Abstract
Existing evidence indicates that SET2, the histone 3 lysine 36 methyltransferase of Saccharomyces cerevisiae, is a transcriptional repressor. Here we show by five main lines of evidence that SET2 is involved in transcriptional elongation. First, most, if not all, subunits of the RNAP II holoenzyme co-purify with SET2. Second, all of the co-purifying RNAP II subunit, RPO21, was phosphorylated at serines 5 and 2 of the C-terminal domain (CTD) tail, indicating that the SET2 association is specific to either the elongating or SSN3 repressed forms (or both) of RNAP II. Third, the association of SET2 with CTD phosphorylated RPO21 remained in the absence of ssn3. Fourth, in the absence of ssn3, mRNA production from gal1 required SET2. Fifth, SET2 was detected on gal1 by in vivo crosslinking after, but not before, the induction of transcription. Similarly, SET2 physically associated with the transcribed region of pdr5 but was not detected on gal1 or pdr5 promoter regions. Since SET2 is also a histone methyltransferase, these results suggest a role for histone 3 lysine 36 methylation in transcriptional elongation.
INTRODUCTION
Post-translational modifications of histones, including acetylation, methylation, phosphorylation and ubiquitylation, regulate chromatin by modulating higher order structure and through presentation of binding sites for recruitment of specific complexes. Recently, considerable interest has been directed towards histone methylations partly because they may serve as epigenetic marks (1). For example, facultative heterochromatin is marked by methylation of histone 3 lysine 9 (H3 K9), which presents a binding site for HP1 and its associated partner, SUV39. Members of the SUV39 class of SET domain proteins methylate H3 K9 itself. Hence this describes a mechanism for the propagation, and potentially, inheritance of a silent chromatin state (2–5). In contrast, emerging evidence for histone 3 lysine 4 (H3 K4) methylation demonstrates roles in active chromatin (6–8). More particularly, trimethylation of H3 K4 correlates with active transcription whereas dimethylation appears to be a determinant of chromatin poised for transcription (8). It therefore appears likely that methylation of H3 K4 plays a role in the relationship between active chromatin and the transcription machinery.
In Saccharomyces cerevisiae, the H3 K4 methyltransferase is the SET1 complex (9–11). Of the five other SET domain proteins in S.cerevisiae (12), only one other, SET2, has been identified as a histone methyltransferase. SET2 methylates H3 K36 (13). The roles of SET2, K36 and K36 methylation in chromatin remain unclear; however, SET2 has been identified as a repressor of gal4 (14) and acts as a repressor in a LexA fusion protein tethering assay (13). Hence SET2 has been proposed to be a transcriptional repressor (13).
As part of a proteomic approach to characterize protein complexes associated with the six SET domain proteins in yeast (9,12), we have purified SET2 from exponentially growing haploid cells. Unexpectedly we found that a fraction of SET2 associates with C-terminal domain (CTD) phosphorylated RNAP II. Phosphorylation of the CTD is characteristic of both the elongating and the SSN3 (also known as SRB10 or UME3) repressed polymerase. However, in the absence of ssn3, SET2 remained associated with CTD phosphorylated RNAP II and was required for expression of gal1 mRNA. SET2 was found on the transcribed regions of the transcriptionally active gal1 and pdr5 genes. This involvement of SET2 in transcriptional elongation implies that methylation of H3 K36 also plays a role in transcriptional elongation.
MATERIALS AND METHODS
Strains, TAP purification and mass spectrometry (MS)
All strains were derived from MGD353-13D (15). C-terminal fusion of the TAP tag (16) to the endogenous locus, purifications and MS were performed as described (12,17). N-terminal TAP tagging of SET2 was performed by directing the TAP tag to the initiating methionine of the SET2 gene, preceded by insertion of the ura3 selection gene flanked with loxP sites into the 5′ non-coding region. After Cre recombinase mediated excision, the 36 bp palindromic loxP site was left as an insertion scar in the 5′ non-coding region. Either the inserted loxP site, the N-terminal TAP tag or both decreased SET2 expression (data not shown) and resulted in lower levels of retrieved SET2, poorer purification and increased non-specific contamination from TAP-SET2 affinity chromatography compared to SET2-TAP (Fig. 1).
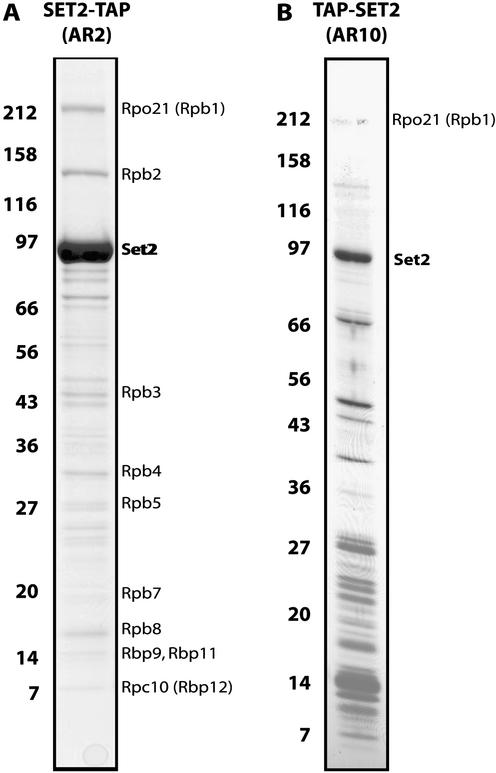
Figure 1.
TAP purification of SET2. Affinity purified SET2 was separated on 7–25% SDS–PAGE and visualized by staining with Coomassie blue. Molecular weight markers indicated on the left are in kilodaltons. All bands present in these gels were identified and only the specific ones are labeled. (A) The TAP tag was fused to the C-terminus of SET2 (SET2-TAP). (B) The TAP tag was fused to the N-terminus of SET2 (TAP-SET2).
Protein assays and immunoblotting
An HR 10/30 Superose 6 size exclusion column (Pharmacia) was loaded with 500 µl of cleared crude cell extract from a TAP-tagged strain and run in glycerol free buffer E (18). Fractions were resolved on an 8% SDS–PAGE gel and analyzed by immunoblotting. Size standards were run in parallel under the same conditions and blots were probed with peroxidase-anti-peroxidase (PAP, Sigma) diluted 1:2000, for detection of the protein A region within the TAP tag. For detection of the phosphorylated or unphosphorylated CTD of RPO21, the monoclonal antibodies H5, H14 and 8WG16 from raw ascites fluid (Covance) and affinity purified CTD4H8 (Upstate) were used at a 1:1000 dilution. Detection of RPO21 via its conserved N-terminus was achieved with the monoclonal ARNA-3 (Research Diagnostics Inc.) at a 1:1000 dilution. Secondary anti-mouse IgM (Sigma) and anti-mouse IgG (Amersham) horseradish-peroxidase conjugates were used according to the manufacturers recommendations for enhanced chemiluminescence. Quantitation of SET2-TAP molecules per cell was performed using the dot blot method described by Borggrefe et al. (19). For detection, the PAP antibody (Sigma) and ECL kit (Amersham Pharmacia Biotech) were used.
6-Azauracil assay
Yeast strains tested were grown in SD-URA at 30°C to log phase (OD600 = 0.6–0.8), washed once and diluted to OD600 = 0.5 with water. From this, a 10-fold dilution series was made and 5 µl were spotted onto 6-azauracil (6-AU; 25 mg/ml, Sigma) containing SD-URA plates. Plates were then incubated at 23, 30 and 37°C for 3 days. All strains tested contained a Kluveromyces ura cassette to convey growth on SD-URA. The cassette was either integrated at the targeted genomic loci or expressed from the pRS306 vector in the wt control.
RNA analysis
Yeast cells were grown in 50 ml YPD and harvested at OD600 = 2.0. RNA extraction was achieved by four intervals of 30 s bead beating and 2 min cooling on ice in an equal volume of acidic phenol:chloroform:isoamylalcohol (125:24:1, Sigma) and breaking buffer (20 mM Tris–Cl pH 7.4, 100 mM KCl, 2 mM MgCl2, 2 mM DTT) followed by two acidic phenol: chloroform extractions. For galactose induction, cells were grown in YP + dextrose to an OD600 = 0.2, washed twice with YP + galactose medium and incubated at 30°C for 180 min before harvesting. Twenty milligrams of total RNA was used for standard northern blotting. DNA hybridization probes were prepared by random prime labeling of 0.6–0.8 kb PCR fragments amplified from the genes using: act1 (F: 5′-AACCCTAAATCAAACAGAGAA-3′; R: 5′-CGGCAGATTCCAAACCCAAAA-3′), gal1 (F: 5′-CATTCATTTGTGCCGTTGCTT-3′; R: 5′-TCGTTCATCAAGGCACCAAAT-3′), hsp26 (F: 5′-TAAGAGGCTACGCACCAAGAC-3′; R: 5′-AACACCATTTGCGTAGTCTGC-3′), suc2 (F: 5′-ATTTACTCCTCTGATGACTTG-3′; R: 5′-GACCAGGGACCAGCATTACTA-3′), tcm1 (F: 5′-GTTGTTGGTGTTGTCG GTTAC-3′; R: 5′-TCAGCTGGGGTTAATCTA-3′), ygp1 (F: 5′-TACGCTAATGGAACAAACAGT-3′; R: 5′-AAGAGTAACCACCGTCATAGA-3′).
Chromatin immunoprecipitation (ChIP)
The ChIP against the SET2-TAP tag was performed as described (20) using rabbit IgG agarose, or empty, beads (Cl-4b, Sigma). Precipitated DNA was analyzed by PCR with multiplex primer sets along the genes of interest. Several concentrations of each template were used over a 30-fold range, and matching lanes were chosen for quantitation and the figures based on the intensity of the signal lying in the linear range. PCR settings: 3 min at 94°C, 24 cycles with 1 min at 94°C, 1 min at 56°C, 2 min at 72°C followed by a final round of 7 min at 72°C. Primer sets and the predicted amplification products given in nucleotides relative to translation start sites are as follows: gal1 A –412 to –252 (F: 5′-AAGACTCTCCTCCGTGCGTC-3′; R: 5′-ATTTGAAGGTTTGTGGGGC-3′), gal1 B 19 to 436 (F: 5′-GAAGAAGTGATTGTACCTGAG-3′; R: 5′-ACCTTTCCGGTGCAAGT TTC-3′), gal1 C 963 to 1280 (F: 5′-TCACAACATTTCCACACCC-3′; R: 5′-ACTCGTTCATCAAGGCACC-3′), gal4 A 555 to 782 (F: 5′-TTGCCCTTCCTGAAAACGGAC-3′; R: 5′ CATCATTAGCGTCGGTGAGTGC-3′), gal4 B 2282 to 2613 (F: 5′-CGTCTGCTTTGTTTGGTGGC-3′; R: 5′-TGGTGGGGTATCTTCATCATC-3′), VI R (F: 5′-CAGGCAGTC CTTTCTATTTC-3′; R: 5′-GCTTGTTAACTCTCCGACAG-3′), pdr5 A –265 to 25 (F: 5′-CTGAGCAATACAA ACAAGGCCTCTCCTA-3′; R: 5′-TATTGTTAAGCTTG GCCTCGGGCATTTT-3′); pdr5 B 1086 to 1344 (F: 5′-CACAGTGGCCATCTATCAATGTTC-3′; R: 5′-GTTCA TTTCCTTCGGGGTCTGTGGTAT-3′); pdr5 C 3967 to 4141 (F: 5′-AGGGGTGCTTTATTTTGGTTGTTC-3′; R: 5′-TAGGCATGGCACTTGGGGTAG-3′).
PCR products were separated on 2.3% MetaPhor agarose gels (Bio Whittaker), stained with Gelstar and quantitated using ImageQuant (Molecular Dynamics). For quantification, signals were first normalized for intensity based on measurement of the bands generated from the input material. Then, the relative increase of precipitated DNA was calculated for each of the assayed regions using the following equation: (SET2-TAP Gal – Control Gal)/(SET2-TAP Glu – Control Glu).
RESULTS
SET2 associates with the RNA polymerase II complex
Chromatographic purification of histone methyltransferases from yeast extracts identified SET2 as a H3 K36 methyltransferase (13). We purified SET2 by C-terminally TAP-tagging the endogenous gene. In agreement with Strahl et al. (13), our purified preparation has H3-specific methyltransferase activity on nucleosomal but not free histone substrates (data not shown). In contrast to TAP purifications of SET1 and SET3 and their associated complexes (9,12), retrieved SET2-TAP was significantly more abundant than any co-purifying protein (Fig. 1A), suggesting that most cellular SET2 is not complexed with other proteins.
Interestingly, 10 other proteins specifically co-purified with Set2-TAP (Fig. 1A). All 10 are components of RNA Polymerase II (RNAP II) and were retrieved in approximate stoichiometric concordance with each other, indicating that the retrieved proteins were complexed together. Although the RNAP II complex consists of 12 proteins (21), we were not able to identify the two missing subunits, RPB6 and RPB10, by MS analysis of SET2-TAP purifications. The absence of these two subunits could represent a new variation of RNAP II or reflect a technical limitation. Both missing proteins present difficulties for identification by mass spectrometry since both are small (RPB6, 17.7 kDa; RPB10, 8.3 kDa), and RPB6 is highly acidic causing it to migrate aberrantly during electrophoresis and understain with both Coomassie blue and silver (P. Cramer, personal communication). Hence it is quite likely that we missed these two subunits for technical reasons.
Apart from SET2 and RNAP II subunits, all of the other bands visible in Figure 1A were identified and found to be common contaminants of the TAP tagging method, as documented by us (17). We have TAP tagged more than 50 chromatin proteins so far and identified more than 200 specifically associated proteins. In only one other case has any RNAP II subunit been co-purified (our unpublished results). Hence RNAP II subunits are not common contaminants for TAP purifications. Taken together, these data suggest that most cellular SET2 is not complexed with other proteins; however, some SET2 is specifically associated with RNAP II.
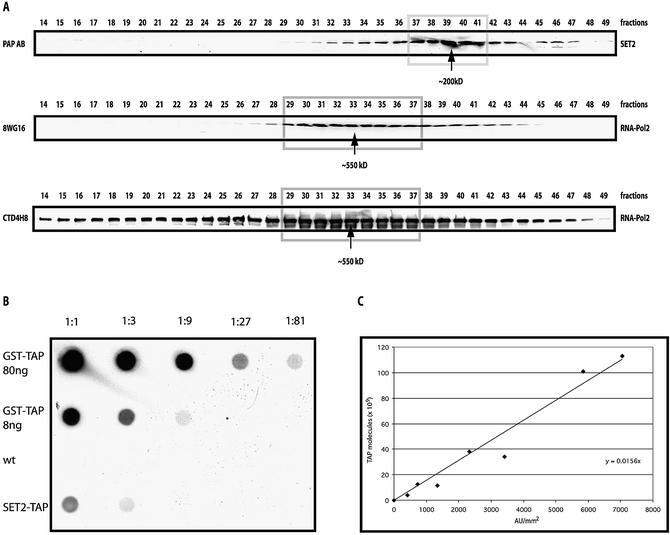
To examine this conclusion, two experiments were undertaken. First, the association between SET2 and RNAP II was re-evaluated by tagging SET2 at its N-terminus and purifying again. Although the N-terminal tagging appeared to reduce the SET2 expression level and led to poorer purification with more contaminants, the largest two subunits of RNAP II were again retrieved at levels substoichiometric to SET2 (Fig. 1B). Second, we looked for two forms of SET2 by Superose 6 size exclusion separation of total cell extract, expecting to see a major peak of uncomplexed SET2 and a minor, larger peak of SET2 complexed with RNAP II. In agreement with Strahl et al. (13), the majority of SET2 was found at ∼200 kDa (Fig. 2A). Since SET2 is 84.5 kDa (here 105 kDa due to the TAP tag), we agree with Strahl et al. that most cellular SET2 appears to be homodimeric. However, a significant portion was found trailing towards higher apparent complex sizes in a shoulder encompassing fractions 30–35. To determine whether the elution peaks of SET2 and RNAP II overlapped, the Superose 6 fractions were probed with two antibodies against the RNAP II subunit, RPO21. The 8WG16 antibody detects the unphosphorylated CTD form of RPO21, and CTD4H8 detects the serine 5 phosphorylated CTD form. As expected, both antibodies detected a peak of RNAP II at ∼550 kDa encompassing fractions 30–35. Interestingly, the unphosphorylated RPO21 appeared to be uniformly associated with the known RNAP II holoenzyme complex, whereas phosphorylated RPO21 was significantly more heterogenous, appearing to participate in larger and smaller complexes, and potentially as free protein. Hence both N-terminal TAP tagging and sizing column analysis strengthen the conclusion that a fraction of SET2 associates with a form of RNAP II.
Figure 2.
Biochemical properties of cellular SET2-TAP. Total cell extracts were fractionated on a Superose 6 sizing column before analysis by western blotting using a secondary antibody (PAP) to the TAP tag or antibodies specific for RPO21 of RNAP II, as indicated. (A) Elution profiles of SET2-TAP and RNAP II detected by PAP (SET2-TAP) or antibodies specific for unphosphorylated (8WG16) and phosphorylated (CTD4H8) RPO21. Molecular weights, assuming globular Stokes radii, were estimated from parallel size standards. (B) Estimation of cellular abundance of SET2-TAP by dot blotting. A standard curve was made using highly purified GST–TAP that had been quantitated by Bradford. Either 80 or 8 ng were spotted followed by a three-fold dilution series as indicated. Ten milligrams of total yeast proteins isolated from wild-type or SET2-TAP strains were spotted at the left as indicated, followed by three-fold dilutions. (C) Graph of the standard curve. The two highest points (80 and 27 ng) fall below the line due to saturation in the assay and have been omitted. Given 200 pg total protein per cell, we estimate 35 000 SET2 molecules/cell.
The number of SET2 molecules per cell was estimated using the dot blot method of Borggrefe et al. (19). A standard curve was made using purified glutathione S-transferase (GST)–TAP and the number of SET2-TAP molecules per cell estimated as 35 000 (Fig. 2B). Since Borggrefe et al. estimated 30 000 RNAP II molecules per cell and, by densitometry, we observe ∼5% of SET2 associated with RNAP II (Figs 1A and 2A), we infer that at least 5% of total RNAP II is associated with SET2.
SET2 specifically associates with the CTD phosphorylated form of RNAP II
The CTD of RPO21, the largest subunit of RNAP II, is a heptapeptide repeat of the sequence YSPTSPS (52 repeats in humans, 26 in yeast). It is subject to regulated phosphorylation and de-phosphorylation during the transcription cycle (22,23). Phosphorylation of serines 5 and 2 within the repeat partly indicates the stage of activity of RNAP II within the cycle, correlating with the elongation phase of transcription (24).
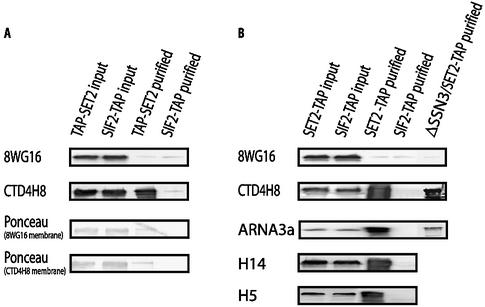
To ascertain whether SET2 associates with a specific post-translational modification of the CTD, TAP purifications from the N-terminal TAP-SET2 were probed with antibodies recognizing either the unphosphorylated or serine 5 phosphorylated CTD. SIF2-TAP, which does not associate with RNAP II (12), served as a negative control, and crude extracts were included for input and positive controls. The upper panel in Figure 3A shows the detection result for the 8WG16 antibody, which recognizes the unphosphorylated CTD of RPO21 (25). Unphosphorylated RPO21 was detected in the crude extract but not in the TAP-SET2 purified eluates, even though Ponceau staining of the membrane (Fig. 3A) indicated blotted RPO21 in the TAP-SET2 lane. In contrast, phosphorylated CTD serine 5 was detected strongly by the CTD4H8 antibody in the TAP-SET2 lane (Fig. 3A). As expected, no evidence for RNAP II association with SIF2-TAP was observed.
Figure 3.
SET2 associates with the elongating, CTD phosphorylated, RPO21. (A) Total cell extracts from the N-terminal tagged TAP-SET2 or SIF2-TAP strains were either directly used (input) or affinity purified (purified) before electrophoresis and blotting. The blots were probed with antibodies detecting unphosphorylated (8WG16, Covance) and serine 5 phosphorylated (CTD4H8, Upstate) RPO21. Ponceau staining of the same blots showing the RPO21 bands on both membranes. (B) As for (A) except that C-terminal tagged SET2 was used from either a wild-type background or a Δssn3 background as indicated. Also, parallel blots were probed with antibodies against RPO21, whether phosphorylated or not, (ARNA3a, Research Diagnostics), serine 2 phosphorylated (H14, Covance) and serine 5 phosphorylated (H5, Covance) RPO21.
The same results for RPO21 phosphorylation status were obtained with C-terminal SET2-TAP purification (Fig. 3B, upper two panels). Additionally, three more RPO21-specific antibodies were employed (Fig. 3B). As expected, ARNA3a, which recognizes RPO21 regardless of its phosphorylation state, identified RPO21 in the SET2-TAP but not SIF2-TAP purification. Specific association of CTD serine phosphorylated RNAP II with SET2 was also observed with two different antibodies, one recognizing phosphorylated serine 2 (H5) and the other phosphorylated serine 5 (H14; 26).
SET2 remains associated with CTD phosphorylated RPO21 in the absence of SSN3
Although RNAP II is phosphorylated at serines 5 and 2 of the CTD during transcriptional elongation, the phosphorylation of the CTD tail can also reflect repression by the SSN3/SSN8 (also known as SRB10/11 or UME5/3) cyclin-dependent kinase prior to transcriptional engagement (21,27). Hence the association of SET2 with CTD phosphorylated RPO21 does not prove association with the elongating form of RNAP II since it could be either the CTD phosphorylated elongating RNAP II, the SSN3 repressed RNAP II or both.
To decide this point, ssn3 was deleted in the SET2-TAP strain and the association with RPO21 examined by western blotting after SET2-TAP purification. Again, RPO21 phosphorylated at serine 5 of the CTD was still specifically retrieved (Fig. 3C). This reinforces the conclusion that SET2 associates with the elongating form of RNAP II. We did not retrieve the same amount of RPO21 in this experiment compared to the SET2-TAP purification of Figure 1A. Deletion of ssn3 in this strain background caused the cells to flocculate and the overall recovery of SET2-TAP was impaired. Hence we cannot exclude the possibility that SET2 associates not only with the elongating polymerase but also with the SSN3 phosphorylated, non-elongating, polymerase.
Evidence for a functional relationship between SET2 and SSN3
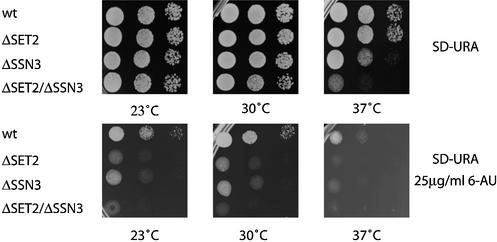
Further examination of the SET2/SSN3 issue unexpectedly uncovered evidence for a functional interaction. Whereas deletion of set2 had no effect on growth at any temperature tested, impaired growth of the Δssn3 strain at 37°C was enhanced by additional deletion of set2 (Fig. 4, top panels). Sensitivity to 6-AU is a common test for mutations that selectively impair elongation (28). As seen in Figure 4, lower panels, deletions of either set2 or ssn3 caused enhanced sensitivity to 6-AU. These data suggest not only a functional interaction between SSN3 and SET2 but also roles for both SET2 and SSN3 in elongation. Alternatively, it may be that the 6-AU test is not a precise indicator for elongation functions.
Figure 4.
Sensitivities of strains carrying deletions of set2 and ssn3 to heat and 6-AU. Yeast strains carrying deletions of set2, ssn3 or neither were spotted onto minimal media plates with or without 6-AU and cultured at three temperatures, as indicated.
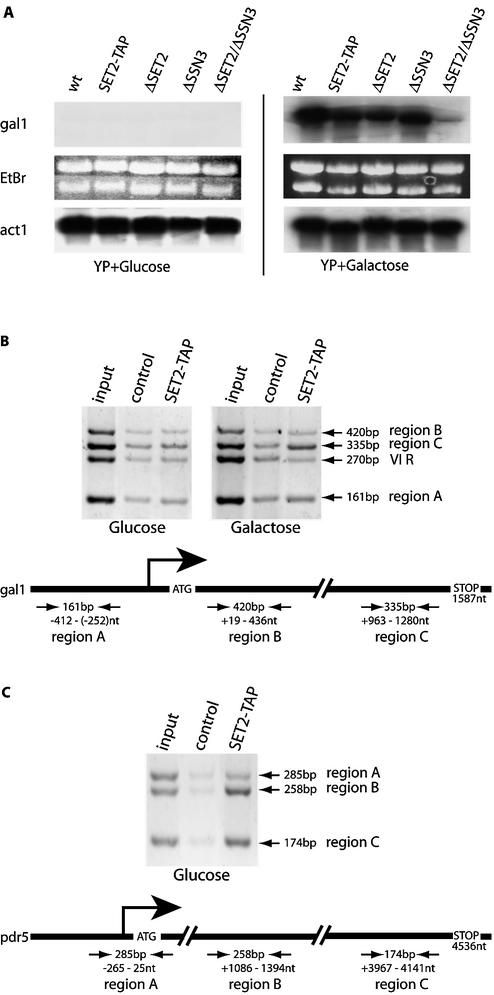
The lack of set2 showed no obvious phenotype in this yeast strain (Fig. 4; data not shown). To look more closely for a Δset2 phenotype, we examined a panel of genes for perturbations of mRNA levels compared to wild-type expression levels. Most mRNA levels were unaffected; however, a mild, ∼2-fold, reduction in gal1 mRNA levels after galactose induction was observed (Fig. 5A). Interestingly, evidence for regulation of gal1 by SSN3 has been published (29). Consequently we examined the combined effect of deletion of both set2 and ssn3 on gal1 mRNA levels to find that galactose-induced expression was abrogated (Fig. 5A).
Figure 5.
SET2 is a transcriptional elongation factor. (A) Northern blots of total RNA prepared from wt, SET2-TAP, Δset2, Δssn3 or Δset2/Δssn3 strains, grown in glucose- or galactose-containing media as indicated, were hybridized with probes from the gal1 or act1 genes. In the center, the ethidium bromide stained gel for the gal1 blot is shown. (B) ChIPs were performed with extracts, prepared according to Hecht and Grunstein (20), using rabbit IgG agarose beads to immunoprecipitate SET2-TAP or empty agarose beads (control). Input represents signals from the total extract without immunoprecipitation. Three primer pairs to amplify regions of the gal1 gene are depicted below on a diagram of the gene, with the sizes of the amplification products indicated. The positions of the amplified regions with respect to the position of the initiating methionine are also indicated. Primer pairs for a telomeric region of chromosome VI (VI R) were included. (C) As for (B) except that the pdr5 gene was investigated.
Physical association of SET2 with two transcribed genes
The association of SET2 with the elongating polymerase and its role in gal1 mRNA expression suggested that it may physically participate in transcriptional elongation of this gene. We examined this possibility by ChIP using the TAP tag of SET2-TAP. Immunoprecipitated DNA was subject to multiplex PCR using primers to regions of the gal1 gene as shown in Figure 5B. To ensure that the ChIP PCR signals lay within the linear range, input DNA was varied across a 30-fold range and only 25 cycles were employed. SET2-TAP was found in the 3′ half of the gene after, but not before, galactose induction of transcription (Fig. 5B). After quantification and analysis according to the equation given in Materials and Methods, which accounts for background and control signals, the association of SET2-TAP with this 3′ section of the active gal1 gene was increased >10-fold. In contrast, no other band in the SET2-TAP lanes varied >1.6-fold. Hence no evidence for SET2 association with upstream or 5′ transcribed regions, before or after galactose induction, could be detected (Fig. 5B). The association of SET2 with transcriptional elongation, and potentially with the later stages of transcriptional elongation, was further examined on the constitutively active pdr5 gene. This gene was chosen because it is amongst the longest transcribed regions in S.cerevisiae, being 4536 bp long. No SET2-TAP at the promoter region was evident; however, association with both the center and 3′ ends of the gene was enhanced 4-fold over control (Fig. 5C). In contrast, we could not find SET2 on the constitutively active act1 gene (data not shown). Both act1 and pdr5 are transcribed at high rates. Holstege et al. (30) reported that act1 mRNA has a half life of 17 min and a production rate of 45.5 per hour and pdr5 mRNA has a half life of 14 min and a production rate of 30.6 per hour. The absence of detectable SET2 on act1 may indicate specificity of SET2 for certain genes and not others. Alternatively it may relate to the size of the act1 gene (1436 bp, including an intron of 308 bp) or the sensitivity of the chromatin immunoprecipitation method.
DISCUSSION
The association of CTD phosphorylated RPO21 with SET2 was published recently by Li et al. (31). We confirm and extend these observations by showing that (i) SET2 is associated with most of, if not the entire, RNAP II holoenzyme, not just the largest two subunits; (ii) no unphosphorylated RPO21 co-purifies with SET2; (iii) the association remains in the absence of ssn3, hence excluding the possibility that the SET2–RPO21 association was solely an aspect of SSN3 mediated repression; and (iv) SET2 physically associates with two actively transcribed genes. Thus we have demonstrated the conclusion that Li et al. infer, namely that SET2 associates with the elongating form of RNAP II. Furthermore we find that SET2 is required for gal1 expression in the absence of SSN3 and hence conclude that SET2 is functionally involved in transcriptional elongation.
By affinity purification, sizing column estimation and quantitation of molecules per cell, we conclude that there are at least two pools of cellular SET2. Quantitation of the Coomassie staining intensities apparent in Figure 1A indicate that about 1/20th of total SET2 is associated with RNAP II, whilst the remainder appears to be an uncomplexed homodimer. Notably, Li et al. (31) showed that alkaline phosphatase treatment of extracts dissociates SET2 from RPO21, presumably by dephosphorylation of the CTD tail. Hence the stoichiometry of SET2:RPO21 association could be underestimated if phosphatases were active during purification. Whilst this remains possible, the number of SET2 or RNAP II per cell (35 000 or 30 000, respectively; 19), and the fact that SET2 exclusively associates with a fraction of RPO21 that is extensively CTD phophorylated (Fig. 3), indicates that either not all cellular SET2 is associated with RNAP II or that a large number (possibly 10 or more) of SET2 molecules interact with one CTD phosphorylated RPO21. We consider the former possibility significantly more likely, but also note that there is no evidence as to whether SET2 interacts with RPO21 as a monomer, dimer or multimer. Notably we observe association of SET2 with the 3′ end of the active gal1 gene, but not at the 5′ end of the transcribed region. As documented by Cho et al. (24), the CTD tail is first phosphorylated on serine 5 at the start of transcriptional elongation and acquires serine 2 phosphorylation during elongation. Taken together with the in vitro finding of Li et al. (31) that a CTD peptide phosphorylated at serine 2 interacted with SET2, but a serine 5 phosphorylated peptide did not, it seems likely that SET2 associates with a significant fraction of serine 2 phosphorylated, elongating, RNAP II in logarithmically growing haploid yeast and thereby with 3′ regions of at least some actively transcribed genes.
Previous clues as to SET2 function have indicated that it acts as a repressor (13,14). None of the data presented here excludes this possibility since a pool of free SET2 dimer may play such a role. Also our data does not exclude SET2 association with SSN3 repressed RNAP II. Our evidence does, however, permit the conclusion that SET2 facilitates the expression of gal1 mRNA. We can exclude the possibility that SET2 action on gal1 is an artifact based on the absence of SSN3, because it was specifically detected by in vivo crosslinking on the transcribing gene in the presence of SSN3. However, SET2 action in transcriptional elongation of the gal1 gene is redundant or auxiliary to an SSN3-directed activity. Sensitivity of the Δssn3 strain to 6-AU and phenotypic enhancements in the Δssn3/Δset2 strain present some further evidence for the proposition that the role of SET2 in transcriptional elongation is redundant to another activity or activities. This conclusion, together with the observation that SET2 associates with the middle to late stages of transcriptional elongation, is concordant with a role for SET2 to ensure that the elongating polymerase, once it has passed the regulatory check points of promoter loading and clearance, successfully completes transcription.
Since SET2 methylates H3 K36 (13), our data suggest that H3 K36 methylation will be found in actively transcribing genes, with possible increased occurrence towards the 3′ ends of transcribed regions. It will be interesting to examine the occurrence and distribution of H3 K36 methylation on the gal1 gene before and after galactose induction. The position of H3 K36 in the nucleosome is highly relevant to transcriptional elongation. The H3 tail passes between the DNA gyres at the position where DNA wrapping begins and K36 is directly adjacent to the junction of linker to nucleosomal-bound DNA (32). Alterations at this junction could have a substantial impact on the progress of elongating polymerases, both to facilitate or impede elongation. With respect to SET2, several scenarios are plausible. The simplest is that methylation of K36 assists elongation by destabilizing DNA association with nucleosomes at the linker/nucleosome junction. Since the positive charge on lysine remains after methylation, this alteration cannot merely be due to a change of electrostatic interaction but could simply be a steric effect. Possibly methylated K36 can be bound by a factor that destabilizes DNA–histone interaction at the junction or prevents binding of a factor that secures the junction. A variation of this theme involves local unravelling at the junction by SET2 itself to access the K36 substrate site.
Our findings with SSN3 suggest that the model of repression by premature phosphorylation of the CTD (27) may need to be re-examined. Recent evidence with SSN3 shows that its repressive activities can be explained in part by regulation of various transcription factors (33). Possibly SSN3 repression is exerted solely through action on transcription factors and plays a conventional role in CTD phosphorylation for elongation; however, further work is required to evaluate these issues.
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Drechsel for assistance in size exclusion chromatography, and David Stanek, Patrick Cramer, Mathias Treier and Julia Schaft for discussions. K.M.K. was supported by a grant from Boehringer Ingelheim Fonds. K.M.N. was supported by a research project grant (RPG-00-110-01-ME10) from the American Cancer Society.
REFERENCES
- 1.Lachner M. and Jenuwein,T. (2002) The many faces of histone lysine methylation. Curr. Opin. Cell Biol., 14, 286–298. [DOI] [PubMed] [Google Scholar]
- 2.Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- 5.Ringrose L. and Paro,R. (2001) Gene regulation. Cycling silence. Nature, 412, 493–494. [DOI] [PubMed] [Google Scholar]
- 6.Noma K., Allis,C.D. and Grewal,S.I. (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein B.E., Humphrey,E.L., Erlich,R.L., Schneider,R., Bouman,P., Liu,J.S., Kouzarides,T. and Schreiber,S.L. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA, 99, 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos-Rosa H., Schneider,R., Bannister,A.J., Sherriff,J., Bernstein,B.E., Emre,N.C., Schreiber,S.L., Mellor,J. and Kouzarides,T. (2002) Active genes are tri-methylated at K4 of histone H3 Nature, 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 9.Roguev A., Schaft,D., Shevchenko,A., Pijnappel,W.W., Wilm,M., Aasland,R. and Stewart,A.F. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J., 20, 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy P.L., Griesenbeck,J., Kornberg,R.D. and Cleary,M.L. (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl Acad. Sci. USA, 99, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan N.J., Dover,J., Khorrami,S., Greenblatt,J.F., Schneider,J., Johnston,M. and Shilatifard,A. (2002) COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem., 277, 10753–10755. [DOI] [PubMed] [Google Scholar]
- 12.Pijnappel W.W., Schaft,D., Roguev,A., Shevchenko,A., Tekotte,H., Wilm,M., Rigaut,G., Seraphin,B., Aasland,R. and Stewart,A.F. (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev., 15, 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strahl B.D., Grant,P.A., Briggs,S.D., Sun,Z.W., Bone,J.R., Caldwell,J.A., Mollah,S., Cook,R.G., Shabanowitz,J., Hunt,D.F. and Allis,C.D. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol., 22, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutfiyya L., Hesman,T. and Johnston,M. (1995) Analysis of the weak GAL4 promoter. Yeast, 11, S220. [Google Scholar]
- 15.Puig O., Rutz,B., Luukkonen,B.G., Kandels-Lewis,S., Bragado-Nilsson,E. and Seraphin,B. (1998) New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast, 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- 16.Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Seraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A., Schaft,D., Roguev,A., Pijnappel,W.W. and Stewart,A.F. (2002) Deciphering protein complexes and protein interaction networks by tandem affinity purification and mass spectrometry: analytical perspective. Mol. Cell. Proteomics, 1, 204–212. [DOI] [PubMed] [Google Scholar]
- 18.Logie C. and Peterson,C.L. (1999) Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol., 304, 726–741. [DOI] [PubMed] [Google Scholar]
- 19.Borggrefe T., Davis,R., Bareket-Samish,A. and Kornberg,R.D. (2001) Quantitation of the RNA polymerase II transcription machinery in yeast. J. Biol. Chem., 276, 47150–47153. [DOI] [PubMed] [Google Scholar]
- 20.Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- 21.Cramer P., Bushnell,D.A., Fu,J., Gnatt,A.L., Maier-Davis,B., Thompson,N.E., Burgess,R.R., Edwards,A.M., David,P.R. and Kornberg,R.D. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 22.Orphanides G. and Reinberg,D. (2000) RNA polymerase II elongation through chromatin Nature, 407, 471–475. [DOI] [PubMed] [Google Scholar]
- 23.Kobor M. and Greenblatt,J. (2002) Regulation of transcription elongation by phosphorylation Biochim. Biophys. Acta, 1577, 261. [DOI] [PubMed] [Google Scholar]
- 24.Cho E.J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson N.E., Aronson,D.B. and Burgess,R.R. (1990) Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem., 265, 7069–7077. [PubMed] [Google Scholar]
- 26.Patturajan M., Schulte,R.J., Sefton,B.M., Berezney,R., Vincent,M., Bensaude,O., Warren,S.L. and Corden,J.L. (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem., 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- 27.Hengartner C.J., Myer,V.E., Liao,S.M., Wilson,C.J., Koh,S.S. and Young,R.A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell, 2, 43–53. [DOI] [PubMed] [Google Scholar]
- 28.Archambault J., Lacroute,F., Ruet,A. and Friesen,J.D. (1992) Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol., 12, 4142–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchin S., Yeghiayan,P. and Carlson,M. (1995) Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl Acad. Sci. USA, 92, 4006–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 31.Li J., Moazed,D. and Gygi,S.P. (2002) Association of histone methyltransferase Set2 with RNA polymerase II plays a role in transcriptional elongation. J. Biol. Chem., 277, 49383–49388. [DOI] [PubMed] [Google Scholar]
- 32.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 33.Chi Y., Huddleston,M.J., Zhang,X., Young,R.A., Annan,R.S., Carr,S.A. and Deshaies,R.J. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev., 15, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]