Abstract
The synthesis of oligonucleotides (ODNs) containing 5-(N-aminohexyl)carbamoyl-2′-O-methyluridine (D) is described, and thermal stability and resistance to enzymatic hydrolysis of the ODNs are compared with ODNs containing 5-(N-aminohexyl)carbamoyl-2′-deoxyuridine (H). The ODNs containing D and the complementary RNA demonstrated a duplex thermal stabilization of 0.4–3.9°C per modification depending on the position and the number, while the ODNs containing H with the RNA showed slightly less effective thermal stabilization. Further more, the ODNs containing D were found to be more resistant to nucleolytic hydrolysis, not only by snake venom phosphodiesterase (SVPD; a 3′-exonuclease) but also by DNase I (an endonuclease). The half-life of the 17mer containing five molecules of D against nucleolytic hydrolysis by SVPD was 240 times greater than the unmodified 17mer ODN, which is 1.8 times greater than the ODN containing 5Hs in the same sequence. Against DNase I, the same ODN containing 5Ds was 24 times greater stable than the unmodified 17mer and 15 times more stable than the ODN containing 5Hs. We also examined whether the duplexes formed by the ODNs containing D and the complementary RNAs could be a substrate of Escherichia coli RNase H. It was revealed that a minimum of five contiguous unmodified 2′-deoxyribonucleosides between Ds was required to constitute a substrate of E.coli RNase H. Thus, the ODN with Ds and at least five contiguous unmodified 2′-deoxyribonucleosides between Ds was found to be a candidate for a novel antisense molecule.
INTRODUCTION
Antisense oligodeoxynucleotides (ODNs) have been applied extensively to the regulation of cellular and viral gene expression (1–4). They hybridize to mRNA targets by Watson–Crick base pairing and inhibit translation of mRNA in a sequence-specific manner. One of the major problems encountered when using naturally occurring phosphodiester ODNs as antisense molecules is their rapid degradation by nucleases found in cell culture media and inside cells. Therefore, several types of backbone-modified ODNs have been synthesized and used for antisense studies (5). Phosphorothioate ODN, in which one non-bridging phosphate oxygen is replaced by sulfur, is one of the most widely used backbone-modified antisense ODNs. Phosphorothioate ODN is markedly more resistant to hydrolysis by nucleases than unmodified ODNs and a substrate of RNase H that selectively cleaves the RNA strand of a DNA–RNA duplex (2). However, it is known that their binding affinity for RNA is lower than that of an unmodified ODN (6). Furthermore, phosphorothioate ODNs sometimes exhibit non-sequence-specific activity (7–9).
On the other hand, naturally occurring polyamines, such as spermidine and spermine, are known to bind strongly to DNAs (10–12) and stabilize duplex (13,14) and triplex formations (15–17), although the precise mode of binding is not clear. Their enhanced thermal stability is explained by the reduction of the anionic electrostatic repulsion between the phosphate moieties by the cationic polyamines under physiological conditions. Therefore, ODN analogs carrying various polyamines have been synthesized, and some of them were shown to increase the thermal stability of duplexes and triplexes and to be more resistant to nucleases than unmodified ODNs (18–32).
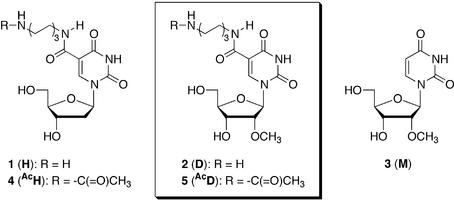
Based on this background, we also synthesized the ODNs containing 5-(N-aminohexyl)carbamoyl-2′-deoxyuridines (1; H) (28–30). We found that the 17mer ODNs containing four residues of H at dispersed positions greatly stabilized duplex formation with both complementary DNA (3.0°C increase per modification as compared to the unmodified DNA) and RNA (1.5°C increase per modification) strands (28,29). Furthermore, the ODNs were markedly more resistant (about 160 times more stable) to nucleolytic hydrolysis by snake venom phosphodiesterase (SVPD; a 3′-exonuclease) than unmodified ODNs and were very stable in a medium containing 10% fetal calf serum (29). However, it was also found that the endonuclease-resistant property of the ODNs containing H was insufficient to allow it to be applied to antisense studies. The half-life of the 17mer ODN containing alternatively 5Hs toward nucleolytic hydrolysis by nuclease S1 (an endonuclease) was 7 times greater than that of the unmodified ODN. However, when increased numbers of unmodified bases were incorporated between the Hs in the ODNs, the nuclease S1-resistant property was greatly reduced (30) (Fig. 1).
Figure 1.
Structures of the modified nucleoside analogs.
It has been reported that a 2′-O-methyl modification of RNA increases the stability of the RNA to nucleolytic hydrolysis by nucleases as well as the thermal stability of the duplex with a complementary RNA (33–35). Thus, we envisioned that a 2′-O-methyl modification of H would further increase the stability of the ODNs to nucleolytic hydrolysis by both exo- and endonucleases and the thermal stability of the duplexes with their complementary RNAs.
In this paper, we report the synthesis of 5-(N-aminohexyl)carbamoyl-2′-O-methyluridine (2; D) and its incorporation into ODNs. The thermal stability of the duplexes composed of the ODNs containing D and their complementary RNAs, the exo- and endonuclease-resistant properties of the ODNs and their ability to elicit RNase H activity were also examined.
MATERIALS AND METHODS
General experimental data
Thin-layer chromatography was done on Merck coated plates 60F254. The silica gel or the neutralized silica gel used for column chromatography were Merck silica gel 5715. The 1H-NMR spectra were recorded with a JEOL EX-270 spectrometer with a tetramethylsilane as an internal standard. Chemical shifts are reported in parts per million (δ), and signals are expressed as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet) or br (broad).
5-Iodo-2′-O-methyluridine (7). A mixture of 2′-O-methyluridine (6) (200 mg, 0.78 mmol), iodine (0.21 g, 0.47 mmol) and ceric ammonium nitrate (CAN; 0.21 g, 0.39 mmol) in AcOH (13 ml) was stirred at 80°C for 1 h. After the solvent was co-evaporated 3 times with H2O, the crude product was crystallized from hot EtOH to give 7 (240 mg, 80%, as a white crystal): 1H-NMR (270 MHz, DMSO-d6) δ 11.68 (br s, 1H, NH-3), 8.52 (s, 1H, H-6), 5.78 (d, 1H, H-1′, J1′,2′ = 4.0 Hz), 5.29 (t, 1H, HO-5′, JHO-5′,5′ = 4.5 Hz), 5.12 (d, 1H, HO-3′, JHO-3′,3′ = 4.0 Hz), 4.10 (ddd, 1H, H-3′, JHO-3′,3′ = J4′,3′ = J2′,3′ = 4.0 Hz), 3.86–3.84 (m, 1H, H-4′), 3.78 (dd, 1H, H-2′, J1′,2′ = J2′,3′ = 4.0 Hz), 3.73–3.53 (m, 2H, H-5′), 3.37 (s, 3H, 2′-OMe); HRMS (FAB, positive) calculated for C10H14N2O6I 384.9898 (MH+), found 384.9884.
5′-O-Dimethoxytrityl-5-iodo-2′-O-methyluridine (8). A mixture of 7 (106 mg, 0.28 mmol), DMTrCl (150 mg, 0.44 mmol) and 4-(dimethylamino)pyridine (DMAP) (2.2 mg, 0.018 mmol) in pyridine (2.5 ml) was stirred at room temperature for 18 h. After the reaction was quenched by the additon of EtOH (0.2 ml), the solvent was co-evaporated twice with toluene. The residue was partitioned between AcOEt and H2O, the organic phase was washed with saturated aqueous NaHCO3 and then brine, and dried (Na2SO4). After evaporation of the solvent, the crude product was purified by column chromatography (neutral-SiO2, CHCl3:MeOH = 10:1) to give 8 (190 mg, 100% as a yellowish foam): 1H-NMR (270 MHz, CDCl3 + D2O) δ 8.15 (d, 1H, H-6, JNH-3,6 = 0.6 Hz), 7.46–6.83 (m, 13H, DMTr), 5.94 (d, 1H, H-1′, J2′,1′ = 3.0 Hz), 4.45 (dd, 1H, H-3′, J2′,3′ = 5.2, J4′,3′ = 12.5 Hz), 4.09–4.06 (m, 1H, H-4′), 3.93 (dd, 1H, H-2′, J2′,3′ = 5.2, J2′,1′ = 3.0 Hz), 3.79 (s, 6H, Ar-OMe), 3.63 (s, 3H, 2′-OMe), 3.48–3.46 (m, 2H, H-5′); HRMS (FAB, positive) calculated for C31H31IN2NaO8 709.1025 (MNa+), found 709.1050.
5′-O-Dimethoxytrityl-5-trifluoroethoxycarbonyl-2′-O-methyluridine (9). A mixture of 8 (105 mg, 0.15 mmol), Et3N (43 µl, 0.31 mmol), CF3CH2OH (110 µl, 1.5 mmol) and Pd(PhCN)2Cl2 (1.2 mg, 3.1 µmol) in CH3CN (1.5 ml) was stirred at 60°C for 6 h under CO atmosphere. The reaction mixture was filtered with a Celite pad to remove the catalysis. After evaporation of the solvent, the residue was partitioned between AcOEt and H2O. The organic layer was washed with brine and then dried (Na2SO4). The crude product was purified by column chromatography (neutral-SiO2, CHCl3:AcOEt = 2:1) to give 9 (95 mg, 91%, as a yellow foam): 1H-NMR (270 MHz, CDCl3 + D2O) δ 8.67 (s, 1H, H-6), 7.43–6.81 (m, 13H, DMTr), 5.88 (d, 1H, H-1′, J2′,1′ = 1.6 Hz), 4.29 (dd, 1H, H-3′, J2′,3′ = 5.1, J4′,3′ = 3.6 Hz), 4.23–4.07 (m, 3H, H-4′, F3CCH2), 3.93 (dd, 1H, H-2′, J2′,3′ = 5.1, J2′,1′ = 1.6 Hz), 3.79 (s, 6H, Ar-OMe), 3.67 (s, 3H, 2′-OMe), 3.53 (dd, 1H, H-5′a, J5′a, 5′b = 11.2, J5′a, 4′ = 2.2 Hz), 3.41 (dd, 1H, H-5′b, J5′a, 5′b = 11.2, J4′, 5′b = 4.0 Hz); HRMS (FAB, positive) calculated for C34H33F3N2NaO10 709.1948 (MNa+), found 709.1985.
5′-O-Dimethoxytrityl-5-(N-trifluoroacetylaminohexyl)carbamoyl-2′-O-methyluridine (10). 1,6-Diaminohexane (52 mg, 0.45 mmol) was added to a solution of 9 (102 mg, 0.15 mmol) and N,N-diisopropylethylamine (39 µl, 0.22 mmol) in DMF (1.5 ml), and the mixture was stirred for 17 h at room temperature. The mixture was concentrated in vacuo and taken in AcOEt, which was washed with H2O and brine. The organic layer was dried (Na2SO4) and concentrated to dryness. The residue was dissolved in DMF (1.5 ml). Et3N (0.21 ml, 1.5 mmol) and CF3COOEt (0.18 ml, 1.5 mmol) were added to the solution, and the mixture was stirred at room temperature for 3 h. The mixture was concentrated in vacuo and taken in AcOEt, which was washed with H2O and brine. The organic layer was dried (Na2SO4) and concentrated to dryness. The residue was purified by column chromatography (neutral-SiO2, CHCl3:AcOEt = 1:1) to give 10 (73 mg, 61%, as a white foam): 1H-NMR (270 MHz, CDCl3) δ 9.00 (s, 1H, NH-3), 8.54 (s, 1H, H-6), 8.50 (t, 1H, CF3CONH, JNH,CH2 = 5.8 Hz), 7.46–6.81 (m, 13H, DMTr), 6.55 (s, 1H, CH2NHCO), 5.87 (d, 1H, H-1′, J2′,1′ = 3.2 Hz), 4.16–4.09 (m, 1H, H-3′), 4.05–4.00 (m, 1H, H-4′), 3.90 (dd, 1H, H-2′, J2′,1′ = 3.2, J2′,3′ = 5.5 Hz), 3.78 (s, 6H, Ar-OMe), 3.53 (s, 3H, 2′-OMe), 3.45 (d, 2H, H-5′, J4′,5′ = 4.8 Hz), 3.39–3.27 (m, 4H, CF3CONHCH2, CH2NHCO), 2.59 (dd, 1H, 3′-OH, J3′,OH = 7.5, J4′,OH = 2.6 Hz), 1.57–1.37 [m, 8H, (CH2)4]; HRMS (FAB, positive) calculated for C40H45F3N4NaO10 821.2985 (MNa+), found 821.2972.
3′-O-[(2-Cyanoethyl)(N,N-diisopropylamino)phosphinyl]-5′-O-dimethoxytrityl-5-(N-trifluoroacetylaminohexyl)carbamoyl-2′-O-methyluridine (11). 2-Cyanoethyl-N,N-diisopropylchlorophosphoroamidite (44 µl, 0.25 mmol) was added to a solution of 10 (102 mg, 0.13 mmol) and N,N-diisopropylethylamine (39 µl, 0.19 mmol) in CH2Cl2 (1.3 ml), and the reaction mixture was stirred for 2 h at room temperature. The mixture was partitioned between AcOEt and saturated aqueous NaHCO3, then the organic layer was washed with brine and dried (Na2SO4). After concentration of the organic phase, the residue was purified by column chromatography (neutral-SiO2, hexane:AcOEt = 1:1) to give 11 (89 mg, 71%, as a white foam): 31P-NMR (CDCl3) δ 151.60, 150.97; HRMS (FAB, positive) calculated for C49H62F3N6NaO11P 1021.4064 (MNa+), found 1021.4020.
5′-O-Dimethoxytrityl-3′-O-succinyl-5-(N-trifluoroacetylaminohexyl)carbamoyl-2′-O-methyluridine (12). A solution of 10 (311 mg, 0.39 mmol), succinic anhydride (80 mg, 0.78 mmol) and DMAP (9.5 mg, 78 µmol) in pyridine (4 ml) was stirred overnight at 50°C. The reaction was quenched by the addition of H2O (1 ml) at room temperature and the solvent was evaporated to dryness. The residue was partitioned between AcOEt and H2O, and the organic layer was washed with saturated aqueous KH2PO4 and brine. The separated organic phase was dried (Na2SO4) and concentrated. The residue was purified by column chromatography (neutral-SiO2, CHCl3:MeOH = 20:1) to give 12 (319 mg, 91%, as a white foam): 1H-NMR (270 MHz, CDCl3) δ 9.85 (s, 1H, NH-3), 8.59–8.53 (m, 2H, H-6, CF3CONH), 7.44–6.78 (m, 13H, DMTr), 6.62 (s, 1H, CH2NHCO), 5.83 (d, 1H, H-1′, J2′,1′ = 4.6 Hz), 5.08 (dd, 1H, H-3′, J2′,3′ = J4′,3′ = 5.3 Hz), 4.21 (dd, 1H, H-4′, J4′,5′ = 9.9, J4′,3′ = 5.3 Hz), 4.11 (dd, 1H, H-2′, J2′,1′ = 4.6, J2′,3′ = 5.3 Hz), 3.77 (s, 6H, Ar-OMe), 3.48–3.26 (m, 9H, 2′-OMe, H-5′, CF3CONHCH2, CH2NHCO), 2.67–2.62 (m, 4H, OCOCH2CH2COO), 1.56–1.36 (m, 8H, (CH2)4); HRMS (FAB, positive) calcd for C44H49F3N4NaO13 921.3146 (MNa+), found 921.3163.
Synthesis of the controlled pore glass support (13). Aminopropyl controlled pore glass (642 mg, 57.3 µmol, 89.2 µmol/g) was added to a solution of 12 (206 mg, 0.23 mmol) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (53 mg, 0.27 mmol) in anhydrous DMF (3 ml), and the mixture was kept for 72 h at room temperature. After the resin was washed with anhydrous pyridine, a capping solution (4 ml, 0.1 M DMAP in pyridine:Ac2O = 9:1) was added and the whole mixture was kept for 8 h at room temperature. The resin was washed with EtOH and acetone, and dried in vacuo. The amount of loaded nucleoside 12 to the solid support is 41.9 µmol/g from calculation of released dimethoxytrityl cation by a solution of 70% HClO4:EtOH (3:2, v/v).
Synthesis of oligonucleotides. ODNs were synthesized on a DNA/RNA synthesizer (Applied Biosystem Model 392) by the phosphoramidite method (36). Each ODN linked to the resin was treated with concentrated NH4OH at 55°C for 12 h. Then the released ODNs protected by a DMTr group at the 5′-end were chromatographed on a C-18 silica gel column with a linear gradient of CH3CN from 0 to 30% in 0.1 M TEAA buffer (pH 7.0). The fractions were concentrated and the residue was treated with aqueous 80% AcOH at room temperature for 20 min, then the solution was concentrated and the residue was co-evaporated with H2O. The residue was dissolved in H2O, and the solution was washed with AcOEt, then the H2O layer was concentrated to give deprotected ODNs. The crude ODNs were dissolved in 0.1 M TEAA buffer and purified by reverse-phase HPLC to give pure ODNs.
Thermal denaturation study. Each sample containing an appropriate ODN and a complementary RNA in a buffer of 10 mM sodium cacodylate (pH 7.0) containing 10 mM NaCl was heated, then cooled gradually to room temperature and used for the thermal denaturation study. Thermally induced transitions were monitored at 260 nm on a Beckman DU-650 spectrometer. The sample temperature was increased 0.5°C/min.
Partial hydrolysis of ODN with SVPD. Each ODN labeled with 32P at the 5′-end (10 pmol) was incubated with SVPD (20 ng) in the presence of Torula RNA (0.15 OD units at 260 nm) in a buffer containing 37.5 mM Tris–HCl (pH 8.0), 8 mM MgCl2 and 5 mM DTT (total 20 µl) at 37°C. At appropriate periods, aliquots of the reaction mixture were separated and added to a solution of EDTA (5 mM, 10 µl), then the mixtures were heated for 3 min at 90°C. The solutions were analyzed by electrophoresis on 20% polyacrylamide gel containing 7 M urea.
Partial hydrolysis of ODN with DNase I. Each ODN labeled with 32P at the 5′-end (10 pmol) was incubated with DNase I (10 or 5 U) in the presence of Torula RNA (0.26 OD units at 260 nm) in a buffer containing 40 mM Tris–HCl (pH 7.5) and 5 mM MgCl2 (total 20 µl) at 37°C. At appropriate periods, aliquots of the reaction mixture were separated and added to a solution of EDTA (5 mM, 10 µl), then the mixtures were heated for 3 min at 90°C. The solutions were analyzed by gel electrophoresis as described above.
RNA hydrolysis by E.coli RNase H. A mixture of ODN (240 pmol) and the complementary RNA (200 pmol) was annealed by heating at 90°C for 5 min and then cooled to 30°C, in the presence of labeled RNA with 32P at the 5′-end (2 pmol) in a buffer containing 10 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 50 mM NaCl, 0.5 mM DTT and 0.01% BSA (total 20 µl). The mixture was incubated with 0.42 U of E.coli RNase H (Takara Shuzo Co., Ltd) at 30°C. Aliquots (1 µl) of the reaction mixture were taken after 5, 15, 30, 60 and 120 min. The samples were added to a solution of 10 M urea in 50 mM TBE buffer and cooled on ice. The hydrolyzed products were analyzed by 20% polyacrylamide gel containing 8 M urea (37). Radioactivity of each band was estimated by a Bio-imaging analyzer (Bas 2000, Fuji Co., Ltd).
Kinetic analysis. The kinetic studies of RNA hydrolysis by E.coli RNase H were carried out under the conditions described above. Reaction times were adjusted to give extents of reaction of 10% or less. The Michaelis constant (Km) and the maximum rate of reaction (Vmax) were obtained by the Hanes–Woolf plot.
RESULTS AND DISCUSSION
Synthesis
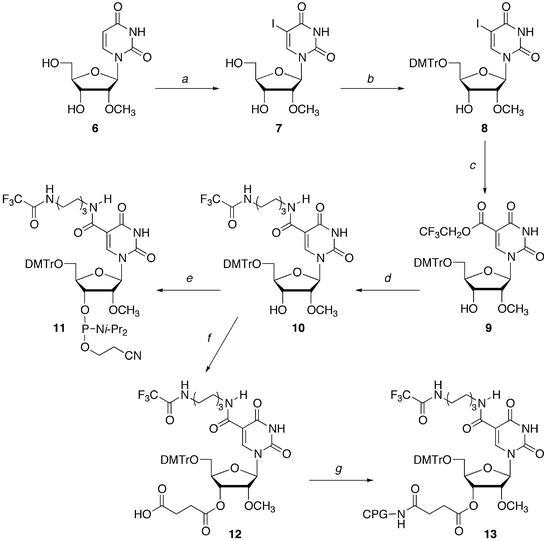
The synthesis of the ODNs containing D was accomplished in a DNA/RNA synthesizer by using the suitably protected 5-(N-aminohexyl)carbamoyl-2′-O-methyluridine phosphoroamidite 11. The synthesis of 11 is shown in Scheme 1. Treatment of 2′-O-methyluridine (6) with iodine and CAN gave 5-iodo-2′-O-methyluridine (7) in 80% yield (38). Compound 7 was reacted with DMTrCl to give 5′-O-DMTr-5-iodo-2′-O-methyluridine (8) quantitatively. Palladium- catalyzed carbonylation of 8 with carbon monoxide in 2,2,2-trifluoroethanol gave the 5-trifluoroethylcarbonyl derivative 9 in 91% yield (29). After introducing the aminohexyl linker into 9, the terminal amino group of the linker was protected with a trifluoroacetyl group to give 10 in 61% yield. Nucleoside 10 was converted to the protected nucleoside phosphoramidite 11 by the standard procedure. To incorporate D into the 3′-end of ODNs, 3′-succinate 12 was prepared from 10, and 12 was linked to controlled pore glass to give the solid support 13 containing 12 (41.9 µmol/g).
Scheme 1. (a) I2, CAN, CH3CO2H, 80°C; (b) DMTrCl, pyridine, room temperature; (c) CF3CH2OH, Pd(PhCN)2Cl2, CO, Et3N, CH3CN, 60°C; (d) (1) H2N(CH2)6NH2, iPr2NEt, DMF, room temperature, (2) CF3CO2Et, Et3N, DMF, room temperature; (e) 2-cyanoethyl-N,N-diisopropylphosphoamidochloride, iPr2NEt, CH2Cl2, room temperature; (f) succinic anhydride, DMAP, pyridine, 50°C; (g) aminopropyl controlled pore glass, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride, DMF, room temperature.
All ODNs used in this study were synthesized in a DNA/RNA synthesizer. The average coupling yield of 11 was 89% using a 0.2 M solution of the amidite derivative in CH3CN and a 1 h coupling time. The fully protected ODNs were deprotected and purified by the standard method. Each ODN produced showed a single peak in reverse-phase HPLC analysis. Furthermore, these ODNs were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS), and the observed molecular weights supported their structures (Table 1).
Table 1. Sequences of ODNs used in this studya.
| ODN | |
|---|---|
| ODN 14 | 5′-TATGTATTTTTATCTGT-3′ |
| ODN 15-II or III | 5′-XATGTATTTTTATCTGT-3′ |
| ODN 16-II or III | 5′-TATGTATTTTTATCTGX-3′ |
| ODN 17-II or III | 5′-TATGTATTXTTATCTGT-3′ |
| ODN 18-II or III | 5′-TATGTATTXTTATCTGX-3′ |
| ODN 19-II or III | 5′-TAXGTATTTXTATCTGX-3′ |
| ODN 20-II or III | 5′-TAXGTATXTTTAXCTGX-3′ |
| ODN 21-II, III, IV, V or VI | 5′-TAXGTAXTTXTAXCTGX-3′ |
| RNA-22 | 3′-AUACAUAAAAAUAGACA-5′ |
| ODN 23-I, II, III or IV | 5′-XgXaXCGTXgXcXgXgY-3′ |
| ODN 24-I, II, III, IV or VI | 5′-XgXaXCGTTXgXcXgXG-3′ |
| ODN 25-I, II, III or IV | 5′-XgXaXCGTTGXgXcXgY-3′ |
| ODN 26-I, II, III or IV | 5′-XgXaXCGTTGTXgXcXG-3′ |
| RNA-27 | 3′-ACAUAGCAACAGACACA-5′ |
| RNA-28 | 3′-ACAUAGCAAACAGACAC-5′ |
| RNA-29 | 3′-ACAUAGCAACACAGACA-5′ |
| RNA-30 | 3′-ACAUAGCAACAACAGAC-5′ |
aI series (X = Y = T, lower-case letters = 2′-deoxyribonucleosides); II series (X = Y = H, lower-case letters = 2′-deoxyribonucleosides); III series (X = D, Y = H, lower-case letters = 2′-O-methylribonucleosides); IV series (X = M, Y = T, lower-case letters = 2′-O-methylribonucleosides); 21-V (X = AcH); VI series (X = AcD, lower-case letters = 2′-O-methylribonucleosides).
UV melting studies of duplexes
Stable duplex formation with mRNA is one of the most important factors in antisense study. The thermal stability of the duplexes between these ODNs and a complementary RNA, 5′-r[ACAGAUAAAAAUACAUA]-3′ (22), was studied by thermal denaturation in a buffer of 0.01 M sodium cacodylate (pH 7.0) containing 0.01 M NaCl. Each profile of the thermal denaturation showed a single transition corresponding to a helix-to-coil transition (data not shown). Melting temperatures (Tms) are listed in Table 2. The free energy (ΔG°) values of the duplexes were determined by calculations based on the slope of a 1/Tm versus ln(CT/4) plot, where CT is the total concentration of single strands. The negative free energy (–ΔG°37) values are also listed in Table 2. The Tm and –ΔG°37 values of the all modified ODNs containing H or D were greater than those of the unmodified ODN 14 (Tm = 33.8°C; –ΔG°37 = 11.5 kcal/mol). Thus, H and D were found to thermally and thermodynamically stabilize the ODN/RNA duplexes. The stability of the duplexes was dependent on the position, number and kind of modified nucleosides. As compared with the ODNs containing one residue of H or D at their 3′-ends, the ODNs containing one residue of H or D at their 5′-ends or the centers more efficiently stabilized the duplexes with the complementary RNA. The duplexes became more thermally stable as the number of H or D increased. The ΔTm1 values [Tm (each ODN) – Tm (the control ODN 14)] for the ODNs containing five residues of H or D at dispersed positions were +13.4 (for ODN 21-II) or +15.2°C (for ODN 21-III), respectively.
Table 2. Hybridization dataa.
| ODN | ODN / RNA-22 | ||||||
|---|---|---|---|---|---|---|---|
| 0.01 M NaCl Tm (°C) | –ΔG°37 kcal/mol | Δ Tm1b (°C) | ΔTm1/unitc (°C) | Δ Tm2d (°C) | 1 M NaCl Tm (°C) | ΔTm3e (°C) | |
| 14 | 33.8 ± 0.4 | 11.5 | 56.0 | +22.2 | |||
| 15-II | 36.6 ± 0.3 | 12.7 | +2.8 | +2.8 | – | ||
| 16-II | 34.8 ± 0.4 | 12.5 | +1.0 | +1.0 | – | ||
| 17-II | 37.5 ± 0.1 | 12.4 | +3.7 | +3.7 | – | ||
| 18-II | 37.9 ± 0.4 | 12.0 | +4.1 | +2.1 | 57.1 | +19.2 | |
| 19-II | 43.0 ± 0.1 | 14.7 | +9.2 | +3.1 | 59.3 | +16.3 | |
| 20-II | 44.4 ± 0.3 | 14.6 | +10.6 | +2.7 | 58.2 | +13.8 | |
| 21-II | 47.2 ± 0.2 | 14.9 | +13.4 | +2.7 | 58.8 | +11.6 | |
| 21-V | 30.4 | –3.4 | 49.5 | +19.1 | |||
| 15-III | 36.3 ± 0.3 | 12.9 | +2.5 | +2.5 | –0.3 | – | |
| 16-III | 34.2 ± 0.0 | 11.2 | +0.4 | +0.4 | –0.6 | – | |
| 17-III | 37.2 ± 0.2 | 12.2 | +3.4 | +3.4 | –0.3 | – | |
| 18-III | 38.1 ± 0.0 | 12.4 | +4.3 | +2.2 | +0.2 | 58.1 | +20.0 |
| 19-III | 45.5 ± 0.1 | 14.5 | +11.7 | +3.9 | +2.5 | 61.2 | +15.7 |
| 20-III | 47.0 ± 0.0 | 15.4 | +13.2 | +3.3 | +2.6 | 60.9 | +13.9 |
| 21-III | 49.0 ± 0.4 | 16.5 | +15.2 | +3.0 | +1.8 | 60.9 | +11.9 |
| 21-VI | 30.6 | –3.2 | 50.9 | +20.3 |
aExperimental conditions are described in Materials and Methods.
bΔTm1 = Tm (each ODN) – Tm (control ODN 14).
cΔTm1/unit = Tm1 (each ODN)/the number of modification.
dΔTm2 = Tm (each ODN containing D) – Tm (each ODN containing the same number of H).
eΔTm3 = Tm (1 M NaCl) – Tm (0.01 M NaCl).
A duplex stabilization effect per modification (ΔTm1/unit) is included in Table 2. Although there is only a slight difference between the position and the numbers incorporated, the ODN containing D (+0.4 to +3.9°C) is more thermally stable than the ODN containing H (+1.0 to +3.7°C). To further compare the abilities of H and D to stabilize ODN/RNA duplex, the ΔTm2 values [Tm (each ODN containing D) – Tm (the control ODN containing the same number of H)] values were also calculated (Table 2). Although the ΔTm2 values for the ODNs containing one residue of H or D were –0.3 or –0.6°C, the values for the ODNs containing more than 2 residues of H or D were +0.2 to +2.6°C. Thus, nucleoside D was found to more efficiently stabilize the duplex formation with the RNA than H, except for the ODNs 15, 16 and 17 containing one modified nucleoside.
Thermal denaturation was also performed under higher ionic strength (1 M NaCl) to confirm the effects of the ammonium ions at the end of the alkyl chains of H and D on the thermal stabilities of the duplexes. Then, the ΔTm3 values [Tm (1 M NaCl) – Tm (0.01 M NaCl)] were compared (Table 2). The values for the duplexes containing H or D became smaller than those for the control duplex (ΔTm3 = +22.2°C). The values for the duplexes containing H or D decreased when the number of H or D incorporated into the ODNs increased. Furthermore, the values (19.1°C for ODN 21-V and 20.3°C for ODN 21-VI, respectively) for the duplexes containing AcH or AcD were similar to or slightly lower than those of the control duplex. Thus, the terminal ammonium ions in H and D help to neutralize the phosphate negative charges.
The thermal stabilities of DNA/RNA duplexes containing nucleoside analogs with electronegative substituents at the 2′-α positions are often discussed on the basis of the gauche effect between the electronegative substituents and O4′ of the furanose ring. The electronegative substituent at the 2′-α position shifts the conformational equilibrium of the sugar to 3′-endo. Shifting the conformation of the DNA strand to 3′-endo puts it in a more RNA-like conformation and causes the hybridization properties to more resemble those of an RNA/RNA geometry (35,39).
The conformation of the sugar moiety of D was studied by 1H NMR. The fractional population of the N-conformer of 10 was calculated by the formula, %N = (10 – J1′,2′) × 10 (40). The J1′,2′ value of 10 was 3.2 Hz, whereas that of the corresponding 2′-deoxyuridine derivative, 5′-O-DMTr-5-(N-trifluoroacetylaminohexyl)carbamoyl-2′-deoxyuridine, was 6.5 Hz. Thus, the N% value was estimated to be 68 and 35%, respectively. Therefore, the high thermal stability of the ODN/RNA duplexes containing D was due to the conformation of the sugar moiety of D as well as to the neutralization of the phosphate negative charges by the terminal ammonium ions of the linkers.
Nuclease resistance
The susceptibility of the ODNs to nucleolytic digestion was examined. Two kinds of nuclease, SVPD and DNase I, were used in this study as models for a 3′-exonuclease and an endonuclease, respectively.
The ODNs containing the modified nucleosides were labeled at the 5′-end with 32P and incubated with SVPD or DNase I. The reactions were then analyzed by polyacrylamide gel electrophoresis under denaturing conditions.
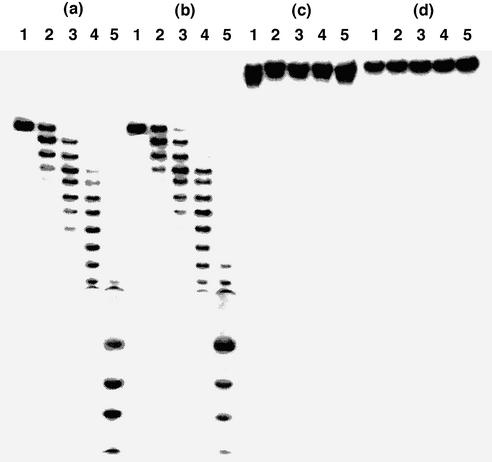
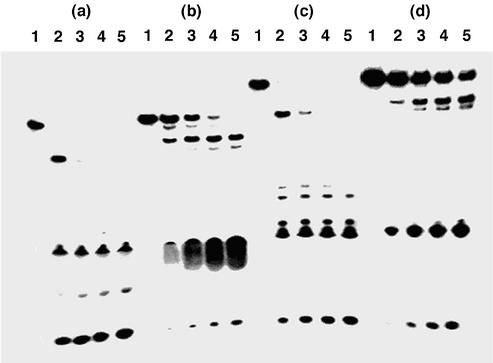
Figure 2 shows the results with SVPD. Although the control 14 and the ODN 21-IV containing five residues of 2′-O-methyluridine (3; M) were hydrolyzed randomly within 30 min of incubation, the phosphodiester linkages at the 5′-sides of the modified nucleosides were resistant to SVPD. After 2 h, no enzymatic degradation of the ODNs containing five residues of H or D was observed at all. The half-lives (t1/2s) of the control ODN 14 and the ODN 21-IV containing M were 5.0 and 5.8 min, whereas those of the ODNs containing 21-II and 21-III were 11 and 20 h, respectively. Therefore, the t1/2 of the 17mer containing 5Ds against SVPD was 240 times longer than the unmodified 17mer ODN, which is 1.8 times greater than the ODN containing 5Hs in the same sequence.
Figure 2.
Polyacrylamide gel electrophoresis of 5′-32P-labeled ODNs hydrolyzed by SVPD: (a) ODN 14; (b) ODN 21-IV; (c) ODN 21-II; (d) ODN 21-III. ODNs were incubated with SVPD for 0 min (lane 1), 10 min (lane 2), 30 min (lane 3), 60 min (lane 4) or 120 min (lane 5). Experimental conditions are described in Materials and Methods.
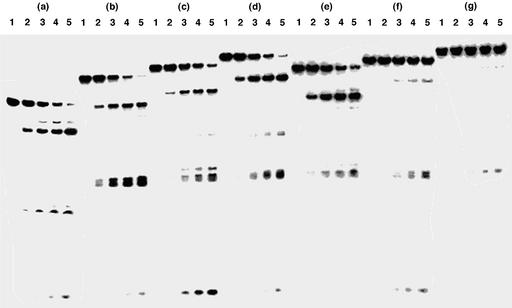
The phosphodiester linkages around the modified nucleoside D were also highly resistant to the endo-hydrolysis by DNase I (Fig. 3). For the most part, the t1/2 of the ODNs containing H did not depend on the number of H (30), while those containing D were dependent on the number of D. The t1/2s of the ODNs containing D increased as the number of D increased. The t1/2s of the control ODN 14 was 7.3 min, whereas the t1/2s of the ODNs 19-II, 20-II, 21-II, 19-III, 20-III and 21-III were 8.3, 13, 12, 14, 90 and 174 min, respectively. The t1/2 of the 17mer ODN 21-II containing five molecules of H was almost equal to that of the control ODN 14, while that of the 17mer ODN 21-III containing five molecules of D was 24 times greater than that of the control ODN. From these results, it can be confirmed that the ODNs containing D were highly resistant against hydrolysis not only by the 3′-exonuclease but also by the endonuclease.
Figure 3.
Polyacrylamide gel electrophoresis of 5′-32P-labeled ODNs hydrolyzed by DNase I: (a) ODN 14; (b) ODN 19-II; (c) ODN 20-II; (d) ODN 21-II; (e) ODN 19-III; (f) ODN 20-III; (g) ODN 21-III. ODNs were incubated with DNase I for 0 min (lane 1), 5 min (lane 2), 15 min (lane 3), 30 min (lane 4) or 60 min (lane 5). Experimental conditions are described in Materials and Methods.
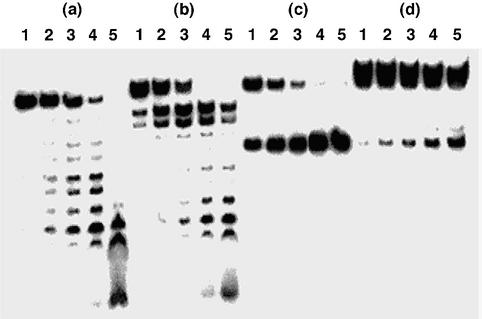
Next, we examined the effects of the terminal ammonium ions of the aminoalkyl chains on the resistance of the ODNs to the endonuclease. We compared the susceptibilities of the ODNs 21-V containing AcH and 21-VI containing AcD to nucleolytic digestion by DNase I with those of the ODNs 14 and 21-IV containing M (Fig. 4). The t1/2s of the ODNs 14, 21-IV, 21-V and 21-VI were 0.73, 5.5, 0.69 and 16 min, respectively. The relative t1/2s of the ODNs 21-IV, 21-V and 21-VI to that of the unmodified ODN 14 were 7.5, 0.95 and 22, respectively. On the other hand, the relative t1/2 of ODN 21-III containing D to the ODN 14 was 24. Thus, the high endonuclease resistant property of the ODNs containing D was not due to the electrostatic interaction between the terminal ammonium ion of the aminoalkyl chain of D and the enzyme, but mainly due to the steric hindrance of the 2′-O-methyl group and the aminoalkyl chain in D.
Figure 4.
Polyacrylamide gel electrophoresis of 5′-32P-labeled ODNs hydrolyzed by DNase I: (a) ODN 14; (b) ODN 21-IV; (c) ODN 21-V; (d) ODN 21-VI. ODNs were incubated with DNase I for 0 min (lane 1), 5 min (lane 2), 15 min (lane 3), 30 min (lane 4) or 60 min (lane 5). Experimental conditions are described in Materials and Methods.
Degradation by RNase H
It has been postulated that the antisense activity of antisense ODNs is due, at least in part, to cleavage of the RNA strand of a DNA/RNA duplex by RNase H. We therefore examined whether the ODN/RNA heteroduplex between an ODN containing D and its complementary RNA could elicit RNase H activity. It has been reported that E.coli RNase H requires at least four contiguous unmodified 2′-deoxyribonucleoside residues to elicit cleavage of the RNA strand (33), and a minimum of five contiguous unmodified 2′-deoxyribonucleosides was required for efficient substrates of HeLa RNase H (41). In this study, we used the ODNs 23–26, with 3–6 contiguous unmodified 2′-deoxyribonucleoside residues between the modified nucleosides M, H and D as DNA parts of the DNA/RNA duplexes.
The duplexes consisting of the ODN 23, 24, 25 or 26 and the complementary RNA 27, 28, 29 or 30 labeled with 32P at the 5′-end were incubated with E.coli RNase H, and the products were analyzed by polyacrylamide gel electrophoresis. The RNA in the duplex composed of the ODN 23-III with three contiguous unmodified 2′-deoxyribonucleoside residues between Ds was hardly hydrolyzed by the enzyme after 2 h of incubation (Table 3). On the other hand, the RNA in the duplex composed of the ODN 24-III with four contiguous unmodified 2′-deoxyribonucleoside residues between Ds was found to be hydrolyzed by the enzyme at the single site, as shown in Figure 5d. Under the same conditions, the RNA in the duplex consisting of the ODN 24-IV with four contiguous unmodified 2′-deoxyribonucleoside residues between Ms was also cleaved by the enzyme at the single site (Fig. 5c), while the RNA in the duplex composed of the ODN 24-II with four contiguous unmodified 2′-deoxyribonucleoside residues between Hs was hydrolyzed almost randomly (Fig. 5b).
Table 3. Half-lives (t1/2s) and relative t1/2s of the ODN/RNA duplexes against hydrolysis by E.coli RNase H.
| ODN | RNA | t1/2 (min) | Relative t1/2 |
|---|---|---|---|
| ODN 23-I | RNA 27 | 12 | 1 |
| ODN 23-II | 44 | 3.7 | |
| ODN 23-III | 900 | 73 | |
| ODN 23-IV | 590 | 49 | |
| ODN 24-I | RNA 28 | 15 | 1 |
| ODN 24-II | 13 | 0.86 | |
| ODN 24-III | 200 | 13 | |
| ODN 24-IV | 7.7 | 0.51 | |
| ODN 25-I | RNA 29 | 3.9 | 1 |
| ODN 25-II | 3.3 | 0.85 | |
| ODN 25-III | 6.2 | 1.6 | |
| ODN 25-IV | 3.6 | 0.92 | |
| ODN 26-I | RNA 30 | 5.8 | 1 |
| ODN 26-II | 3.7 | 0.64 | |
| ODN 26-III | 1.8 | 0.31 | |
| ODN 26-IV | 5.4 | 0.93 |
Figure 5.
Polyacrylamide gel electrophoresis of 5′-32P-labeled RNA 28 hydrolyzed by E.coli RNase H in the presence of complementary ODNs: (a) ODN 24-I; (b) ODN 24-II; (c) ODN 24-IV; (d) ODN 24-III. RNAs were incubated with E.coli RNase H in the presence of complementary ODNs for 5 min (lane 1), 15 min (lane 2), 30 min (lane 3), 60 min (lane 4) or 120 min (lane 5). Experimental conditions are described in Materials and Methods.
The t1/2s of the enzymatic hydrolysis of the RNAs in the ODN/RNA duplexes against RNase H and relative t1/2s of the RNAs in the duplexes containing the modified nucleosides M, H or D to those in the unmodified duplexes are summarized in Table 3. The relative t1/2 of the ODN 24-IV/RNA 28 duplex with four contiguous unmodified 2′-deoxyribonucleoside residues between Ms was 0.51, whereas that of the ODN 24-III/RNA 28 duplex with four contiguous unmodified 2′-deoxyribonucleoside residues between Ds was 13. On the other hand, the relative t1/2 of the ODN 25-III/RNA 29 duplex with five contiguous unmodified 2′-deoxyribonucleoside residues between Ds was 1.6. Thus, a minimum of five contiguous unmodified 2′-deoxyribonucleosides between Ds was required for efficient substrates of E.coli RNase H.
Kinetic analysis for the RNA degradation by RNase H
In order to investigate the RNA hydrolysis by RNase H in detail, we next carried out the kinetic analysis for the RNA hydrolysis by the enzyme. The ODNs, 24-III, 24-IV and 24-VI, with four contiguous unmodified 2′-deoxyribonucleoside residues between the modified nucleosides M, D and AcD, which afforded the single cleavage site, were used in this study. The Michaelis constant (Km) and the maximum rate of reaction (Vmax) were determined experimentally, and the turnover number (kcat) and the kcat/Km values were then calculated. The kinetic parameters are summarized in Table 4.
Table 4. Kinetic parameters of the hydrolysis of RNA-28 in the duplex by E.coli RNase H.
| ODN | Km (µM) | Vmax (µM/min) | kcat (1/s) | kcat/Km (1/s/µM) |
|---|---|---|---|---|
| ODN 24-IV | 3.9 | 0.30 | 5.0 | 1.3 |
| ODN 24-III | 9.6 | 0.011 | 0.19 | 0.02 |
| ODN 24-VI | 10.0 | 0.17 | 2.8 | 0.28 |
The Km value of the ODN/RNA duplex composed of the ODN 24-IV containing M was 3.9 µM, whereas the values of the ODNs 24-III containing D and 24-VI containing AcD were 9.6 and 10.0 µM, respectively. The Km values of the duplexes containing D or AcD were only ∼3-fold greater than Km value of the duplex containing M. On the other hand, the kcat value of the duplex composed of the ODN 24-IV containing M was 5.0 sec–1, whereas the kcat values of the ODNs 24-III containing D and 24-VI containing AcD were 0.19 and 2.8 sec–1, respectively. The kcat value of the duplex containing D was 1/30 of that containing M, whereas the kcat value of the duplex containing AcD was about half that containing M. These results indicate that the aminohexyl chain of the modified nucleoside D does not influence the affinity of the ODN/RNA duplex for the enzyme, but rather the rate of the hydrolysis of RNA by the enzyme.
CONCLUSION
In this paper, we have described the synthesis and properties of the ODNs containing D. We found that the ODNs containing D thermally stabilized the duplexes with the complementary RNA markedly. Furthermore, the ODNs containing D were found to be highly resistant to nucleolytic hydrolysis not only by SVPD (a 3′-exonuclease) but also by DNase I (an endonuclease). The t1/2 of the 17mer ODN containing five molecules of D against nucleolytic hydrolysis by DNase I was 24 times greater than that of the unmodified 17mer. Susceptibility of the duplexes formed by the ODNs containing D and the complementary RNA against nucleolytic hydrolysis by E.coli RNase H was examined in detail. It was revealed that the aminohexyl chain of D does not influence the affinity of the ODN/RNA duplex to the enzyme, but rather the rate of the hydrolysis of RNA by the enzyme. A minimum of five contiguous unmodified 2′-deoxyribonucleosides between Ds was found to be required for efficient substrates of E.coli RNase H. Thus, the oligonucleotide with Ds and at least five contiguous unmodified 2′-deoxyribonucleosides between Ds will be a candidate for a novel antisense molecule.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Y. Misawa (Hokkaido University) for technical assistance and Ms H. Matsumoto, A. Maeda, S. Oka and M. Kiuchi (Center for Instrumental Analysis, Hokkaido University) for technical assistance with NMR, MS and elemental analysis. This investigation was supported in part by a Grant-in-Aid for Creative Scientific Research (13NP0401) from the Japan Society for Promotion of Science. This paper constitutes part 221 of Nucleosides and Nucleotides (for part 220 in this series, see 42).
REFERENCES
- 1.Uhlmann E. and Peyman,A. (1990) Antisense oligonucleotides: a new therapeutic principle. Chem. Rev., 90, 544–584. [Google Scholar]
- 2.Milligan J.F., Matteucci,M.D. and Martin,J.C. (1993) Current concepts in antisense drug design. J. Med. Chem., 36, 1923–1937. [DOI] [PubMed] [Google Scholar]
- 3.Branch A.D. (1998) A good antisense molecule is hard to find. Trends Biochem. Sci., 23, 45–50. [DOI] [PubMed] [Google Scholar]
- 4.Stein C.A. (1999) Keeping the biotechnology of antisense in context. Nat. Biotechnol., 17, 209. [DOI] [PubMed] [Google Scholar]
- 5.Mesmaeker A.D., Altmann,K.-H., Waldner,A. and Wendeborn,S. (1995) Backbone modifications in oligonucleotides and peptides nucleic acid systems. Curr. Opin. Struct. Biol., 5, 343–355. [DOI] [PubMed] [Google Scholar]
- 6.Kibler-Herzog L., Zon,G., Uznanski,B., Whittier,G. and Wilson,M.D. (1991) Duplex stabilities of phosphorothioate, methylphosphonate and RNA analogs of two DNA 14-mers. Nucleic Acids Res., 19, 2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guvakova M.A., Yakubov,L.A., Vlodavsky,I., Tonkinson,J.L. and Stein,C.A. (1995) Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors and remove it from low affinity binding sites on extracelluar matrix. J. Biol. Chem., 270, 2620–2627. [DOI] [PubMed] [Google Scholar]
- 8.Rockwell P., O’Connor,W.J., King,K., Goldstein,N.I., Zhang,L.M. and Stein,C.A. (1997) Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc. Natl Acad. Sci. USA, 94, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal S. and Zhao,Q. (1998) Antisense therapeutics. Curr. Opin. Chem. Biol., 2, 519–528. [DOI] [PubMed] [Google Scholar]
- 10.Tabor C.W. and Tabor,H. (1976) 1,4-Diaminobutane (putrescine), spermidine and spermine. Annu. Rev. Biochem., 45, 285–306. [DOI] [PubMed] [Google Scholar]
- 11.Tabor C.W. and Tabor,H. (1984) Polyamines. Annu. Rev. Biochem., 53, 749–790. [DOI] [PubMed] [Google Scholar]
- 12.Etter M.C. (1990) Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res., 23, 120–126. [Google Scholar]
- 13.Tabor H. (1962) Protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry, 1, 496–501. [DOI] [PubMed] [Google Scholar]
- 14.Thomas T.J. and Bloomfield,V.A. (1984) Ionic and structural effects on the thermal helix-coil transition of DNA complexed with natural and synthetic polyamines. Biopolymers, 23, 1295–1306. [DOI] [PubMed] [Google Scholar]
- 15.Hampel K.J., Crosson,P. and Lee,J.S. (1991) Polyamines favor DNA triplex formation at neutral pH. Biochemistry, 30, 4455–4459. [DOI] [PubMed] [Google Scholar]
- 16.Thomas T. and Thomas,T.J. (1993) Selectivity of polyamines in triplex DNA stabilization. Biochemistry, 32, 14068–14074. [DOI] [PubMed] [Google Scholar]
- 17.Musso M. and Van Dyke,M.W. (1995) Polyamine effects on purine-purine-pyrimidine triple helix formation by phosphodiester and phosphorothioate oligodeoxyribonucleotides. Nucleic Acids Res., 23, 2320–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung C.-H., Breslauer,K.J. and Stein,S. (1993) Polyamine-linked oligonucleotides for DNA triple helix formation. Nucleic Acids Res., 21, 5489–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prakash T.P., Barawkar,D.A., Kumar,V.A. and Ganesh,K.N. (1994) Synthesis of site-specific oligonucleotide-polyamine conjugates. Bioorg. Med. Chem. Lett., 4, 1733–1738. [Google Scholar]
- 20.Barawkar D.A., Kumar,V.A. and Ganesh,K.N. (1994) Triplex formation at physiological pH by oligonucleotides incorporating 5-Me-dC-(N4-spermine). Biochem. Biophys. Res. Commun., 205, 1665–1670. [DOI] [PubMed] [Google Scholar]
- 21.Barawkar D.A., Rajeev,K.G., Kumar,V.A. and Ganesh,K.N. (1996) Triplex formation at physiological pH by 5-Me-dC-(N4-spermine) [X] oligodeoxynucleotides: non protonation of N3 in X of X*G:C triad and effect of base mismatch/ionic strength on triplex stabilities. Nucleic Acids Res., 24, 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid N. and Behr,J.-P. (1995) Recognition of DNA sequences by strand replacement with polyamino-oligonucleotides. Tetrahedron Lett., 36, 1447–1450. [Google Scholar]
- 23.Sund C., Puri,N. and Chattopadhyaya,J. (1996) Synthesis of C-branched spermine tethered oligo-DNA and the thermal stability of the duplexes and triplexes. Tetrahedron, 52, 12275–12290. [Google Scholar]
- 24.Hashimoto H., Nelson,M.G. and Switzer,C. (1993) Formation of chimeric duplexes between zwitterionic and natural DNA. J. Org. Chem., 58, 4194–4195. [Google Scholar]
- 25.Hashimoto H., Nelson,M.G. and Switzer,C. (1993) Zwitterionic DNA. J. Am. Chem. Soc., 115, 7128–7134. [Google Scholar]
- 26.Ozaki H., Nakamura,A., Arai,M., Endo,M. and Sawai,H. (1995) Novel C5-substituted 2′-deoxyuridine derivatives bearing amino-linker arms: synthesis, incorporation into oligodeoxyribonucleotides and their hybridization properties. Bull. Chem. Soc. Jpn, 68, 1981–1987. [Google Scholar]
- 27.Ono A., Dan,A. and Matsuda,A. (1993) Synthesis of oligonucleotides carrying linker groups at the 1′-position of sugar residues. Bioconjug. Chem., 4, 499–508. [DOI] [PubMed] [Google Scholar]
- 28.Ono A., Haginoya,N., Kiyokawa,M., Minakawa,N. and Matsuda,A. (1994) A novel and convenient post-synthetic modification method for the synthesis of oligodeoxyribonucleotides carrying amino linkers at the 5-position of 2′-deoxyuridine. Bioorg. Med. Chem. Lett., 4, 361–366. [Google Scholar]
- 29.Haginoya N., Ono,A., Nomura,Y., Ueno,Y. and Matsuda,A. (1997) Synthesis of oligodeoxyribonucleotides containing 5-(N-aminoalkyl)carbamoyl-2′-deoxyuridines by a new postsynthetic modification method and theri thermal stability and nuclease-resistance properties. Bioconjug. Chem., 8, 271–280. [DOI] [PubMed] [Google Scholar]
- 30.Ueno Y., Kumagai,I., Haginoya,N. and Matsuda,A. (1997) Effects of 5-(N-aminohexyl)carbamoyl-2′-deoxyuridine on endonuclease stability and an ability of oligodeoxynucleotide to activate RNase H. Nucleic Acids Res., 25, 3777–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueno Y., Mikawa,M. and Matsuda,A. (1998) Synthesis and properties of oligodeoxynucleotides containing 5-[N-[2-[N,N-bis(2-aminoethyl)amino]ethyl]carbamoyl]-2′-deoxyuridine and 5-[N-[3-[N,N-bis(3-aminopropyl)amino]propyl]carbamoyl]-2′-deoxyuridine. Bioconjug. Chem., 9, 33–39. [DOI] [PubMed] [Google Scholar]
- 32.Kanazaki M., Ueno,Y., Shuto,S. and Matsuda,A. (2000) Highly nuclease-resistant phosphodiester-type oligodeoxynucleotides containing 4′α-C-aminoalkylthymidines form thermally stable duplexes with DNA and RNA. A candidate for potent antisense molecules. J. Am. Chem. Soc., 122, 2422–2432. [Google Scholar]
- 33.Inoue H., Hayase,Y., Iwai,S. and Ohtsuka,E. (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotides splints and RNase H. FEBS Lett., 215, 327–330. [DOI] [PubMed] [Google Scholar]
- 34.Inoue H., Hayase,Y., Imura,A., Iwai,S., Miura,K. and Ohtsuka,E. (1987) Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res., 15, 6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesnik E.A., Guinosso,C.J., Kawasaki,A.M., Sasmor,H., Zounes,M., Cummins,L.L., Ecker,D.J., Cook,P.D. and Freier,S.M. (1993) Oligodeoxynucleosides containing 2′-O-modified adenosine: synthesis and effects on stability of DNA:RNA duplexes. Biochemistry, 32, 7832–7838. [DOI] [PubMed] [Google Scholar]
- 36.Gait M.J. (ed.) (1984) Oligonucleotides Synthesis: A Practical Approach. IRL Press, Oxford, UK.
- 37.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor University Press, Cold Spring Harbor, NY.
- 38.Asakura J. and Robbins,M.J. (1990) Cerium (IV)-mediated halogenation at C-5 of uracil derivatives. J. Org. Chem., 55, 4928–4933. [Google Scholar]
- 39.Nishizono N., Sumita,Y., Ueno,Y. and Matsuda,A. (1998) Effects of 2′-O-(trifluoromethyl)adenosine on oligodeoxynucleotide hybridization and nuclease stability. Nucleic Acids Res., 26, 5067–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seio K., Wada,T., Sakamoto,K., Yokoyama,S. and Sekine,M. (1996) Chemical synthesis and conformational properties of a new cyclouridylic acid having an ethylene bridge between the uracil 5-position and 5′-phosphate group. J. Org. Chem., 61, 1500–1504. [Google Scholar]
- 41.Wu H., Lima,W.F. and Crooke,S.T. (1999) Properties of cloned and expressed human RNase H1. J. Biol. Chem., 274, 28270–28278. [DOI] [PubMed] [Google Scholar]
- 42.Nomura M., Shuto,S. and Matsuda,A. (2003) Synthesis of the cyclic and acyclic acetal derivatives of 1-(3-C-ethynyl-β-d-ribo-pentofuranosyl)cytosine, a potent antitumor nucleoside. Design of prodrugs to be selectively activated in tumor tissues via a bioreduction—hydrolysis mechanism. Bioorg. Med. Chem. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.