Abstract
Using a powerful computer-assisted analysis strategy, a large-scale search of small nucleolar RNA (snoRNA) genes in the recently released draft sequence of the rice genome was carried out. This analysis identified 120 different box C/D snoRNA genes with a total of 346 gene variants, which were predicted to guide 135 2′-O-ribose methylation sites in rice rRNAs. Though not exhaustive, this analysis has revealed that rice has the highest number of known box C/D snoRNAs among eukaryotes. Interestingly, although many snoRNA genes are conserved between rice and Arabidopsis, almost half of the identified snoRNA genes are rice specific, which may highlight further the differences in rRNA methylation patterns between monocotyledons and dicotyledons. In addition to 76 singletons, 70 clusters involving 270 snoRNA genes were also found in rice. The large number of the novel snoRNA polycistrons found in the introns of rice protein-coding genes is in contrast to the one-snoRNA-per-intron organization of vertebrates and yeast, and of Arabidopsis in which only a few intronic snoRNA gene clusters were identified. Furthermore, due to a high degree of gene duplication, rice snoRNA genes are clearly redundant and exhibit great sequence variation among isoforms, allowing generation of new snoRNAs for selection. Thus, the large snoRNA gene family in plants can serve as an excellent model for a rapid and functional evolution.
INTRODUCTION
In eukaryotes, the mature 18S, 5.8S and 25/28S rRNAs of the cytoplasmic ribosomes are produced by processing and modifying precursor rRNA (pre-rRNA) in the nucleolus (1,2). This complex course of rRNA biogenesis involves many small nucleolar RNAs (snoRNAs) enriched in the nucleolus in the form of small nucleolar ribonucleoprotein particles (snoRNPs) (3,4). A small number of the snoRNAs, such as the abundant U3 and U14 and the unique RNase MRP, are required for specific cleavage of pre-rRNA. However, the majority of them are involved in 2′-O-ribose methylation and pseudouridylation of rRNAs (5). All of the snoRNAs characterized to date, with the exception of RNase MRP, fall into two families, box C/D and box H/ACA snoRNAs, which can be distinguished on the basis of common sequence motifs and structural features (6–8). The box C/D snoRNAs display two conserved motifs, the 5′-end C box (5′-UGAUGA-3′) and the 3′-end D box (5′-CUGA-3′), usually flanked by short inverted repeats. Adjacent to the box D or to an additional internal box D′ there is a 10–21 nt sequence complementarity to rRNAs acting as guide for site-specific 2′-O-ribose methylation of rRNAs. The target is invariably located 5 nt upstream of the D or D′ box (9,10). In addition to the H box (ANANNA) in the hinge region and an ACA motif 3 nt from the 3′-end of the molecule, box H/ACA snoRNAs are characterized by a common hairpin–hinge–hairpin–tail secondary structure which is essential for guiding pseudouridylation of rRNAs (11,12). These two snoRNA families now appear much larger than anticipated, and recent studies have broadened their substrate repertoire to other cellular RNAs, including snRNAs, tRNAs and possibly even mRNAs, suggesting various other functions of snoRNAs in addition to ribosome biogenesis (13–17).
The genomic organization of snoRNA genes displays a great diversity in different eukaryotes. In vertebrates, almost all snoRNAs are nested within introns of host genes (7), but a few snoRNA genes, such as U3, are transcribed independently. In yeast, in addition to a few intronic snoRNAs, the majority of snoRNAs are single genes with their own promoter; clustered snoRNA genes driven by a promoter upstream have also been reported in a couple of cases (18,19). In plants, although early biochemical data had revealed their highly methylated rRNAs (20,21), only very recently was the first comprehensive study of box C/D snoRNA genes carried out with the Arabidopsis genome (22–24). An experimental RNomics study has identified 140 snmRNAs in Arabidopsis, many of which correspond to box H/ACA snoRNA (25). One of the important results from research on Arabidopsis is the discovery of multiple snoRNA gene clusters and new mode of expression as polycistrons in plants (26,27). The findings also show a large number of modified residues on rRNAs, including sites novel to plants, and sites situated on new expansion domains of the plant rRNAs.
Knowledge of plant snoRNA genes continues to expand, particularly with the progress of the Rice Genome Project. The genome of rice is four times larger and more complex than that of Arabidopsis. In our initial study of the rice genomic DNA sequences in the database, we discovered a new mode of organization, i.e. intronic snoRNA gene clusters in plant (28,29). A more comprehensive study of snoRNA gene content and organization from rice as well as other plants will be important to clarify the plant rRNA methylation profile and to understand the strategy of adopting novel modification sites in plant rRNAs and its biological meaning. In this regard, we took advantage of the recently published draft sequence of the rice genome (Oryza sativa L. ssp. indica) (30,31) to perform a large-scale search for snoRNAs with the powerful computer-assisted analysis strategy. A large number of rice snoRNA genes have been identified and compared with those of other organisms, especially Arabidopsis whose snoRNA gene database is available (32). Detailed analysis has also been performed on gene organization and sequence variation of rice snoRNAs to find clues about the mechanisms of the duplication and functional evolution of snoRNA genes in plants.
MATERIALS AND METHODS
Database search and sequences analysis
The rice (O.sativa L. ssp. indica) genome scaffolds available at http://btn.genomics.org.cn/rice were searched for potential box C/D snoRNAs in the following ways. (i) A eukaryote snoRNA search program was used to identify putative snoRNA genes with box C/D, a terminal stem with at least three base pairings and, in most cases, an rRNA complementary sequence (18,19). (ii) Rice homologues of the known snoRNA genes from A.thaliana and Zea mays were identified. Flanking sequences of the snoRNA candidates were also examined for other possible box C/D snoRNAs. About 1 kb of flanking sequences of the snoRNA candidates was searched further for additional non-canonical C/D candidates. (iii) A BLAST program (33) was used to search gene variants of all novel snoRNA genes to establish the number of snoRNA isoforms. Intronic snoRNA and their host gene were defined according to the annotation of the rice genome and our analyses of the expressed sequence tag (EST) database in GenBank. Sequence alignment of snoRNA isoforms was performed with Clustal X 1.8.
Construction and screening of the rice cDNA library from small nuclear RNAs
Nuclei were isolated from 5 g of rice yellow seedlings as described (34) and the nuclear pellet was used for RNA preparation by guanidine thiocyanate/phenol–chloroform extraction (35). Nuclear RNAs were first polydenylated using poly(A) polymerase (Takara). Synthesis of the first strand of cDNA was performed with 5 µg of poly(A)+-tailed RNA in a 20 µl reaction mix containing 0.5 µg of primer HindIII(T)16 and 200 U of MMLV reverse transcriptase (Promega) for 45 min at 42°C. The reaction mixture was separated on a denaturing 8% polyacrylamide gel (8 M urea, 1× TBE buffer). cDNAs with sizes ranging from 50 to 300 nt were excised and eluted from the gel. The cDNAs were tailed with poly(dG) at the 3′-end by using terminal deoxynucleotidyl transferase (Takara), and then amplified by PCR with primers HindIII(T)16 and BamHI(C)16, and cloned into plasmid pTZ18 as described previously (36). The cDNA library was screened by PCR with the P47 and P48 universal primer pair. Only the recombinant plasmids carrying fragments longer than 100 bp were selected for sequencing, which was performed with an automatic DNA sequencer (Applied Biosystems, 377) using the Big Dye Deoxy Terminator cycle-sequencing kit (Applied Biosystems).
Mapping of ribose methylations by reverse transcription
Ribose-methylated nucleotides of rice rRNA were determined by primer extension at low dNTP concentrations as described previously (29). In brief, two reverse transcription reactions were performed in parallel using 15 µg of total RNA from 0.5 cm rice seedlings and a dNTP concentration of either 4 µM or 1.5 mM. Each extension product was run alongside a sequencing ladder prepared with the same specific primer and a plasmid carrying the corresponding rice rDNA coding sequence as template.
Oligonucleotides
Oligonucleotides were synthesized and purified by Sangon Co. (Shanghai, China). The following oligonucleotides were used in the construction and screening of the rice cDNA library: HindIII(T)16, 5′-CCCCAAAGCTTTTTTTTTTTT TTTT-3′; and BamHI(C)16, 5′-GGAATTCGGATC CCCCC CCCCCCCCCCCC-3′. Rice rRNA ribose-methylated nucleotides were assayed by reverse transcription with the following primers: P18S28, 5′-GCCATTCGCAGTTTCACAGT-3′; P18S162, 5′-GCGTCAGCCTTTTATCTA-3′; P18S282, 5′-CGAAAGTTGATAGGGCAGAA-3′; P18S368, 5′-CTGCT GCCTTCCTTGGATGT-3′; P18S454, 5′-ATTACCAGACA CTAAAGCG-3′; P18S582, 5′-CC CAAGGTCCAACTACG AGC-3′; P18S647, 5′-AGGTCGGTGCCTGCCGTGAG-3′; P18S682, 5′- GAGCACTCTAATTTCTTCAA-3′; P18S809, 5′-GCCAACACAATAGGACCG-3′; 18S1060, 5′-TTGATT TCTCATAAGGTGC-3′; P18S1156, 5′-TGAGTCAAATTA AGCCGCAG-3′; P18S1272, 5′-AACCAGACAAATCGC TCCAC-3′; P18S1466, 5′-AATTTCCCAAGATTACCCG-3; P18S1576, 5′-GTACAAAGGGCAGGGACG-3′; P18S1776, 5′-CAATGATCCTTCCGCAG-3′; P18N3R, 5′-GTAATTT GCGCGCCTGCTGC-3′; 18N7R, 5′-TCCGTCAATTC CTTTAAGTT-3′; 18N9R, 5′-CTAAGGGCATCACAGAC CTG-3′; 18N10R, 5′-TAGCGACGGGCGGTGTGTAC-3′; and 18N11R, 5′-TGATCCTTCCGCAGGTTCACC-3′. The primers used in reverse transcription of rRNA and rDNA sequencing were 5′-end labeled with [γ-32P]ATP (Yahui Co.) and submitted to purification according to standard laboratory protocols as described previously (37).
RESULTS
Identification of 120 box C/D snoRNA genes from O.sativa
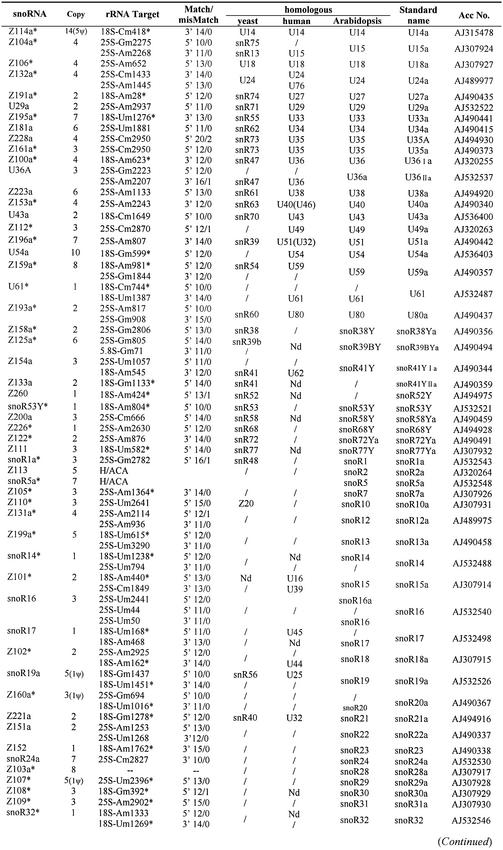
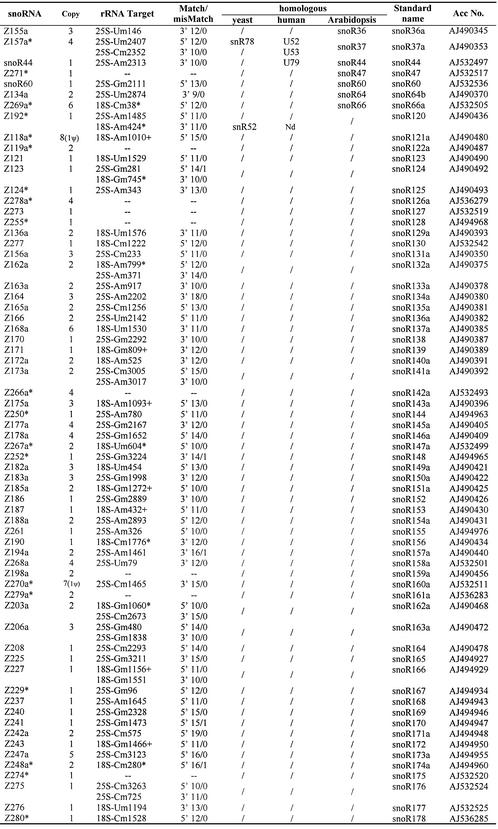
To search for box C/D snoRNAs in the rice genome, we used the eukaryotic snoRNA search program that previously has been proved to be effective in dealing with the yeast and plant genomic DNA sequences (18,22,29). The program was improved by information on Arabidopsis snoRNAs and its rRNA methylation pattern. Then it was used to scrutinize the high quality rice (O.sativa L. ssp. indica) genome scaffolds (i.e. non-overlapping contigs linked together in the correct order and orientation) (30). The program searched for candidates that display, within segments of at most 200 nt, box C (RUGAUGA), box D (CUGA), an rRNA complementarity of at least 10 nt, the usually imperfect box D′ if the rRNA complementarity is not directly adjacent to box D, and terminal short inverted repeats. Among all these elements, we considered primarily the quality of the potential snoRNA–rRNA duplexes (in terms of length of the duplex, number and position of mismatches and of UG pairings). Once a novel snoRNA candidate was found, its flanking sequences were also examined for other possible box C/D snoRNAs, and a BLAST search was performed to find gene variants and clarify the copy number of isoforms. Taken together with our previous results (29), a total of 346 putative snoRNA gene variants representing 118 different box C/D snoRNA genes and two box H/ACA type, snoR2 and snoR5, have been identified from rice (Table 1).
Table 1. Novel rice (O.sativa L. ssp. indica) snoRNA genes.
Only one copy of each snoRNA species is listed, and the number of all snoRNA isoforms is given. snoRNA genes marked with an asterisk are identified experimentally from the cDNA library, and rRNA targets with an asterisk correspond to sites mapped in reverse transcription analysis [this study and Liang et al. (29)]. rRNA targets marked with a small cross correspond to the sites which gave a negative signal in the reverse transcription assay. Match/mismatch indicates the quality of the rRNA antisense element in snoRNA genes. System names are according to the plant snoRNA nomenclature proposed by Brown et al. (24,32). Nd, methylated site but no snoRNA identified; /, non-methylated site and no known snoRNA.
Judging from the sequence complementarity to rRNA and the corresponding 2′-O-methylation target sites (9,10), 58 of the 120 snoRNA genes have homologues in other organisms (see Table 1). These include 15 snoRNA genes well conserved among plants, yeast and human, and 55 between rice and Arabidopsis. These 58 snoRNA genes with one or two rRNA complementarities are predicted to guide 71 2′-O-ribose methylation sites. To our surprise, the remaining 50 snoRNA genes did not have any homologue even in Arabidopsis, and are probably rice specific. They were predicted to guide 64 2′-O-ribose methylation sites. Consistent with all other eukaryotes, the majority of the methylation sites are clustered on the phylogenetically conserved regions of rice rRNAs. A dozen methylation sites are also scattered on the rRNA expansion domains, as demonstrated in Arabidopsis (23). In addition, 10 rice box C/D snoRNAs with no target site in rRNAs, including two homologues to Arabidopsis, were identified mainly through analyses of flanking sequences of guide snoRNAs and a cDNA library screen (see below). Their function in the nucleus remains to be elucidated.
According to the plant snoRNA nomenclature proposed by Brown et al. (24,32), the rice snoRNA genes (initially named as Z100, etc.) have been given standard names to provide a consistent identity with yeast and human orthologues. In principle, when the complementary region(s) of an identified gene corresponded to a vertebrate or vertebrate/yeast snoRNA, the rice snoRNA was given the vertebrate name (e.g. U14). If the complementary sequence corresponded only to a yeast snoRNA, then it was given the yeast number followed by ‘Y’ (e.g. snoR38Y). Novel rice genes were named in succession with the serial number of the Arabidopsis genes (see Table 1). In a few cases, new terms were added to indicate some structural characteristics. For example, when two rice snoRNA genes each had one of the two antisense elements of a yeast or human gene, they were named after the yeast or human gene but with a different suffix, I or II, for similar to the upstream or the downstream antisense element, respectively (e.g. U36I and U36II, snoR41Y I and snoR41Y II).
Experimental detection of rice snoRNAs and mapping of rRNA methylation
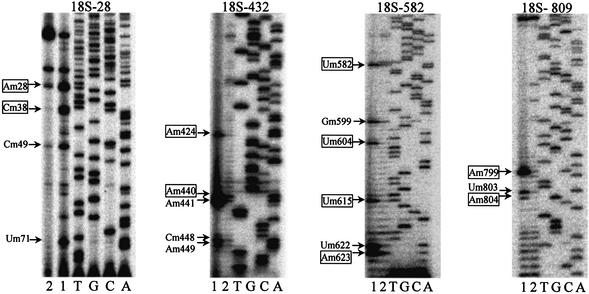
The expression of the rice snoRNA genes was investigated by screening the cDNA library from small nuclear RNAs. Fifty-three snoRNA-related sequences have been obtained, and 20 typical sequences are shown in Table 2. The sequences obtained are identical to those of their genes, and most of them correspond to the snoRNA genes with multiple copies and are encoded by snoRNA gene clusters (see Table 3). Among the 53 snoRNA-related sequences, five do not correspond to a predicted snoRNA. These include snoRNAs devoid of rRNA complementarity, such as snoR127, snoR128, snoR142, snoR161 and snoR163, which are not linked with any standard snoRNA guide. The experimentally confirmed snoRNA genes are indicated in Table 1 by asterisks. Meanwhile, primer extension analyses were also applied to mapping rice rRNA methylation sites predicted by the snoRNA genes. Since most of the 2′-O-ribose methylation sites in rice 25S rRNA are being determined (38), a range of primers was designed for a systematic mapping of rice 18S rRNA. Primer extension analyses were carried out with total RNA from rice seedlings, and 2′-O-methylation sites in rRNA were revealed by pauses of reverse transcription at a lower concentration of dNTPs (39). Altogether, 29 ribose methylation sites, which were predicted by the snoRNA genes, have been mapped in rice 18S rRNA and are indicated in Table 1. Examples of mapping gels are shown in Figure 1. Because of the limitation of this technique, 15 predicted sites could not be detected in the primer extensions, mainly due to the effects of strong secondary structures of rRNAs hampering the detection of low dNTP-dependent reverse transcription pauses. It is worth noting that seven predicted rRNA methylation sites were analysed and gave a negative signal in the reverse transcription assay, which could reflect a distinct function of the cognate snoRNA, possibly as an RNA chaperone for pre-rRNA folding or an RNA methylation guide expressed differentially. The primer extension analyses also revealed some new methylation sites in rice rRNAs, yet the corresponding snoRNA genes have not been found (Fig. 1 and our unpublished results).
Table 2. Examples of box C/D snoRNAs identified from the cDNA library of small nuclear RNAs.
Boxes C, D and D′ are underlined, and the antisense elements are shaded.
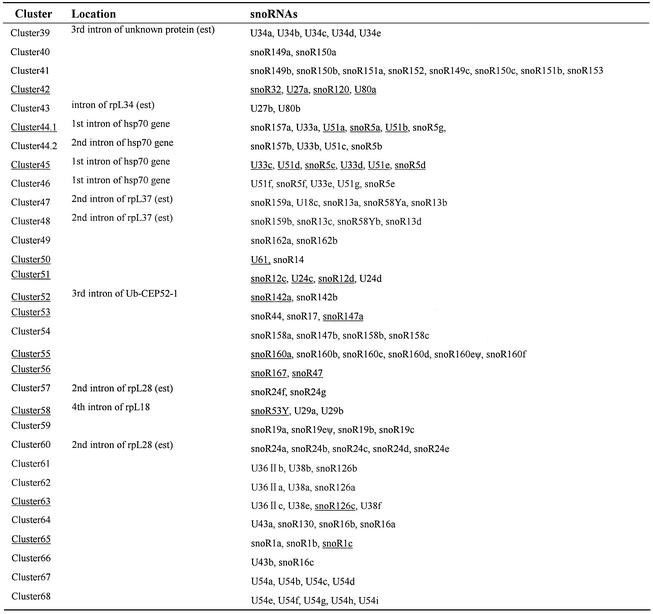
Table 3. Location and constitution of the rice (O.sativa L. ssp. indica) snoRNA gene clusters.
Intronic clusters are denoted according to the annotation of the rice genome draft and our analyses of rice EST databases. snoRNAs underlined are identified experimentally. The clusters containing the snoRNA(s) identified experimentally are also underlined.
Figure 1.
Mapping of 18S rRNA 2′-O-ribose methylation sites. Lane 1, primer extension at 4 µM dNTP; lane 2, control reaction at 1.5 mM dNTP; lanes A, C, G and T, the rDNA sequence ladder. The sites of ribose methylation in rRNA were revealed by reverse transcription pauses at low dNTP concentrations. Arrows indicate potential methylation sites, and boxed sites are predicted by the novel snoRNAs of this study.
Genomic organization of rice snoRNA genes
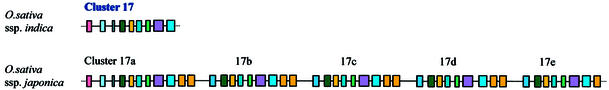
Gene cluster is the main genomic organization of snoRNA gene in rice. Remarkably, in addition to 76 singletons, 270 snoRNA genes are organized into 70 clusters (Table 3). Although most clusters are composed of 2–5 homologous or heterologous snoRNA genes, some complex clusters are observed (such as cluster 15 that contains 12 box C/D snoRNA genes and two box H/ACA genes). Larger and more complex snoRNA gene clusters may also exist in the rice genome. For instance, cluster 17 was made up of nine different snoRNA genes and found at the 3′-end of a scaffold in the genome draft of O.sativa L. ssp. indica which appeared incomplete. We examined a homologous genomic clone (accession no. AY013245) from another rice subspecies, O.sativa L. ssp. japonica, and, to our surprise, found a huge cluster composed of 42 snoRNA genes, which corresponds to five tandem duplications of cluster 17 in the genome draft of O.sativa L. ssp. indica (Fig. 2). This finding may indicate that the japonica genome is larger than that of indica because of an expansion by insertion of transposable elements in some chromosomes (40).
Figure 2.
Five tandem duplications of cluster 17 in rice subspecies O.sativa L. ssp. japonica. snoRNA genes are represented by different coloured boxes. In detail: cluster 17, snoR12a–U24a–snoR29eψ–snoR30c–snoR31c–snoR10c–snoR77Yc–U49c–snoR2e; cluster 17a, snoR12a–U24a–snoR29aψ–snoR30a– snoR31a–snoR10a–snoR77Ya–U49c–snoR2a–U14a–U14b; cluster 17b, snoR29b–snoR30b–snoR31b–snoR10b–snoR77Yb–U49b–snoR2b–U14c–U14d; cluster 17c, snoR29c–snoR30c–snoR31c–snoR10c–snoR77Yc–U49c–snoR2c–U14e–U14f; cluster 17d, snoR29d–snoR30d–snoR31d–snoR10d–snoR77Yd– U49d–snoR2d–U14g–U14h; cluster 17e, snoR29e–snoR30e–snoR31e–snoR10e–snoR77Ye–U49e–snoR2e–U14i.
In our previous study, we described five intronic snoRNA gene clusters in rice (29). In the current study, we have demonstrated further that 25 of 65 snoRNA gene clusters are intronic (see Table 3). It is possible that the number of intronic snoRNA gene cluster will increase after complete annotation of all genes in the rice genome. Multiple snoRNA gene clusters were found in the introns of protein-coding genes, most of which are involved in nucleolar organization or ribosomal biogenesis, such as ribosomal proteins rpS9, rpL13a, rpL18, rpS20, rpL23A, rpL28, rpL30, rpL34, rpL37 and rpL40. Varied host genes, such as those for Hsp70, NADH dehydrogenase, small nuclear nucleoprotein G, receptor-like protein kinase and, particularly, a hypothetical protein-coding gene with a small open reading frame (ORF) were also found. The novel intronic snoRNA polycistrons found in preponderance in rice protein-coding genes are characteristic of the genomic organization of the rice snoRNA gene.
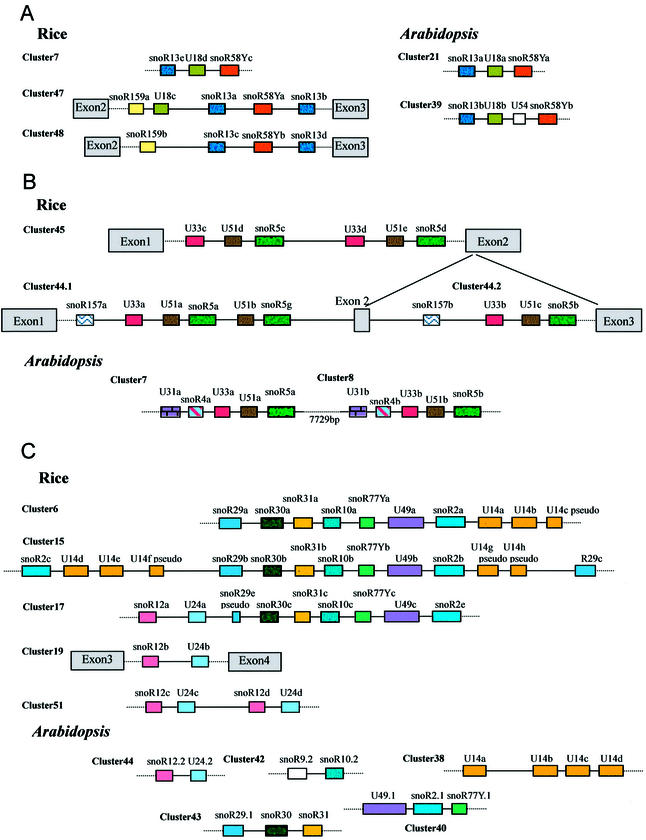
Notably, the rice clusters, intronic or non-intronic, are made mainly of snoRNA genes that are conserved between rice and Arabidopsis. Comparative analyses between the two plants revealed that many of the clusters are related in gene content and gene organization. For example, clusters 3, 39 and 60 are three homogeneous clusters in both rice and Arabidopsis. Many heterogeneous clusters were also related in both plants [see Table 3 and Brown et al. (24)]. Typically, three tightly linked snoRNA genes (R13, U18 and snoR58) in rice cluster 7 have an identical counterpart in cluster 21 of Arabidopsis (Fig. 3A), implying an ancient linkage before the divergence of monocotyledon from dicotyledon. This ancestral cluster has undergone gene duplication and recombination to generate novel clusters, clusters 47 and 48 in rice and cluster 39 in Arabidopsis, respectively. In the novel cluster 39 of Arabidopsis, an additional snoRNA gene, U54, was inserted between U18 and snoR58Y, whereas more extensive changes, including the removal or deletion of U18, have taken place in clusters 47 and 48 of rice.
Figure 3.
(Overleaf) Schematic diagrams of snoRNA gene linkage conserved in rice and Arabidopsis. snoRNA genes are represented schematically by coloured boxes. All are drawn to scale except the exons. (A) snoR13, U18 and snoR58Ya are three linked snoRNA genes in both rice and Arabidopsis. (B) Rice cluster 45 originated from a local duplication of three linked snoRNA genes (U33–U51–snoR5) in the first intron of an hsp70 gene. Rice clusters 44.1 and 44.2 both contain a U33–U51–snoR5 linkage and are nested in the two introns of another hsp70 gene. In Arabidopsis, two non-intronic clusters, clusters 7 and 8, contain the U33–U51–snoR5 linkage with U31 and snoR4. (C) Rice clusters 6, 15 and 17 have a core structure of seven snoRNA genes (snoR29–snoR30–snoR31–snoR10–snoR77Y–U49–snoR2). Related small clusters in rice and Arabidopsis are also shown.
Many intronic snoRNA clusters (clusters 1, 3, 4, 21, 22, 39, 52, 57 and 60) consist of duplicated copies of the same snoRNA. However, cluster 45, first identified in the intron of rice hsp70 gene (28), is composed of four box C/D snoRNA genes and two box H/ACA snoRNA genes, which evidently originated from a local duplication of the three linked snoRNA genes (U33–U51–snoR5). The rice hsp70 gene also has multiple copies and, in this study, three intronic snoRNA gene clusters were found in two other rice hsp70 genes (Fig. 4B). All these contain the three linked snoRNA genes (U33–U51–snoR5) with one or more novel snoRNA gene around. Interestingly, two non-intronic clusters containing the set of U33–U51–snoR5 genes were also found in Arabidopsis [Fig. 3B and Brown et al. (24)]. These observations showed strong conservation of the linkage of some snoRNA genes during the course of plant evolution. This point was supported further by the analyses of large clusters in rice, such as clusters 6, 15 and 17 (Fig. 3C). In these clusters, seven closely linked genes (snoR29–snoR30–snoR31–snoR10–snoR77Y– U49–snoR2) make up a common core structure with the addition of other snoRNA genes up- or downstream. Interestingly, the large rice clusters seemed to have formed by linkage of multiple small clusters that were found in rice and Arabidopsis (Fig. 3C).
Figure 4.
Sequence alignment of snoRNA isoforms. (A) Alignment of U36I isoforms showing sequence variations. Structural elements are boxed and the identical element is shaded. (B) Alignment of snoR39BY isoforms. The upstream antisense element (UAE) identical among all snoR39BY isoforms is shaded; differences in their downstream antisense element (DAE) are indicated in red. The methylation sites predicted by rRNA–snoRNA duplexes are marked with their coordination in rRNAs.
Redundancy and sequence variation of rice snoRNA genes
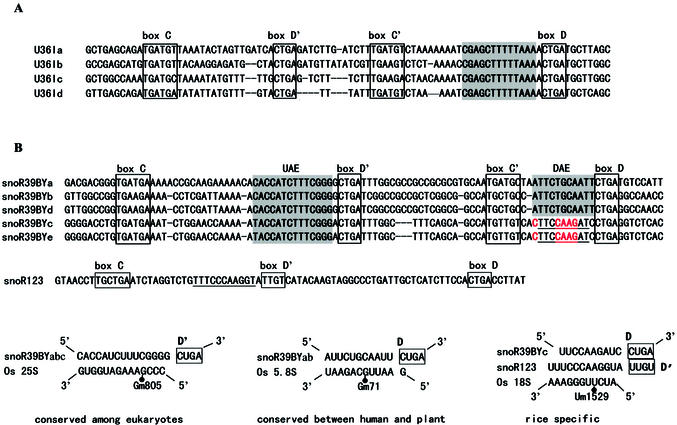
Generally, rice snoRNA genes are clearly redundant. At least two-thirds of the rice snoRNA genes have two or more isoforms. Remarkably, U14, a highly conserved snoRNA among eukaryotes, has 14 copies including five truncated sequences identified as pseudogenes. A rice-specific snoRNA gene, snoR121, exhibits seven isoforms with one pseudogene. Sequence alignments of the rice snoRNA isoforms revealed numerous sequence changes, including small insertions or deletions, which occurred frequently in the less important regions, and occasionally in the conserved elements such as the complementary sequences, box C, D, C′, D′ and the terminal repeats which are required for the stability and function of snoRNAs. Despite showing sequence variation to some extent, the identified snoRNA isoforms possess at least a common rRNA complementary sequence targeting methylation of the same residue (Fig. 4A).
The accumulation of mutations in snoRNA isoforms would lead to partial alteration of snoRNA’s function in loss or gain of an rRNA complementary sequence. As shown in Figure 4B, rice snoR39BY possesses two rRNA complementary sequences and exhibits five isoforms. The isoforms have in common the upstream antisense element targeting methylation of rice 25S rRNA at Gm805. However, substitution of four nucleotides in the downstream antisense element of snoR39BYc and e resulted in loss of the guide sequence for methylation of rice 5.8S rRNA at Gm71. The mutations may generate a new complementary sequence, which was a homologue to snoR123, with the potential to target methylation of rice 18S rRNA at Um1529. Similar situations were also observed in many other cases such as snoR137, U33 and snoR41YI isoforms (data not shown).
Sequence variation of snoRNA genes, in some cases, might be coordinative to the mutation or polymorphism in rRNA sequence in order to preserve a phylogenetically conserved methylation site in diverse organisms. As shown in Figure 5, when compared with yeast, two mutations adjacent to the methylated site Am545 of rice 18S rRNA have taken place. Correlated mutations have also occurred in the rRNA complementary sequence of rice snoR41YI, which could compensate for the change in rRNA and maintain a perfect rRNA–snoRNA duplex for guiding rRNA methylation at the same site as that of yeast. A similar example was observed for rice snoR68Y where a compensatory mutation occurred in the 25S rRNA–snoRNA duplex (Fig. 5).
Figure 5.
Compensatory mutations in the rRNA–snoRNA duplex to maintain the conserved rRNA methylation sites in rice and yeast. Compensatory mutations in rRNA–snoRNA duplexes are indicated by arrows.
DISCUSSION
Rice has the highest content of box C/D snoRNAs among known eukaryotes
We took advantage of the recently released draft sequence of the rice genome (O.sativa L. ssp. indica) to identify systematically box C/D snoRNA genes by using computer-assisted searches (19,41,42). Our search identified 120 different box C/D snoRNA genes with a total of 346 gene variants from rice. These genes were predicted to guide 135 methylation sites in rRNA, agreeing well with early biochemical data, which had revealed more than 120 2′-O-ribose-methylated residues in higher plants (20,21). A computational screen of the yeast genome has revealed 41 snoRNA genes responsible for 51 sites among 55 rRNA methylated modifications (19). In human, about 107 2′-O-methylated residues were characterized (2). Recently the analysis of the Arabidopsis genome identified 97 different box C/D snoRNAs with a total of 175 different gene variants, predicting 118 rRNA methylated sites (27). As our search for box C/D snoRNA was performed under stringent criteria, some rice snoRNAs might remain hidden in the rice genome. Though not exhaustive, our search has already revealed that rice has the highest number of known snoRNA species among all organisms.
Comparative analyses showed that many snoRNA genes are conserved among rice and other eukaryotes, and especially between rice and Arabidopsis. However, almost half of the identified snoRNA genes appear to be rice specific. This result demonstrates the high diversity of snoRNAs in the two flowering plants despite the high similarity in their rRNA sequences. It also implies a more complicated rRNA methylation pattern in plants than had been previously thought.
It has been shown that all sites of rRNA ribose methylation are specified through the same snoRNA guide process (19,43). However, the rRNA methylation pattern in diverse organisms and the mechanism(s) that produce it remain to be described. Higher plants provide a good system for us to study rRNA modification, not only because of the richness in rRNA modification, but also because of a highly divergent modification pattern in plants, as indicated in this study. The diversity of snoRNA genes and the rRNA methylation pattern in various plants should be investigated for further understanding of the functions of rRNA methylation and the mechanisms of evolution with which plants adopt novel modification sites in their rRNAs.
Diversity of snoRNA gene organization in plants
Recently, characterization of multiple snoRNA gene clusters in Arabidopsis has shown that the polycistron is a predominant gene organization of plant snoRNA (22–24). Compared with Arabidopsis, there are larger and more complex clusters in rice due to a higher content of snoRNA genes. It has been shown that duplication and transposition are still active in rice (30), which might continue to multiply snoRNA genes and complicate their organization in the rice genome. The huge snoRNA gene cluster 17 in O.sativa L. ssp. japonica is the result of this tendency.
Remarkably, the outstanding feature of rice snoRNA gene organization is the prevalence of intronic snoRNA clusters in the rice genome, which was identified initially from rice hsp70 and four other protein-coding genes (28,29). The intronic gene clusters imply a novel gene organization and gene expression, i.e. the transcription of snoRNAs as a polycistron in an mRNA precursor of the host gene and probably a splicing-independent processing pathway involving endonucleolytic cleavage as well as exonucleolytic trimming to release all individual snoRNAs (26). This novel polycistronic organization is clearly different from intronic snoRNA genes found in both yeast and vertebrates, where the mode of one snoRNA per intron is strictly maintained when a host gene encodes numerous snoRNAs (44). To our knowledge, the intronic snoRNA gene cluster as a novel polycistronic organization is unique to plants and not found in any other organism. It is worth noting that although the intronic snoRNA polycistron is now a characteristic of plants, there are only a few intronic snoRNA clusters in Arabidopsis (27,29). This probably reflects the small and compact genome size (∼120 Mb) of Arabidopsis (45), where most introns are too small (∼170 bp on average) to accommodate clustering snoRNAs. It is therefore likely that the prevalence of the intronic snoRNA gene cluster would be related to the large genome in monocotyledonous plants, such as rice and other cereals.
All snoRNA guides for rRNA methylation are intron encoded in vertebrates, emphasising the expression coordination between host genes of snoRNAs and rRNA genes, and the absolute linkage of splicing with rRNA modification. However, plant methylation guide snoRNAs display two distinct genomic organizations, intronic and non-intronic polycistron, implying two different mechanisms of snoRNA expression. In particular, a splicing-independent processing pathway was suggested in regulation of the production of snoRNAs under varying extreme environmental conditions which plants generally have to tolerate, and extreme conditions when splicing may be shut down (27).
Sequence variation of snoRNA isoforms and functional evolution
Overall genomic analyses of rice demonstrate that plants have the highest number of known snoRNA genes among eukaryotes. This result is consistent with the highly repeated genome of monocotyledonous plants. In rice, ∼42% of the genome was in exact 20 nt oligmer repeats, and at least 24% of the rice genome was identifiably of transposon origin (30). Frequent genetic rearrangements, including unequal crossover, gene conversion and duplication, during polyploidisation are largely responsible for the multiplication and redundancy of snoRNA genes in plants. A large number of snoRNA isoforms generated by duplication in rice display large sequence variation. In many cases, the function remains unchanged despite the extensive sequence divergence among snoRNA isoforms, implying a selection constraint for fixation of a snoRNA gene. However, the high diversity of snoRNA genes in plants demonstrates that functional evolution is in action. Accumulation of mutations in the rRNA complementary region may alter the function of rice snoRNA isoforms. New snoRNA isoforms with novel functions have appeared through duplication and mutation, as reported in Arabidopsis (23,24). In fact, functional evolution needs only a few changes in a snoRNA gene. A single nucleotide deletion or insertion in rRNA complementary sequence will create a non-functional gene or a new one, as in the case of Arabidopsis Z45 variants (22) and some rice snoRNA isoforms (data not shown). It is worth noting that the duplicated snoRNA genes in rice may have the same function but could well have a different or specific expression pattern. At present, few plant snoRNA gene promoters and differential expression have been investigated.
It is well known that the compensatory mutation occurring in rRNA sequences is largely responsible for the conservation of the secondary structure of rRNA among all organisms (46). Remarkably, the compensatory mutations also take place between rRNA and snoRNA to maintain rRNA methylations at phylogenetically conserved sites. This shows that functional evolution in coordination has occurred between snoRNAs and rRNAs, since the two types of RNAs are highly related in post-transcriptional modification.
The box C/D snoRNA gene is a large non-coding RNA family possessing a bi-modular structure with two functionally independent but structurally related halves (7,8). Our analyses of rice snoRNA isoforms indicate that the two functional regions vary in rhythm, i.e. in many cases, one of the two rRNA antisense elements in snoRNA genes is extremely conserved while the other exhibits a high degree of variation. This results in having more snoRNA genes with a single rRNA antisense element. Due to their structural characteristics, and being free from the constraint of an ORF in protein genes, multiple box C/D snoRNA gene isoforms in plants exhibit a large sequence variation that could, under selection, lead to an ever increasing diversity of plant snoRNA genes. Thus plant snoRNA genes can serve as an excellent model for rapid and functional gene evolution.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the technical assistance of Xiao-Hong Chen and Zhang-Peng Huang. We also thank Dr Alan Yen for helpful discussion, and Professor Mohssen Ghadessy for revising the text of the manuscript. This research was supported by the National Natural Science Foundation of China (key project 30230200) and by the Fund for Distinguished Young Scholars from the Ministry of Education of China.
REFERENCES
- 1.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 2.Maden B.E.H. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–301. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 35, 897–934. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W. and Pogaèiæ,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 5.Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 6.Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small nucleolar RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 7.Bachellerie J.P., Cavaille,J. and Qu,L.H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNA guides revisited. In Garrett,R.A., Douthwaite,S., Liljas,A., Matheson,A., Moore,P.B. and Noller,H. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203.
- 8.Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 10.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation rRNA. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 11.Ganot P., Bortolin,M.L. and Kiss,T. (1997) Related site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 12.Ni J., Tie,A.L. and Fournier,M.J. (1997) Small nucleoloar RNAs direct site-specific synthesis of pseudouidine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 13.Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by another small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 14.Jady B. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiss T. (2002) Small nucleolar RNAs: an abundant group of non-coding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 17.Bachellerie J.P., Cavaille,J. and Hüttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. [DOI] [PubMed] [Google Scholar]
- 18.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-ferrer,M. and Bachellerie,J.P. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 20.Lau R.Y., Kennedy,T.D. and Lane,B.G. (1974) Wheat embryo ribonucleates: III. Modified nucleotide constituents in each of the 5.8S, 18S and 26S ribonucleates. Can. J. Biochem., 52, 1110–1123. [DOI] [PubMed] [Google Scholar]
- 21.Cecchini J.P. and Miassod,R. (1979) Studies on the methylation of cytoplasmic ribosomal RNA from cultured higher plant cells. Eur. J. Biochem., 98, 203–214. [DOI] [PubMed] [Google Scholar]
- 22.Qu L.H., Meng,Q., Zhou,H. and Chen,Y.Q. (2001) Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res., 29, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barneche F., Gaspin,C., Guyot,R. and Echeverría,M. (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- 24.Brown J.W., Clark,G.P., Leader,D.J., Simpson,C.G. and Lowe,T. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 25.Marker C. Zemann,A., Terhorst,T., Kiefmann,M., Kastenmayer,J.P., Green,P., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol., 12, 2002–2013. [DOI] [PubMed] [Google Scholar]
- 26.Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1997) Clusters of multiple different small nucleolar RNA in plants are expressed as and processed from polycistronic pre-snoRNA. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown J.W., Echeverria,M. and Qu,L.H. (2003) Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci., 8, 42–49. [DOI] [PubMed] [Google Scholar]
- 28.Qu L.H., Zhong,L., Shi,S.H., Lu,Y.J., Fang,R. and Wang,Q. (1997) Two snoRNAs are encoded in the first intron of the rice hsp70 gene. Prog. Nat. Sci., 7, 371–377. [Google Scholar]
- 29.Liang D., Zhou,H., Zhang,P., Chen,Y.Q., Chen,X., Chen,C.L. and Qu,L.H. (2002) A novel gene organization: intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res., 30, 3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 31.Goff S.A., Ricke,D., Lan,T.H., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 32.Brown J.W., Echeverria,M., Qu,L.H., Lowe,T., Bachellerie,J.P., Hüttenhofer,A., Kastenmeyer,J.P., Shaw,P. and Marshall,D. (2003) Plant snoRNA database. Nucleic Acids Res., 31, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 34.Guilfoyle T.J. (1995) Isolation and characterization of plant nucleic. Methods Cell Biol., 50, 101–112. [DOI] [PubMed] [Google Scholar]
- 35.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 732–735. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H., Chen,Y.Q., Du,Y.P. and Qu,L.H. (2002) The Schizosaccharomyces pombe mgU6-47 snoRNA is required for the methylation of U6 snRNA at 41. Nucleic Acids Res., 30, 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Li W., Jiang,K., Jin,Y.X. and Wang,D.B. (2003) Detection of rice 25S rRNA 2′-O-ribose methylation sites. Acta Biochem. Biophys. Sin., in press. [PubMed] [Google Scholar]
- 39.Maden B.E.H., Corbett,M.E., Heeney,P.A., Pugh,K. and Ajuh,P.M. (1995) Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie, 77, 22–29. [DOI] [PubMed] [Google Scholar]
- 40.Feng Q., Zhang,Y., Hao,P., Wang,S., Fu,G., Huang,Y., Li,Y., Zhu,J., Liu,Y., Hu,X. et al. (2002) Sequence and analysis of rice chromosome 4. Nature, 420, 316–320. [DOI] [PubMed] [Google Scholar]
- 41.Omer A.D., Lowe,T.W., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 42.Gaspin C., Cavaille,G., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 43.Bachellerie J.P. and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- 44.Tycowski K.T., Shu,M.D. and Steitz,J.A. (1996) A mammalian gene with introns instead of exons generating stable RNA products. Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- 45.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 46.Michot B., Qu,L.H. and Bachellerie,J.P. (1990) Evolution of large-subunit rRNA structure. The diversification of divergent D3 domain among major phylogenetic groups. Eur. J. Biochem., 188, 219–229. [DOI] [PubMed] [Google Scholar]