Abstract
Information overload is a significant problem for modern medicine. Searching MEDLINE for common topics often retrieves more relevant documents than users can review. Therefore, we must identify documents that are not only relevant, but also important. Our system ranks articles using citation counts and the PageRank algorithm, incorporating data from the Science Citation Index. However, citation data is usually incomplete. Therefore, we explore the relationship between the quantity of citation information available to the system and the quality of the result ranking. Specifically, we test the ability of citation count and PageRank to identify “important articles” as defined by experts from large result sets with decreasing citation information. We found that PageRank performs better than simple citation counts, but both algorithms are surprisingly robust to information loss. We conclude that even an incomplete citation database is likely to be effective for importance ranking.
Introduction
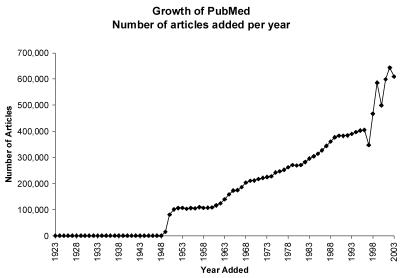
MEDLINE, the premier bibliographic database of biomedical literature, is growing at an accelerating rate. In 2003, over 600,000 new articles were indexed into PubMed, the National Library of Medicine’s interface onto MEDLINE (Figure 1) which currently contains over 15 million entries [1]. To illustrate the magnitude of this avalanche, if only 1% of the new articles are relevant to a family physician, he should read an average of over 16 new articles every single day of the year. However, even 10% of this rate is difficult for a practicing clinician. Therefore, we must help users focus on the “must read” articles.
Figure 1.
Growth of PubMed measured in articles added per year (data retrieved from PubMed itself)
Synthetic literature is the current, partial solution to the problem of information overload. It includes review articles, books, guidelines, annotated bibliographies, meta-analysis and, in general, literature based on other literature. Unfortunately, synthetic literature is far from perfect as the review process takes time, energy, and funds. Therefore, it is not surprising that this literature often lags behind the most current state of knowledge [2].
Traditional information retrieval systems seek to return results relevant to a particular query. A problem with this strategy is that there are simply too many relevant results for common queries. For example, a search for “heart attack” retrieved 101,163 results on January 4, 2005. MEDLINE does not have indicators of article quality, and PubMed presents results in (approximate) reverse chronological order [3]. This does not help users identify the “must read” articles. Future information retrieval systems should identify results that are important as well as relevant.
The concept of relevance is very loosely defined [4]. It usually stands for “pertinence to the matter at hand.” [5] Importance is also a poorly-defined concept, but may be thought of as information that is highly valuable to the user or the field. Just as with relevance, experts can legitimately disagree regarding the relative importance of a given article. There is no absolute gold standard measure of importance, but some proxies for importance can be agreed upon. Certain journals, for example, are highly regarded, and articles published in them will garner attention.
Citation counts are among the accepted measures of article quality and importance within the scientific community [6]. An article that has been cited many times is thought to be more important than an article that has never been cited. However, not all authors share this view [7].
Citation-based importance measures are also attractive not only because of their natural acceptance in the biomedical sciences, but because they are successful in the World Wide Web (WWW).
There are structural similarities between the WWW and the biomedical literature. A simple model of the WWW describes web pages as nodes and hyperlinks as directional links between them. Articles and citations from one article to another in the biomedical literature can be thought of as nodes and links, respectively. Link analysis has been used to build successful WWW search engines such as Google [8]. These structural similarities suggest that information retrieval techniques from the WWW could be applied to biomedical literature with useful results.
The PageRank algorithm is Google’s measure of importance [9]. It leverages citations, but includes the importance of the citing article as a factor in the calculation. A technical discussion of PageRank is beyond the scope of this paper, and the interested reader is referred to [8] and [9] for an introduction.
However, citation analysis poses at least two significant technical challenges. First, maintaining a database of citations from one article (or one web site) to another is difficult. No database is likely to be complete; the appearance of citations lags behind the publication of a paper by months or even years. Second, combining two dissimilar products like PubMed and a citation database introduces another set of problems. A custom mapping layer between them has to be developed, and assessing its performance is non-trivial. Differences in journal names and record formats make finding corresponding articles difficult, even by hand. Therefore, not all valid citations are recognized by the system.
This paper describes a set of experiments to determine whether incomplete citation sets are useful for ranking PubMed results. Specifically, we evaluated the impact of citation loss on the performance of two importance ranking algorithms: PageRank and simple citation counts.
Materials and methods
To evaluate the retrieval of important articles we used the Society of Surgical Oncology’s Annotated Bibliography (SSO-AB), second edition [10]. The SSO-AB contains articles about ten solid tumors. These articles were identified as important by a panel of experts from the Society. We performed one PubMed query for each topic and ranked the results using PageRank and simple citation counts, repeating the experiment with decreasing citation sets. Each PubMed query (Table 1) was designed to retrieve a relatively large result set to simulate a naïve user searching for general information on a topic.
Table 1.
PubMed queries used to retrieve the SSO-AB articles
| SSO topic | PubMed query |
|---|---|
| Breast cancer | breast cancer |
| Gastric cancer | gastric cancer |
| Colorectal cancer | (colon OR rectal) AND cancer |
| Endocrine cancer | (((thyroid OR adrenal OR parathyroid OR islet cell")) AND cancer) OR pheochromocytoma OR insulinoma OR carcinoid OR gastrinoma |
| Esophageal cancer | esophageal cancer |
| Hepatobiliary cancer | (hepatocellular OR biliary) AND cancer |
| Lung cancer | lung cancer |
| Melanoma | melanoma |
| Pancreatic cancer | pancreas cancer |
| Soft tissue sarcoma | soft tissue sarcoma |
We have designed and implemented a system that allows us to explore novel information retrieval strategies for biomedical literature (Figure 2). The system maintains a local, automatically updated copy of PubMed and a partial copy of the Science Citation Index (SCI) [11]. The Science Citation Index is a database of citations from one article to another. The retrieval step uses PubMed but ranking is performed locally (Figure 2) using different algorithms. The two algorithms currently implemented are a simple citation count, and PageRank. A WWW front end is available within the local network so that users may consult the system and provide feedback.
Figure 2.
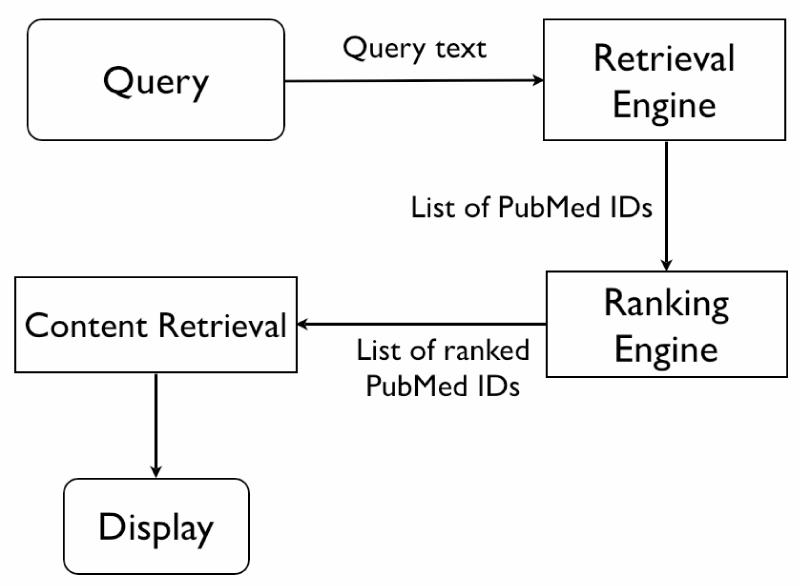
Basic system architecture. A query is passed to PubMed (Retrieval Engine) for processing. The original ordering is discarded and a new ranking is computed locally. The full PubMed entries are retrieved from a local store (Content Retrieval) and displayed.
A mapping layer that maps the SCI onto PubMed was created. The mapping layer allows us to obtain the number of times that a particular article was cited and which articles did the citing.
Queries were performed using the BioPython v1.30 (http://www.biopython.org) module's PubMed access facilities for the Python programming language. All returned PubMed IDs were stored locally. A copy of PubMed as of November 30, 2004 was used for the experiments. The citation data was from the SCI, 1999–2004, third quarter update.
Two algorithms, simple citation count and PageRank, were tested for robustness. A separate data set for each algorithm was generated by starting with the original dataset and randomly deleting 10, 20, 30, 40, 50, 60, 70, 80, 90, and 99% of citations to generate 10 new citation sets.
Each of the resulting citation sets plus the original was used to compute scores and rank the PubMed IDs previously retrieved for each of the ten queries. The PageRank algorithm was computed using a custom Python script according to the details published in [8] and [9]; d was set to 0.85 and 100 iterations were performed. The citation count for each document was the number of citations that could be found for that document. The position of the SSO-AB documents within the ranked result sets was determined and standard 11 step recall/precision curves were generated for each of the ten queries and averaged to give a single curve for each dataset.
Results
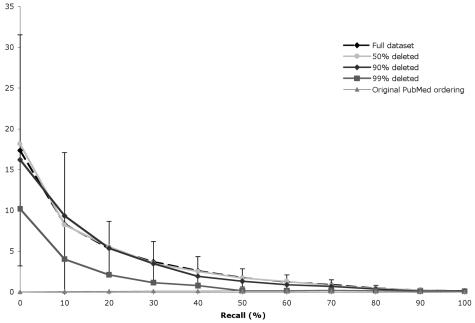
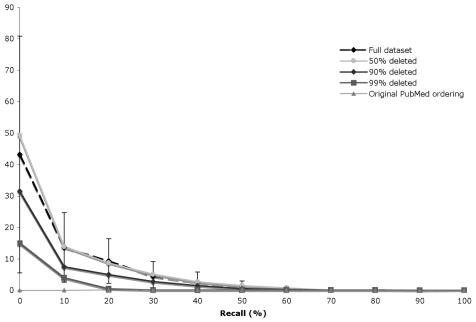
Figures 3 and 4 show standard 11 level recall/precision curves for the simple citation count and PageRank respectively. The curves for 10%-40% and 60%-80% overlap the “full dataset” and 50% curves in both cases, and have been omitted to improve readability. PubMed’s performance is included for reference. Error bars represent ± 1 standard deviation.
Figure 3.
Recall/precision curves for the simple citation count with progressively smaller datasets (intermediate curves omitted for clarity)
Figure 4.
Recall/precision curves for the PageRank algorithm with progressively smaller datasets (intermediate curves omitted for clarity)
Discussion
Surprisingly, we found almost no degradation in the performance of either algorithm, even when removing 90% of the citations. Removing 99% of the citations results in a lower recall/precision curve, but not enough to be statistically significant. These results suggest that even a poor citation set is sufficient for importance ranking. We also found that PageRank is better than the simple citation count at identifying important articles.
Our results suggest that, after a certain amount of citation information, rankings tend to stabilize. While more information is desirable, it does not appear to be indispensable. This mirrors the way recommendations work in the real world; after enough endorsements from authority figures, an article may be judged important regardless of how many extra recommendations it receives.
There are several important limitations that must be considered. First, the experiments were conducted in the domain of surgical oncology. Although the SSO-AB covers ten topics within surgical oncology, most of MEDLINE is outside its scope. While we believe that the results for other topics should be similar, this has yet to be demonstrated. One of the challenges in generalizing our experiments is the absence of a MEDLINE test collection that identifies important, as opposed to relevant, articles. However, we plan future experiments using other collections, such as the American College of Physicians (ACP) Journal Club, which is a larger and more general collection of important articles in internal medicine updated every two months (http://www.acpjc.org/).
The second important limitation is citation lag. A crucial paper published today will have no citations for some time. Therefore, it will be mistakenly excluded from the top of the results. Potential solutions include using other values that approximate importance, such as journal impact factor or calculating expected citation counts.
Different users may have different information needs. We believe that these algorithms will be more useful to reviewers, students, and general practitioners than to researchers looking for the latest information. A potential solution may be to adjust the search behavior based on the user’s self-reported role (e.g., student versus researcher).
To our knowledge, this is the first use of the SSO-AB as a reference collection. However, it was compiled by highly regarded experts in the field with no intention of building an information retrieval test collection. Therefore, the corpus is not likely to be biased in favor of any specific information retrieval strategy. All articles are available on PubMed. The current SSO-AB edition was published in 2001, making it old enough to be cited, but not old enough to be irrelevant. Future experiments will include other “naturally occurring” test collections like the ACP Journal Club which will allow us to evaluate the practical importance of citation lag, as well as to generalize our results.
Conclusions
Information overload requires information retrieval systems to identify important, as well as relevant, documents. Citation analysis appears to be a promising way to prioritize relevant MEDLINE articles retrieved in response to general queries. However, citation databases are difficult to build and maintain. Therefore, even the best databases are not likely to be complete. We found that simple citation count and PageRank seem to perform well even in the setting of very incomplete citation data. We believe that our results should encourage the use of small citation sets for importance ranking research.
Acknowledgements
Supported in part by NLM grant 5 K22 LM008306 and a training fellowship from the W. M. Keck Foundation to the Gulf Coast Consortia through the Keck Center for Computational and Structural Biology. The authors are also grateful to Thomson-ISI for granting use of the Science Citation Index for research purposes.
References
- 1.US National Library of Medicine [homepage on the Internet]. PubMed Milestone - 15 Millionth Journal Citation [published July 7, 2004; cited January 15, 2005]. NLM Technical Bulletin. Available from: http://www.nlm.nih.gov/pubs/techbull/ja04/ja04_technote.html
- 2.Hersh W. Health and Biomedical Information. In: Hersh W, editor. Information Retrieval. New York: Springer; 2002. p. 22–82.
- 3.US National Library of Medicine [homepage on the Internet]. PubMed Help - Display Order [updated March 11, 2005; cited March 16, 2005]. Available from: http://www.ncbi.nlm.nih.gov/entrez/query/static/help/pmhelp.html#DisplayOrder
- 4.Saracevic, T. Information science: Integration in perspectives. In: Ingwersen P, Pors NO editors. Proceedings of the Second Conference on Conceptions of Library and Information Science (CoLIS 2); 14–17 Oct. 1996; Copenhagen (Denmark). Copenhagen: The Royal School of Librarianship; 1996. p. 201–218.
- 5.Dictionary.com [database on the Internet]. The American Heritage® Dictionary of the English Language, Fourth Edition. Houghton Mifflin Company; 2000 [cited March 15, 2005]. Relevance. Available from: http://dictionary.reference.com/search?q=relevance
- 6.Lee KP, Schotland M, Bacchetti P, Bero LA. Association of journal quality indicators with methodological quality of clinical research articles. JAMA. 2002 Jun 5;287(21):2805–8. doi: 10.1001/jama.287.21.2805. [DOI] [PubMed] [Google Scholar]
- 7.Walter G, Bloch S, Hunt G, Fisher K. Counting on citations: a flawed way to measure quality. Med J Aust. 2003 Mar 17;178(6):280–1. doi: 10.5694/j.1326-5377.2003.tb05196.x. [DOI] [PubMed] [Google Scholar]
- 8.Brin S, Page L. The Anatomy of a Large-Scale Hypertextual Web Search Engine. WWW7 / Computer Networks. 1998;30(1–7):107–117. [Google Scholar]
- 9.Page L, Brin S, Motwani R, Winograd T. The PageRank Citation Ranking: Bringing Order to the Web [monograph on the Internet]. Stanford University; 1999 [cited March 15, 2005]. Available from: http://dbpubs.stanford.edu/pub/1999–66
- 10.Pisters PWT, Edge SB editors. Surgical Oncology: Yesterday, Today and Tomorrow, an annotated bibliography of the important literature on common problems in surgical oncology [monograph on the Internet]. The Society of Surgical Oncology Inc.; 2001 [cited January 13, 2005]. Available from: http://www.surgonc.org/sso/biblio/biblio.htm
- 11.Thomson Scientific. Science Citation Index [CD-ROM]. Philadelphia: Thomson ISI; 1999–2004 Jan-Sept.