patients of four primary care practices at the Brigham and Women’s Hospital in Boston, MA who fulfilled one of the following criteria:
ཉ at least one billing code of diabetes mellitus
ཉ at least one serum glucose > 199 mg/dL
ཉ at least one measurement of hemoglobin A1C While none of these criteria has a high specificity for any of the three conditions DITTO sought to identify, together they select a patient population with high prevalence of all three (e.g. about 1/3 with diabetes).
Physician notes were used for processing by DITTO; billing data was used for comparison with the currently employed methods of patient cohort identification (see below in Testing).
Testing
The performance of DITTO was compared to two existing standards: computational ascertainment of diagnosis from billing codes (currently most commonly used method) and manual chart review (the “gold standard”).
At least two ICD-9-CM billing codes of 250.xx, 401.xx, and 278.0x over a two-year period (2002–2003) were required to establish the diagnosis of diabetes, hypertension, and overweight, respectively – a commonly accepted standard16. To ensure comparability between the two methods, only notes over the same two-year period were used for DITTO analysis. The criteria used to establish the documentation of diagnosis of each of the three diseases using DITTO are listed in Table 3. The entire chart (including physician-maintained problem list) was used in manual review to determine whether the patient had the disease during the two-year period of the study.
Table 3.
Criteria Used to Establish Diagnoses Using DITTO Data
| Disease | Criteria |
|---|---|
| Diabetes | WT ≥ 2 |
| Hypertension | (WT≥1 OR BP≥1) AND (WT+BP)≥2 |
| Overweight | WT≥1 OR BMI≥1 OR WE≥1 |
WT: Number of sentences with disease word tags but without negative qualifiers required to establish diagnosis of the disease
BP: Number of blood pressure measurements where either systolic blood pressure ≥140 or diastolic blood pressure ≥89
BMI: Number of sentences with body mass index reported > 24.9
WE: Number of sentences with weight reported greater than that which would result in a BMI > 24.9 in a person who was less than two standard deviations above the mean height for the gender.
For each of the diseases, 150 patients (the number used in previous reports15) were randomly selected out of the total study population for comparison between the three methods. The patient charts were reviewed by one of the authors (AT) who was blinded to the results of either DITTO or billing code analysis. Kappa statistic was used to evaluate agreement between the DITTO analysis and manual chart review17. McNemar’s test was used to determine statistical significance of the difference in performance between DITTO and billing code analysis18.
IRB
The study protocol was reviewed and approved by Partners Human Research Committee.
Results
Out of the total of 52,600 patients seen at the practices whose records were analyzed, records of 7,057 patients were selected into the study data set as described above.
Text analysis of 131,033 physician notes (1.67 GB) took 47 minutes. The estimated processing speed was 1.7×105 notes / hour or 2.3GB of text / hour.
The results of the evaluation of diagnosis identification by DITTO and billing data as compared to the manual chart review on randomly selected 450 patients (150 for each diagnosis) are found in Table 4. Sensitivity of DITTO ranged between 74.2 and 96.2% and was invariably substantially higher than that of billing data analysis. Specificity of DITTO ranged from 85.9% to 100% and was usually the same or comparable to the billing data analysis.
Table 4.
Diagnosis Identification by DITTO vs. Billing Data Analysis
| Disease | Test | DITTO | Billing | p value |
|---|---|---|---|---|
| Diabetes | Sens | 96.2% | 76.9% | 0.024 |
| Spec | 98.0% | 98.0% | ||
| Hypertension | Sens | 90.7% | 74.4% | 0.078 |
| Spec | 85.9% | 92.2% | ||
| Overweight | Sens | 74.2% | 14.4% | <0.001 |
| Spec | 100% | 100% |
Sens = Sensitivity
Spec = Specificity
Kappa statistics for agreement between DITTO-based diagnosis and manual chart review are listed in Table 5. The agreement for diabetes and hypertension had kappa statistics > 0.75 (excellent agreement) while agreement for overweight had a kappa of 0.67 (substantial agreement).
Table 5.
Agreement between DITTO and Manual Chart Review
| Disease | Kappa | p-value that kappa > 0.75 |
|---|---|---|
| Diabetes | 0.94 | < 0.001 |
| Hypertension | 0.77 | 0.053 |
| Overweight | 0.67 | N/A |
Discussion
As described in this report, DITTO is exclusively characterized by the following combination of features:
High accuracy
High speed (10 to 100-fold faster than full NLP systems)
Platform independence (DITTO is implemented in OS-independent Perl without any system calls) and simplicity of local installation
Low cost
Unlike most other tools, which employ a comprehensive full-NLP approach, DITTO is focused on one disease at a time. This makes implementation of complete lexical and grammatical parsing unnecessary, thus allowing to attain significant advantages in speed without jeopardizing accuracy. DITTO requires minimal changes (mostly related to the format of the record separators in the text file containing physician notes and possibly accommodations for local differences in the medical vernacular19) for implementation in a different healthcare facility. It is therefore ideally suited for its task: rapid and accurate identification of large patient cohorts with a documented diagnosis of a particular disease.
Most other studies that have evaluated performance of information extraction from physician notes assessed the success of the extraction from a particular document14, 15, 20 or a sentence13. DITTO was put through a more rigorous as well as more clinically relevant test, where all information in the patient’s chart, including data not available to DITTO (e.g. physician-maintained problem list or notes outside of the 2002–2003 range processed by DITTO) was used to establish documentation of the diagnosis.
Under these constraints DITTO has performed remarkably well, particularly when compared with the currently most commonly used method for identification of patient cohorts – billing code analysis. Deficiencies in DITTO’s performance – for example, lower specificity in identifying patients with hypertension and lower sensitivity in identifying overweight patients – were apparently due to the underlying structure of medical documentation. Diagnoses that are relatively easy to establish and are well documented in the chart (e.g. diabetes) lend themselves to highly accurate detection. On the other hand, blood pressure can be transiently elevated for a number of reasons, and therefore hypertension was frequently mentioned in the charts even though manual review of available evidence did not find sufficient basis for the diagnosis. Finally, obesity is relatively seldom documented at all, leading to a decreased sensitivity of identification.
While a significant improvement over the existing techniques, DITTO has its limitations. It is well known that a large fraction of patients with diabetes and hypertension have not had their diagnosis established medically, and therefore not documented in the notes; these patients would be missed by the tool. Implementation of DITTO to identify a different diagnosis may require a separate validation procedure. DITTO is also not suitable for simultaneous identification of multiple diagnoses; this task is best handled by tools that implement comprehensive ontologies and syntactic and semantical processing. The physician who carried out manual chart reviews was blinded to DITTO results for these patients but not to the DITTO technique (e.g. word tags), which is a limitation of the evaluation procedure in this study.
In summary, DITTO is a highly accurate, rapid tool for identification of patient cohorts with a particular diagnosis documented in the chart. DITTO can be used for investigations of quality of care in a specific healthcare facility, by researchers looking to identify potential subjects for a study, or to provide basis for clinical decision support. DITTO can be adapted to a different healthcare facility or for detection of a different disease. It represents an important advance in the field, and we plan to continue to develop this concept further to improve its performance and functionality. DITTO code is available free of charge by request.
Acknowledgements
This research was supported in part by the NLM Training Grant 5-T15-LM-07092-11 and NLM grant 1U54LM008748-01. We would like to thank Dennis Gurgul for unwaivering IT support and Dr. Leejen Wei for his advice on statistical evaluation.
References
- 1.Saydah SH, Geiss LS, Tierney E, Benjamin SM, Engelgau M, Brancati F. Review of the performance of methods to identify diabetes cases among vital statistics, administrative, and survey data. Ann Epidemiol. 2004 Aug;14(7):507–516. doi: 10.1016/j.annepidem.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Schubart JR, Einbinder JS. Evaluation of a data warehouse in an academic health sciences center. Int J Med Inform. 2000 Dec;60(3):319–333. doi: 10.1016/s1386-5056(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki A, Horiuchi N, Hasegawa K, Uehara M. The proportion of death certificates of diabetic patients that mentioned diabetes in Osaka District, Japan. Diabetes Res Clin Pract. 1993 Jun;20(3):241–246. doi: 10.1016/0168-8227(93)90084-i. [DOI] [PubMed] [Google Scholar]
- 4.Hebert PL, Geiss LS, Tierney EF, Engelgau MM, Yawn BP, McBean AM. Identifying persons with diabetes using Medicare claims data. Am J Med Qual. 1999 Nov–Dec;14(6):270–277. doi: 10.1177/106286069901400607. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein LB. Accuracy of ICD-9-CM coding for the identification of patients with acute ischemic stroke: effect of modifier codes. Stroke. 1998 Aug;29(8):1602–1604. doi: 10.1161/01.str.29.8.1602. [DOI] [PubMed] [Google Scholar]
- 6.Raiford DS, Perez Gutthann S, Garcia Rodriguez LA. Positive predictive value of ICD-9 codes in the identification of cases of complicated peptic ulcer disease in the Saskatchewan hospital automated database. Epidemiology. 1996 Jan;7(1):101–104. doi: 10.1097/00001648-199601000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Haapanen N, Miilunpalo S, Pasanen M, Oja P, Vuori I. Agreement between questionnaire data and medical records of chronic diseases in middle-aged and elderly Finnish men and women. Am J Epidemiol. 1997 Apr 15;145(8):762–769. doi: 10.1093/aje/145.8.762. [DOI] [PubMed] [Google Scholar]
- 8.Bates DW, Evans RS, Murff H, Stetson PD, Pizziferri L, Hripcsak G. Detecting adverse events using information technology. J Am Med Inform Assoc. 2003 Mar–Apr;10(2):115–128. doi: 10.1197/jamia.M1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper GF, Miller RA. An experiment comparing lexical and statistical methods for extracting MeSH terms from clinical free text. J Am Med Inform Assoc. 1998 Jan–Feb;5(1):62–75. doi: 10.1136/jamia.1998.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc. 2001 Jan–Feb;8(1):80–91. doi: 10.1136/jamia.2001.0080080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe HJ, Antipov I, Hersh W, Smith CA, Mailhot M. Automated semantic indexing of imaging reports to support retrieval of medical images in the multimedia electronic medical record. Methods Inf Med. 1999 Dec;38(4–5):303–307. [PubMed] [Google Scholar]
- 12.Berrios DC. Automated indexing for full text information retrieval. Proc AMIA Symp. 2000:71–75. [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep–Oct;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamlin BW, Heinze DT, McDonald CJ. Automated extraction and normalization of findings from cancer-related free-text radiology reports. AMIA Annu Symp Proc. 2003:420–424. [PMC free article] [PubMed] [Google Scholar]
- 15.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002 Jul;224(1):157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 16.HEDIS 2005. Volume 2: Technical Specifications. Vol 2: National Committee on Quality Assurance.; 2005.
- 17.Cohen J. A coefficient of agreement for nominal scales. Educational and psychological measurement. 1960;20:37–46. [Google Scholar]
- 18.Hawass NE. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. Br J Radiol. 1997 Apr;70(832):360–366. doi: 10.1259/bjr.70.832.9166071. [DOI] [PubMed] [Google Scholar]
- 19.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med. 1998 Jan;37(1):1–7. [PubMed] [Google Scholar]
- 20.Heinze DT, Morsch ML, Holbrook J. Mining free-text medical records. Proc AMIA Symp. 2001:254–258. [PMC free article] [PubMed] [Google Scholar]