Abstract
To help maintain a complete, accurate and timely Problem List, we are developing a system to automatically retrieve medical problems from free-text documents. This system uses Natural Language Processing to analyze all electronic narrative text documents in a patient’s record. Here we evaluate and compare 3 different applications of NLP technology in our system: the first using MMTx (MetaMap Transfer) with a negation detection algorithm (NegEx), the second using an alpha version of a locally developed NLP application called MPLUS2, and the third using keyword searching. They were adapted and trained to extract medical problems from a set of 80 problems of diagnosis type. The version using MMTx and NegEx was improved by adding some disambiguation and modifying the negation detection algorithm, and these modifications significantly improved recall and precision. The different versions of the NLP module were compared, and showed the following recall / precision results: standard MMTx with NegEx version 0.775 / 0.398; improved MMTx with NegEx version 0.892 / 0.753; MPLUS2 version 0.693 / 0.402; and keyword searching version 0.575 / 0.807. Average results for the reviewers were a recall of 0.788 and a precision of 0.912.
Introduction
A longitudinal Electronic Medical Record (EMR) is being developed in our institution, with the Problem List as a key component. The latter document is already linked to direct medical knowledge access (i.e. “Infobutton”1), and will be linked to automated data collection forms and order entry. To serve these functions, the Problem List has to be as accurate, complete and timely as possible. But in our institution, this document is usually incomplete and inaccurate, and is often totally unused, especially in the inpatient domain. To address this deficiency, we developed an application using Natural Language Processing (NLP) to harvest potential Problem List entries from the multiple free-text electronic documents available in our EMR. These proposed medical problems drive an application designed for management of the Problem List. Within this application, they are proposed to the physicians for addition to the official (actual) Problem List. The global aim of our project is to automate the process of creating and maintaining a Problem List for hospitalized patients and thereby to help to guarantee the timeliness, accuracy and completeness of this information. This Automated Problem List system is made of two components: a background application that does all the document processing and analysis, and a foreground application that is the Problem List management application. In this experiment, the NLP part of the background application was evaluated, as it looked for 80 different medical problems. These problems were diagnostic in nature, and were selected based on their frequency of use in the field of evaluation (cardiovascular).
Background
A substantial part of the medical record is made of free-text documents that represent patient history and reports of therapeutic interventions or clinical progress2. Decision-support, research, the optimization of database operations, and improvement in medical administration create a need for coded data instead. As a possible answer to this issue, NLP can convert free text into coded data.
Techniques for automatically encoding textual documents from the medical record have been evaluated by several groups. Examples are the Linguistic String Project3, and MedLEE (Medical Language Extraction and Encoding system)4. MedLEE has been recently adapted to extract UMLS concepts from medical text documents, achieving 83% recall and 89% precision5. Other systems automatically mapping clinical text concepts to standardized vocabularies have been reported, like MetaMap6. MetaMap and its Java™ version called MMTx (MetaMap Transfer) were developed by the U.S. National Library of Medicine (NLM). They are used to index text or to map concepts in the analyzed text to UMLS concepts. The mapped concepts returned by the program are ranked, but no negation detection is performed. MetaMap has been shown to identify most concepts present in MEDLINE titles7. It has been used for Information Retrieval8, Information Extraction in biomedical text9 and patient’s electronic messages10, and was shown to extract the most critical findings in 91% of the pathology reports in a study by Shadow et al.11.
Independent negation detection is required when using MMTx, because of its lack of discrimination between present and absent concepts (i.e. negated concepts). In the medical domain, this is important due to the fact that findings and diseases are often described as absent. A few negation detection algorithms have been developed, like NegEx, a computationally simple algorithm using regular expressions, with a sensitivity of 94.5% and a specificity of 77.8% 12.
NLP has been a topic of interest for our Medical Informatics group at the LDS Hospital and the University of Utah (Salt Lake City, Utah) for a number of years. SPRUS (Special Purpose Radiology Understanding System)13 was the first NLP application developed by our group. Later came SymText (Symbolic Text processor)14 and MPLUS15. The latter provides syntactic analysis based on a context-free grammar with a bottom-up chart parser, interleaved with object-oriented semantic analysis using semantic networks. The networks used are Bayesian networks (also called belief networks)16, trained to infer probabilistic relationships between extracted terms and their meaning. This approach has the advantages of being tolerant to noisy data, and of allowing training to refine performances. The alpha version used here and called MPLUS2 is under development and is a recent redesign created to take advantage of a new, accelerated syntactic parsing algorithm. Its goal is to achieve a faster parse time. For the project described in this paper, MPLUS2 has been adapted and trained to extract medical problems from electronic free-text documents17.
Methods
For this experiment, four different versions of the NLP module in the background application were used: two versions using MMTx and NegEx (one with no disambiguation and one with some disambiguation), one version using the alpha version of MPLUS2, and a last version using a keyword search algorithm.
NLP module using MMTx and NegEx
Since we were only interested in 80 different medical problems, and not in the whole UMLS Metathesaurus content, we created a subset of the Metathesaurus adapted to our system. The selection process resulted in a reduction to about 0.25% of the original data set, from more than a million to about 2,500 concepts. This reduction made the NLP module more than 3 times faster, and also made it more accurate. For negation detection, we used an algorithm called NegEx12. We used its improved version called NegEx218. The algorithm was implemented as a Java method connected to a database table listing all negation and conjunction words or phrases.
After a first run of the MMTx/NegEx version without disambiguation, we analyzed all errors. About two thirds of the errors were created by MMTx, and the other third by NegEx. A small portion of MMTx’s errors were linked to a lack of general context, like when detecting cardiac arrest in the description of a heart surgery procedure using cardioplegia, or when detecting problems when risks for surgery discussed with the patient were listed. The lack of local context resulted in errors like detecting mental depression in “ST segment depression”, detecting coma in “Glasgow coma score”, or pulmonary emphysema in “subcutaneous emphysema”. A larger portion of MMTx errors were related to ambiguous acronyms, like angina pectoris detected in “AP chest x-ray”, peptic ulcer detected with “GU”, aortic stenosis detected with “as”, diabetes mellitus detected with “B-mode imaging”, mitral regurgitation with “Mr.”, mental depression with “M.D.”, or pulmonary emphysema with “EMS transported the patient…”. A large proportion of NegEx errors were due to the limited size of the window in which concepts are eventually negated. For example, in the sentence “She denies cerebrovascular accident, renal problems, bleeding problems, or significant varicose veins”, denies would be detected as the negation term and varicose veins would be missed for negation, because the window of 6 words after the negation term would not include it.
To reduce the errors cited above, some disambiguation was added to MMTx. Ambiguous acronyms were replaced by their corresponding full term before passing the sentence to MMTx, and additional disambiguation was done on MMTx’s output. For example, when cardiac arrest was detected in a sentence, and cardioplegia or cardioplegic was present in the same sentence, then the detected cardiac arrest was considered absent (i.e. not a real problem). To reduce some of NegEx’s errors, the size of the window around a negation phrase was made variable, instead of limited to 6 words. It was automatically extended to the end of the sentence or to the next negation or conjunction phrase if closer.
NLP module using MPLUS2
Another version of this tool used the locally developed NLP application called MPLUS2. This application is trainable for different contexts and the semantic part was adapted to accommodate the clinical documents and medical concepts necessary to identify medical problems (e.g. history and physical, surgery report, consultation note, etc…). Training for this tool applies principally to the semantic model. Its semantics are represented as collections of Bayesian Networks representing the relationship between the words and phrases in a sentence and the concepts that they represent. Once the structure of the network is defined, the relationships among the semantic elements are captured as tables of probabilities. The Bayesian Network used by our application was made of 11 nodes and is displayed below (Figure 1), with nodes corresponding to the word(s) or phrase(s) in the text at the bottom, and related concepts at the top.
Figure 1.
Bayesian Network with example values for each node when analyzing the sentence “The patient presents with shortness of breath” in the “History of Present Illness” section of a “Consultation Note”.
To capture these relationships, training cases are needed. We created those training cases using a semiautomated technique. First an application was used to automatically select relevant sentences from a set of 91,483 sentences extracted from 3,000 randomly selected free-text documents (different from the test set) and to create about 1,500 training cases. It used pattern recognition to find sentences with word(s) or phrase(s) recognized as possibly expressing one of the 80 problems we were interested in. The recognized phrase and its corresponding problem, along with the document type, the section title, and the sentence text were then proposed to a human reviewer who extracted the words or sub-phrase expressing the state of the problem, and added the state of the problem (present, possible, or absent) to the case. The different components of the phrase expressing the problem were also extracted (main term and modifiers). The training cases were finally created using the information extracted. Training cases were formatted in a tab-delimited format as required by Netica19, the Bayesian Networks processing application used by MPLUS2. After the training, the tables in the NLP application’s Bayesian Networks contained a statistical representation of the relationships between the words and phrases in the sentences and the problems that we had identified for extraction.
NLP module using keyword search
The keyword search version was similar to the MMTx/NegEx version, except that MMTx was replaced by a pattern matching algorithm coupled to a keyword table. NegEx was therefore also used. The keyword table was composed of phrases and their corresponding UMLS CUI (Concept Unique Identifier). It was built using the UMLS Metathesaurus MRCONSO table and a manually built table linking the 80 selected concepts with all related subconcepts (e.g. Right Bundle Branch Block was mapped to Incomplete Right Bundle Branch Block, Complete Right Bundle Branch Block, and Other or unspecified Right Bundle Branch Block). All phrases corresponding to the 80 concepts and their subconcepts were first retrieved from the MRCONSO table (6,928 phrases). We then did some filtering, removing all phrases containing brackets, angled brackets, commas, forward slashes, squared brackets, dashes, and the words NOS or unspecified (e.g. “Anemia unspecified”, “Anemia <1>”, “Anemia (disorder)”, “Anemia, aplastic”, etc.). After removal of duplicates, the final table contained 4,570 keywords.
Results
The set of test documents was made of 160 randomly selected clinical documents of different types (discharge summaries, radiology reports, pathology reports, progress notes, etc.) Each document was processed four times: once with the keyword search version, once with each MMTx/NegEx version, and once with the MPLUS2 version. A reference standard was created by having each document read by two physician reviewers, and by a third if the two disagreed. Reviewers selected the problems present in the document from our list of 80 medical problems. Reviewers’ overall agreement was almost perfect, with a Cohen’s kappa of 0.9 and a Finn’s R of 0.985 when building the reference standard.
Four standard measures to evaluate the accuracy of a NLP system were used. The first two are the most common: precision (equivalent to positive predictive value here; Equation 1) and recall (equivalent to sensitivity or true positive rate here; Equation 2). Another typical value combining precision and recall – the F-measure (Equation 3) – was also calculated. A β of 1 gives equal weight to precision and recall, and a β higher than 1 gives more weight to the recall. Finally, fallout (equivalent to false positive rate here; Equation 4) was also calculated.
| (1) |
| (2) |
| (3) |
| (4) |
To calculate these values, problems were counted and categorized as correct (COR; concept present in the document and found by NLP), spurious (SPU; concept found by NLP but absent from the document), missing (MIS; concept present in the document but not found by NLP), or noncommittal (NON; concept absent from the document and not found by NLP). Correct problems are true positives, spurious problems are false positives, missing problems are false negatives, and noncommittal problems are true negatives. A medical problem was considered present if mentioned in the text as probable or certain in the present or the past (e.g. “the patient has asthma”; “past history positive for asthma”; “pulmonary edema is probable”), and considered absent if negated in the text or not mentioned at all (e.g. “this test excluded diabetes…”, “he denies any asthma”).
Recall, precision, and fallout were measured and mean and 0.95 confidence intervals were computed. The F-measure was calculated with a β of 2, to give more importance to the recall, the most important feature for our system (Table 1). To support the completeness of the Problem List, our aim is to detect as many medical problems present as possible.
Table 1.
Evaluation results with means and 95% confidence intervals
| Measure | MMTx1 | MMTx2 | MPLUS2 | Reviewers | Keyword |
|---|---|---|---|---|---|
| Recall | 0.775 (0.718–0.833) | 0.892 (0.848–0.934) | 0.693 (0.632–0.755) | 0.788 (0.748–0.827) | 0.575 (0.51–0.64) |
| Precision | 0.398 (0.346–0.45) | 0.753 (0.695–0.81) | 0.402 (0.344–0.459) | 0.912 (0.883–0.94) | 0.807 (0.744–0.87) |
| F-measure (β=2) | 0.652 | 0.86 | 0.605 | 0.845 | 0.61 |
| Fallout | 0.0284 (0.0252–0.0316) | 0.01 (0.0078–0.0128) | 0.039 (0.032–0.046) | 0.001 (0.001–0.002) | 0.0045 (0.0031–0.006) |
| Time (in seconds) | 72.3 | 54.7 | 39 | 132.2 | 1.9 |
MMTx1 : MMTx with no disambiguation & original NegEx
MMTx2 : MMTx with disambiguation & improved NegEx
Time: Average processing time per document
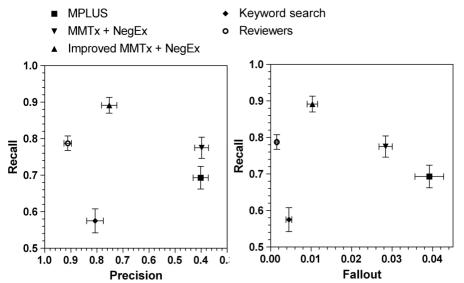
The recall and precision results are represented on a recall - precision graph with mean values and 95% confidence intervals (Figure 1). The true positive rate (equivalent to recall here) - false positive rate (equivalent to fallout here) graph (Figure 1) – also known as ROC graph (Receiver Operating Characteristic) – gives a result very similar to the recall-precision graph.
Figure 1.
Recall-Precision/Fallout graphs
Statistical analysis of the results showed that the revisions of MMTx and NegEx significantly improved recall and precision, and reduced fallout. All differences between the other versions were significant, if not stated otherwise. The alpha version of MPLUS2 had a recall between the keyword search and MMTx/NegEx, but had a precision not different than the standard MMTx/NegEx. Its F-measure was almost identical to the keyword search’s F-measure. The keyword search version of our NLP tool had a lower recall than both MMTx/NegEx and MPLUS2, but had a higher precision and lower fallout. Human reviewers had the highest precision and lowest fallout, but had a recall lower than the improved MMTx/NegEx, and not different than the standard MMTx/NegEx version. The test used for these analyses was Mann-Whitney U-statistic, for non-normality reasons
Discussion
This evaluation showed that our system using MMTx with a custom data subset and disambiguation, and NegEx with a flexible negation window has good recall and satisfying precision. Those measures fulfill our requirements for the NLP module of our Automated Problem List system in a clinical setting. A sufficient recall is required to significantly improve the quality of the Problem List, and a sufficient precision is desirable to avoid proposing a group of problems overloaded with false positives. This study also showed that improvements to MMTx and NegEx improved recall and precision. A simple keyword search algorithm with NegEx instead of MMTx/NegEx significantly lowered the recall, but also reduced the fallout and increased the precision. This recall was clearly insufficient for clinical use of our system, but may be improved by enhancing the keywords table. This version was by far the fastest.
When the alpha version of MPLUS2 was used, recall was lower than MMTx/NegEx and precision much lower than the improved MMTx/NegEx version. These results have to be considered with caution, since the probabilistic semantic analysis of MPLUS2 would clearly need more training, and the probability threshold to consider a problem as present was 0.5 in this evaluation; but this could be set higher to improve precision. Also, we used the latest version of MPLUS2 that is still at an early stage of its development. The ancestor of MPLUS2 and MPLUS – SymText – showed a recall of 0.92 and a precision of 0.94 when extracting pneumonia concepts from radiology reports20, but was too slow for clinical use in our Automated Problem List system. And, finally, human reviewers were much slower than NLP, but had the highest precision, with a lower recall than MMTx/NegEx.
Results of our MMTx/NegEx versions compare favorably with another evaluation of MMTx, where a recall of 53% was reported; however, this result has to be considered cautiously because of small sample size and other reasons21. Our system only extracted a limited set of concepts, and all children of those concepts were matched to the parent ones, therefore improving the recall. In a similar task, MedLEE has been evaluated when extracting UMLS concepts from medical text documents, and achieved 83% recall and 89% precision5.
To improve the generalizability and reduce biases in this evaluation, we attempted to follow published criteria for effective evaluation of NLP systems22. Most criteria listed in the cited publication were respected. The exception was the fact that the developer of the system also designed and led its evaluation. To minimize this issue, documents were randomly selected after the system was frozen for evaluation, and reviewers did their task fully independently. Data collection was fully automated and reviewers were blinded, to avoid assessment biases. Measures were all standardized and mostly highly automated. A recruitment bias was avoided by clearly defining the inclusion/exclusion criteria of patients to randomly select test documents from.
The sample size was sufficient to show significant differences between the different versions of the NLP module.
Finally, this evaluation gave us the information required to select the most appropriate NLP module for clinical use of our Automated Problem List system.
Acknowledgments
We would like to thank Lee Christensen for his work developing and helping to train MPLUS2. This research was supported by a Deseret Foundation (Salt Lake City, Utah) Grant (#444).
References
- 1.Reichert JC, Glasgow M, Narus SP, Clayton PD. Using LOINC to link an EMR to the pertinent paragraph in a structured reference knowledge base. Proc AMIA Symp. 2002:652–6. [PMC free article] [PubMed] [Google Scholar]
- 2.Pratt AW. Medicine, Computers, and Linguistics. Advanced Biomedical Engineering. 1973;3:97–140. [Google Scholar]
- 3.Chi E, Lyman M, Sager N, Friedman C. Database of computer-structured narrative: methods of computing complex relations. In: IEEE, editor. SCAMC 85; 1985; 1985. p. 221–226.
- 4.Friedman C, Johnson SB, Forman B, Starren J. Architectural requirements for a multipurpose natural language processor in the clinical environment. Proc Annu Symp Comput Appl Med Care. 1995:347–51. [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated Encoding of Clinical Documents Based on Natural Language Processing. J Am Med Inform Assoc 2004. [DOI] [PMC free article] [PubMed]
- 6.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 7.Pratt W, Yetisgen-Yildiz M. A Study of Biomedical Concept Identification: MetaMap vs. People. Proc AMIA Symp. 2003:529–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Aronson AR. Query expansion using the UMLS Metathesaurus. In: Proc AMIA Symp; 1997; 1997. p. 485–9. [PMC free article] [PubMed]
- 9.Weeber M, Klein H, Aronson AR, Mork JG, de Jongvan den Berg LT, Vos R. Text-based discovery in biomedicine: the architecture of the DAD-system. Proc AMIA Symp. 2000:903–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan PF, Aronson AR. Towards linking patients and clinical information: detecting UMLS concepts in e-mail. J Biomed Inform. 2003;36(4–5):334–341. doi: 10.1016/j.jbi.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Shadow G, McDonald C. Extracting structured information from free text pathology reports. In: Proc AMIA Symp; 2003; Washington, DC; 2003. p. 584–588. [PMC free article] [PubMed]
- 12.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 13.Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Work in progress. Radiology. 1990;174(2):543–8. doi: 10.1148/radiology.174.2.2404321. [DOI] [PubMed] [Google Scholar]
- 14.Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proc Annu Symp Comput Appl Med Care. 1994:247–51. [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen L, Haug P, Fiszman M. MPLUS: a probabilistic medical language understanding system. Proceedings of the Workshop on Natural Language Processing in the Biomedical Domain. 2002:29–36. [Google Scholar]
- 16.Jensen F. An Introduction to Bayesian Networks: Springer Verlag; 1996.
- 17.Meystre S, Haug PJ. Medical problem and document model for natural language understanding. Proc AMIA Symp. 2003:455–9. [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman WW. NegEx 2. (http://web.cbmi.pitt.edu/chapman/NegEx.html).
- 19.Norsys Software Corp. Netica™ Application. (http://www.norsys.com/netica.html)
- 20.Fiszman M, Chapman WW, Evans SR, Haug PJ. Automatic identification of pneumonia related concepts on chest x-ray reports. Proc AMIA Symp. 1999:67–71. [PMC free article] [PubMed] [Google Scholar]
- 21.Divita G, Tse T, Roth L. Failure Analysis of MetaMap Transfer (MMTx) Medinfo. 2004;2004:763–7. [PubMed] [Google Scholar]
- 22.Friedman C, Hripcsak G. Evaluating natural language processors in the clinical domain. Methods Inf Med. 1998;37(4–5):334–44. [PubMed] [Google Scholar]