Abstract
The BioMediator system developed at the University of Washington (UW) provides a theoretical and practical foundation for data integration across diverse biomedical research domains and various data types. In this paper we demonstrate the generalizability of its architecture through its application to the UW Human Brain Project (HBP) for understanding language organization in the brain. We first describe the system architecture and the characteristics of the four data sources developed by the UW HBP. Second we present the process of developing the application prototype for HBP neuroscience researchers posing queries across these semantically and syntactically heterogeneous neurophysiologic data sources. Then we discuss the benefits and potential limitations of the BioMediator system as a general data integration solution for different user groups in genomic and neuroscience research domains.
INTRODUCTION
The rapid advancement of technology in the past decade has led to a plethora of new biomedical data sources, including Web-accessible public resources as well as private experimental databases developed by individual laboratories1. This wealth of data provides a tremendous opportunity for life scientists to ask questions and solve problems in unprecedented ways2. To harness these community resources and assemble all available information to investigate specific biological problems, researchers must be able to find, extract, merge and synthesize information from multiple data sources dispersed in various locations. These sources, often heterogeneous in format and architecture, span a broad spectrum of knowledge domains, from molecular cell biology, genomics, to physiology and neuroscience3. With the enormous amounts and variety of data available, integration of biological data has become a major challenge facing researchers and institutions that wish to explore these rich deposits of information.
Considerable effort and significant progress have been made in data integration systems in the biomedical domain4, 5. Some examples include the EnsEMBL Database Project6, Kleisli7, OPM8, and TAMBIS9. Most of the existing data integration systems are designed and applied specifically to integrating genomic and proteomic data sources in the realm of molecular biology. However, biological data sets are not confined to only genomics and proteomics research. For example, in response to the Human Brain Project (HBP), numerous databases have been created storing multiple types of neuroscience data ranging from structural and functional images to electrophysiological signals to behavioral data10. Furthermore, the data sources created for various biomedical research domains are often completely different from each other in both content and representation. The specificity driven by the needs of a research project makes it ideal to have “one-off” solutions; however such systems don’t scale well for multiple groups of research scientists. Therefore it is highly desirable to have a generalizable data integration system that can be applied across different biomedical research domains and data types.
The BioMediator system (www.biomediator.org) developed at UW provides a theoretical and practical foundation for data integration across diverse biomedical domains and various biomedical data types via a knowledge base driven centralized federated database model. A prototype of the BioMediator system has been successfully implemented for biologists to query across heterogeneous data sources for genetics research using molecular and genomic data11, 12, replicating to some extent the work done by previous integration projects such as TAMBIS and Kleisli. In this paper, we demonstrate the generalizability of the BioMediator architecture for data integration in a different research domain – neuroscience, using neurophysiologic databases developed by UW HBP for understanding language localization in the brain.
RELATED WORK
First-generation bioinformatics solutions for data integration typically employ specific, non-generalizable, non-modular approaches to translate data from one format into another. This means writing programs to parse, extract and transform necessary data for each particular application. The second-generation of data integration solutions provide a more structured environment for code-reuse and more flexible, scalable, robust integration. They can roughly be divided into two major categories according to access and architecture: the data warehousing approach, and the federated approach.3, 5
The data warehouse approach copies data sources into a centralized system with a global data schema and an indexing system for integration and navigation. They require reliable operation and maintenance, and fairly stable underlying databases. Examples of the warehousing approach include UCSC Genome Browser13, EnsEMBL Database Project6, and AllGenes14.
The federated approaches do not require a centralized persistent database, and thus the underlying data sources remain autonomous. The federated systems maintain a common data model and rely on a schema mapping to translate heterogeneous database schema into the target schema for integration5. The advantage of the federated approach is its flexibility, scalability and modularity. Examples of federated systems include TAMBIS9, ACEDB15, Kleisli7, OPM8 and DiscoveryLink16.
Each of these “general purpose” data integration systems has its own strength, however it hasn’t been shown whether these systems can be effectively applied in research domains other than molecular biology. Although the SenseLab17 project at Yale University has been successful in integrating multidisciplinary neuroscience data at the genetic, protein, cellular and circuit levels, it is not a “general purpose” system in that all data sources were pooled into a single EAV/CR database system, and therefore cannot be easily reconfigured for use by diverse research groups. Further discussion of the BioMediator architecture can be found in the Architecture section of our 2004 IIWeb paper12.
METHODS
To demonstrate that the BioMediator system can in fact be easily used for answering queries across diverse data types other than molecular, and data sources for research in a different biomedical domain other than genetics/genomics, we have implemented a prototype application using the existing BioMediator integration system and used the UW HBP databases as the test bed for our application.
BioMediator Architecture
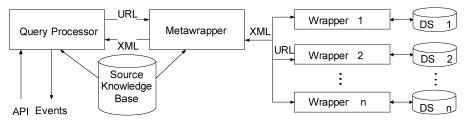
The BioMediator system uses a distributed approach and a mediated schema stored in a frame based knowledge base for querying across multiple structured and semi-structured data sources. The system consists of four components: source knowledge base (SKB)18, query processor, metawrapper, and wrappers (Figure 1). The SKB, stored in Protégé19, contains the mediated schema along with a catalogue of all possible data sources and the mediated schema elements generated by each of those sources. The query processor provides an API for launching and managing queries posed against the mediated schema. The metawrapper employs mapping rules (expanded mapping directives) to semantically translate incoming queries and outgoing result sets between the mediated namespace and the wrappers' native namespaces. Finally, wrappers interface directly with the native namespaces of the data sources, posing queries and returning XML results. A more detailed description of the BioMediator architecture is presented in the Design & Implementation section of our 2002 AMIA paper20.
Figure 1.
BioMediator system architecture
Several features make BioMediator an excellent tool for data integration in the life sciences. First, users can easily modify and extend the SKB (mediated schema and translation rules) through Protégé’s graphical user interface as their needs and schemas evolve. Second, each user can create custom mediated schemata that describe his/her view of the “universe” and pose queries against it. Third, to support user driven schema evolution, the data source wrappers are generalized in that all of the available data fields are exposed, whether or not they map to a given mediated schema. When changes are made to a mediated schema, previously invisible fields can be mapped to the new schema with no additional programming. And finally, the system provides support for exploratory search behavior in which the users issue a declarative query, browse the results in a constrained fashion and then initiate new queries to explore related topics11, 12.
UW Human Brain Project
The aim of UW HBP is to develop tools that help neuroscientists understand language organization in the brain. The hypothesis is that variations in cortical surface anatomy may be associated with variations in language ability10, 21. In order to make such inferences, research scientists need to extract information from various forms of raw and processed neuroscience data and knowledge dispersed across disparate sources. Specifically, four distinct data sources have been identified as particularly useful in answering questions and testing hypotheses about language organization in the brain:
CSM (Cortical Stimulation Mapping) This is a patient-oriented relational database stored in MySQL that records data obtained at the time of neurosurgery for epilepsy22. These data represent the cortical locations of language processing in the brain (detected by noting errors made by the patient during electrical stimulation of those areas).
Image Manager This is a MySQL relational database storing collections of images. Each image has associated with it one or more annotations, which consists of a closed polygon specified by a sequence of image coordinates and an anatomical name23.
FMA (Foundational Model of Anatomy) This is an ontology representing a large semantic network of the entire human anatomy. It is accessed by OQAFMA, which is a query tool that accepts queries written in a database query language called StruQL, and returns results in XML24. In our integration system, FMA serves as a reference for anatomical names that links CSM and Image Manager, because the anatomical regions stored in CSM (e.g., middle part of the superior temporal gyrus) are more specific than those in stored in Image Manager, which are annotated with higher-level anatomical names (e.g., superior temporal gyrus).
fMRI This is a knowledge base stored in Protégé that contains processed functional image data as well as processing protocol parameters.
The HBP data sources are particularly suitable for our experiment. They represent the data and knowledge of the neuroscience domain, which is a completely different research field from genetics; furthermore, the representation and organization of physiological data are considerably dissimilar from molecular data. Yet being able to query across all of them pose a similar integration challenge: information and data are located in semantically and syntactically heterogeneous sources.
HBP Schema and Wrappers Development
To examine the flexibility of BioMediator as an integration platform for the neuroscience domain, we followed the methodology outlined by Donelson11.
Obtain the natural language queries through user interviews. After meeting with the UW HBP members, a list of sample queries was collected across CSM, ImageManager and fMRI. For example:
Q1. Find the names of anatomical structures over all patients, in which a CSM error of type 2 (semantic paraphasia) occurred. For each of these names find all images annotated with this name from Image Manager.
Q2. Find all patients in which a CSM error occurred in part of "superior temporal qyrus" and all images from Image Manager annotated with the anatomical name. And for each patient, retrieve his/her fMRI records.
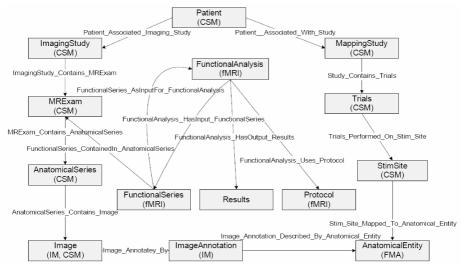
By examining the natural language queries in conjunction with the four HBP data sources, fashion a mediated schema (Figure 2) capable of answering the neuroscientists' queries. This customized schema contains only the entities and relationships of interest to this experiment.
Implement wrappers to all four data sources and add semantic mapping rules to the source knowledge base (SKB) to support the new mediated schema.
Execute queries and examine results. We used the sample queries obtained in step 1 to test the integrated HBP data system. The generated results were examined manually by members of the HBP group to ensure their correctness.
Figure 2.
HBP mediated schema
RESULTS
In less than one and half months, we were able to build a working BioMediator prototype application tailored to the needs of neuroscience researchers of the UW HBP following a sequence of four steps as described in previous section (step 1 = 2 days, step 2 = 1 week, step 3 = 3 weeks, and step 4 = 1 week). The system successfully queries across four semantically and syntactically heterogeneous neurophysiologic data sources to help identify various cortical regions associated with specific language errors. Minimal programming was needed in the entire process. In fact, the only programming involved was implementation of lightweight data source wrappers. And the task itself is very straightforward since the wrappers only need to syntactically translate incoming queries (from native URL name space, e.g., http://wrapper?key=value, to the native query format, e.g., SQL), and outgoing result sets (from native format to XML) without changing the native semantics.
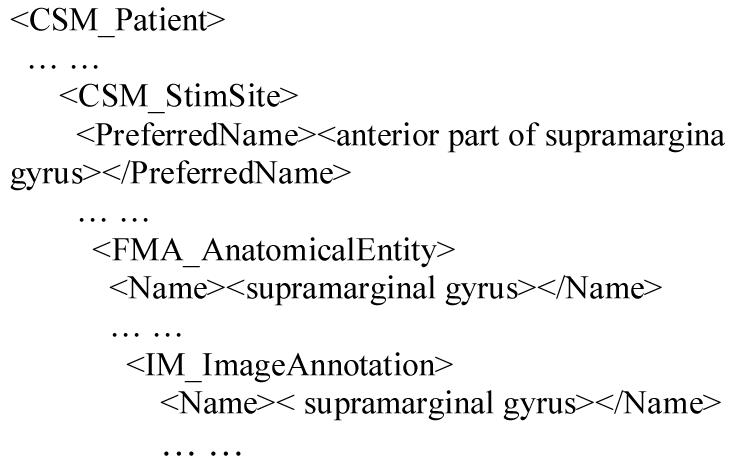
There are two user interfaces for posing queries across HBP data sources: a JSP interface and a TouchGraph25 browser. The JSP user interface takes input values for selected search parameters (e.g., CSM error type or stimulation site) and returns result in nested XML tree. Fragments of the XML output for query Q1 are listed below. The system must query FMA to determine parts of the supramarginal gyrus, and then query ImageManager for relevant images.
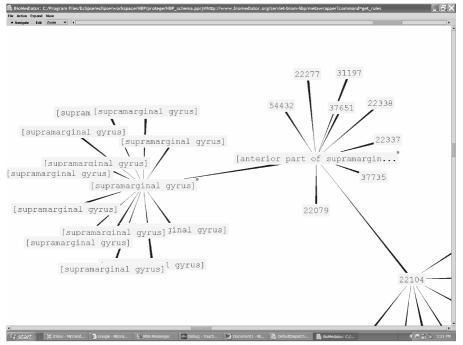
The TouchGraph25 browser is a general-purpose interface that allows users directly interact with the results graph. Figure 3 shows a zoomed view of the results graph displaying clustered nodes of ImageAnnotation and StimSite after executing query Q1.
Figure 3.
TouchGraph result display for query Q1 showing the stimulation site and images annotated with “supramarginal gyrus”. Image identifiers (e.g., 22277) pertaining to the “anterior part of [the] supramarginal gyrus” appear in the cluster to the right.
DISCUSSION
The modular design of BioMediator provides a flexible and reconfigurable platform for integrating structured and semi-structured scientific data from diverse biomedical domains. Components of BioMediator are easily substituted; hence an instance of the system can be quickly configured for use by multiple user groups with differing research interests. In most cases, only the source knowledge base, wrappers to data sources, and the user interface would need to be created or modified to support new user groups. The source knowledge base can be easily modified and extended through the Protégé GUI, and we have begun development on a GUI tool to make creating bidirectional mapping rules easier. Based on our study, we believe that with appropriate documentation and training researchers will be able to develop mediated schemata customized to their needs and bind these to sources in the library of wrappers. The impact this has on biomedical research requires a formal and extensive user evaluation in the future, which will help us to better understand the usability of the system and assess how well it meets the needs of researchers.
Through this experiment we’ve also encountered an issue that needs to be addressed in order to make the system work more optimally for HBP researchers. This issue is related to the choice of query processor used in the existing BioMediator integration model. BioMediator was originally built using a more traditional query processor11. A PQL26 query was posed against the mediated schema. The query was then translated to XQuery (a query language that accommodates semi-structured data)27 by a reformulator and executed by an XQuery engine (first Tukwila, and later Qexo)12. Similar to XQuery, PQL is a path based query language. It allows users to specify constraints on any entity in the query, analogous to declarative queries in SQL. Therefore the users have more control over what they will see in the result.
In current version of BioMediator, we replaced the XQuery engine with a browser engine that allows users to specify broad queries with head constraints and global path constraints only12. This feature is more suitable for exploratory search than constrained search. With exploratory search, which is more common in inductive genetics research, the users often don’t know exactly what they are looking for. Instead they’d start with an initial query such as: “Given the name of a genetic disease, determine all gene/protein pairs associated with that disease.” The users then can browse the results and issue new queries to explore related topics.
However, in addition to the exploratory queries (as, for example query Q1 and Q2), the more common types of HBP queries require establishing conditions on multiple entities simultaneously. For example, “Find all patients with Verbal IQ < 80 in which a CSM error of type 2 occurred in some part of the superior temporal gyrus,” includes constraints on three entities: patient (Verbal IQ < 80), stimulation (error of type 2), and anatomical location (part of the superior temporal gyrus). The current browser engine considers each constraint independently and therefore returns more results than necessary.
To allow HBP researchers to filter on the query output, we added post-processing so that the users can at least specify which entities of interest to be displayed in the final result sets. However post-processing is not efficient because this work is performed at the end of the query process, consequently prolonging total system response time. Although efficient query processing is not a requirement, the ability to respond in a satisfactory timeframe is an important attribute. To improve performance, an alternative solution would be to replace the current browser engine with an XQuery-based query engine and use a reformulator for translating user input from the JSP user interface into XQuery. With this approach, HBP researchers would be able to pose more refined and precise queries and therefore use the system more effectively to ask questions they need answers to.
In the near future, one of our goals is to extend the current BioMediator application to include analytic tools that allow researchers to perform various statistical tests on HBP neuroscience data as well as other types of biomedical data. Also, new tools for better generalizing access to and secured sharing of private databases (e.g., clinical patient data) and experimental data (e.g., expression array data) will be explored. With enhanced functionality and improved efficiency, the BioMediator system has the potential to become a powerful tool that facilitates collaboration between clinical research and biomedical informatics within a single unifying framework.
ACKNOWLEDGEMENTS
We would like to thank J. Barbero and S. E. Thiebaud for their contribution. Funding was provided by NLM (T15LM07442), NHGRI (R01HG02288), Human Brain Project (MH/DC02310) and BISTI (P20LM007714).
REFERENCES
- 1.Stevenes R, Goble C, Baker P, Brass A. A classification of tasks in bioinformatics. Bioinformatics. 2001;17:180–188. doi: 10.1093/bioinformatics/17.2.180. [DOI] [PubMed] [Google Scholar]
- 2.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–72. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 3.Lacroix Z, Critchlow T. Bioinformatics: Managing Scientific Data. San Francisco: Morgan Kaufmann Publishers; 2003.
- 4.Karasavvas KA, Baldock R, Burger A. Bioinformatics integration and agent technology. Journal of Biomedical Informatics. 2004;37(3):205–219. doi: 10.1016/j.jbi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Sujansky W. Methodological Review: Heterogeneous Database Integration in Biomedicine. Journal of Biomedical Informatics. 2001;34:285–298. doi: 10.1006/jbin.2001.1024. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al. The Ensembl genome database project. Nucleic Acids Research. 2002;30(1):38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung S, Wong L. Kleisli: a new tool for data integration in biology. Trends Biotechnol. 1999;17(9):351–5. doi: 10.1016/s0167-7799(99)01342-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen I, Markowitz V. An Overview of the Object-Protocol Model (OPM) and OPM Data Management Tools. Information Systems 1995;20(5).
- 9.Stevens R, Baker P, Bechhofer SGN, Jacoby A, Paton N, et al. TAMBIS: transparent access to multiple bioinformatics information sources. Bioinformatics. 2000;16(2):184–185. doi: 10.1093/bioinformatics/16.2.184. [DOI] [PubMed] [Google Scholar]
- 10.Brinkley JF, Rosse C. Imaging Informatics and the Human Brain Project: the Role of Structure, Review. Yearbook of Medical Informatics 2002:111–128. [PubMed]
- 11.Donelson L, Tarczy-Hornoch P, Mork P, Dolan C, JA M, M B, et al. The BioMediator System as a Data Integration Tool to Answer Diverse Biologic Queries. In: Medinfo; 2004; 2004. p. 768–72. [PubMed]
- 12.Shaker R, Mork P, Brockenbrough JS, Donelson L, Tarczy-Hornoch P. The BioMediator System as a Tool for Integrating Biologic Databases on the Web. In: VLDB-IIWeb; 2004; Toronton, Canada; 2004. p. 77–82.
- 13.Karolchik D, Baertsch R, Diekhans M, Furey T, Hinrichs A, Lu Y, et al. The UCSC Genome Browser Database. Nucleic Acids Research. 2003;31(1):51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Computational Biology and Informatics Laboratory. All Genes: A Web Site Providing Access to an Integrated Database of Known and Predicted Human and Mouse Genes. Center for Informatics, University of Pennsylvania. http://www.allgenes.org
- 15.Stein L, Thierry-Mieg J. Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Research. 1998;8(12):1308–15. doi: 10.1101/gr.8.12.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hass L, Schwarz P, Kodali P, Kotlar E, Rice JWS. DiscoveryLink: A System for Integrated Access to Life Sciences Data Sources. IBM Systems Journal. 2001;40(2):489–511. [Google Scholar]
- 17.Miller PL, Nadkarni P, Singer M, Marenco L, Hines M, Shepherd G. Integration of multidisciplinary sensory data: a pilot model of the Human Brain Project approach. J Am Med Inform Assoc. 2001 Jan-Feb;8(1):34–38. doi: 10.1136/jamia.2001.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mork P, Halevy A, Tarczy-Hornoch P. A Model for Data Integration Systems of Biomedical Data Applied to Online Genetic Databases. AMIA Symp. 2001:473–7. [PMC free article] [PubMed]
- 19.Protege home page. Stanford. http://www.protege.stanford.edu/
- 20.Shaker R, Mork P, Barclay M, Tarczy-Hornoch P. A Rule Driven Bi-Directional Tranlslation System Remapping Queries and Result Sets Between a Mediated Schema and Heterogeneous Data Sources. In: AMIA Symposium; 2002; San Antonio, TX; 2002. p. 692–696. [PMC free article] [PubMed]
- 21.Corina DP, Gibson EK, Martin R, Poliakov A, Brinkley J, Ojemann GA. Dissociation of action and object naming: Evidence from cortical stimulation mapping. Human Brain Mapping. 2005;24(1):1–10. doi: 10.1002/hbm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojemann JS, Ojemann J, Lettich E, Berger M. Cortical Language Localizations in Left, Dominant Hemisphere. J Neurosurg. 1989;71:316–26. doi: 10.3171/jns.1989.71.3.0316. [DOI] [PubMed] [Google Scholar]
- 23.Brinkley JF, Jakobovits RM, Rosse C. An online Image management system for anatomy teaching. In: American Medical Informatics Association Fall Symposium; 2002; 2002. p. 983.
- 24.Rosse C, Brinkley JF. A reference ontology for bioinformatics: the foundational model of anatomy. J. Biomedical Informatics. 2003;36(6):501–517. doi: 10.1016/j.jbi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 25.TouchGraph. TouchGraph LLC. http://www.touchgraph.com/
- 26.Mork P, Shaker R, Halevy A, Tarczy-Hornoch P. PQL: A Declarative Query Language over Dynamic Biological Schemata. AMIA Symp. 2002. [PMC free article] [PubMed]
- 27.XQuery: An XML Query Language. W3C. http://www.w3.org/TR/2005/WD-xquery-20050211/