Abstract
The fast and reliable estimation of the genome sizes of various species would allow for a systematic analysis of many organisms and could reveal insights into evolutionary processes. Many methods for the estimation of genome sizes have already been described. The classical methods are based on the determination of the phosphate content in the DNA backbone of total DNA isolated from a defined number of cells or on reassociation kinetics of high molecular weight genomic DNA (c0t assay). More recent techniques employ DNA-specific fluorescent dyes in flow cytometry analysis, image analysis or absorption cytometry after Feulgen staining. The method presented here is based on the absolute quantification of genetic elements in a known amount (mass) of genomic DNA by real-time quantitative PCR. The method was evaluated on three different eukaryotic species, Saccharomyces cerevisiae (12.1 Mb), Xiphophorus maculatus (550 Mb) and Homo sapiens sapiens (2.9 Gb), and found to be fast, highly accurate and reliable.
INTRODUCTION
The genome size describes the DNA content in picograms per haploid genome and is often called the ‘C value’ or ‘Γ’, where it describes the number of base pairs.
The size of small genomes with up to only a few megabase pairs, e.g. the genomes of prokaryotes or unicellular eukaryotes, can be estimated by pulsed field gel electrophoresis, in which the lengths of restriction fragments or whole chromosomes are measured directly (1–3). In early biochemical analyses used for the determination of the C value, the phosphate content of DNA isolated from a defined number of cells was determined (4); the results tend to overestimate the genome sizes due to the detection of phosphate from other sources, especially from nuclear RNA and nucleotides. Another classical method is based on the analysis of reassociation kinetics, using hydroxyl apatite chromatography to separate single-stranded and double-stranded DNA (dsDNA) (5,6). With flow cytometry-based methods, the DNA content of individual nuclei is analyzed by fluorescence measurements after staining with propidium iodide (7–12) or ethidium bromide (13) or by absorbance photometry and image analysis after Feulgen staining (14). The latter methods require normalization using a reference, normally diploid human lymphocytes or chicken reticulocytes. The DNA contents per cell of these reference cells are believed to be 7.0 and 2.5 pg, respectively.
Here, we describe a method to determine the C value based on the absolute quantification of a single copy gene in a genomic DNA sample by real-time PCR (15–17). The amount (mass) of sample used in this analysis is determined by UV absorption spectrometry. The sample must contain pure DNA without significant contamination with RNA, which cannot be discriminated from DNA in the absorption spectrum. Precautions must be taken therefore to eliminate all RNA contamination before measuring the DNA concentration by UV absorption spectrometry. The C value can then be easily calculated by dividing the mass of sample DNA by the copy number determined for single-copy genes. The method was evaluated for three different eukaryotic species, the sequenced strain 368 FY1679 of Saccharomyces cerevisiae, the platyfish Xiphophorus maculatus and Homo sapiens sapiens.
MATERIALS AND METHODS
Biological samples
Cells from the sequenced strain of S.cerevisiae, no. 368 FY 1679, genotype MATa/α ura3-52/ura3-52 GAL2/GAL2 trp1-63/+ leu2-Δ1/+ his3-Δ200/+, were obtained from J.H. Hegemann (Institute of Microbiology, Heinrich-Heine-University, Düsseldorf, Germany). For the isolation of human blood lymphocyte DNA, blood was taken from healthy Caucasian individuals in 10 ml monovettes containing K-EDTA to avoid coagulation (Sarstedt, Nürnbrecht, Germany). Fish of the species X.maculatus were provided by A. Anders and J. Michel (Institute of Genetics, Justus-Liebig-University, Giessen, Germany).
DNA isolation
Genomic DNA of yeast was isolated from 3 ml of an overnight culture using the QIAamp DNA Mini Kit ‘Tissue’ (Qiagen, Hilden, Germany), following the instructions of the manufacturer, including an exhaustive RNase A (Qiagen) digestion step (incubation of 200 µl sample with 400 µg RNase for 10 min at 37°C) to degrade cellular RNA prior to the purification step on the QIAamp spin column, in which mono-, oligo- and poly(deoxy)ribonucleotides (<200 nt) are removed. DNA from human blood was isolated using the QIAamp DNA Mini Kit ‘Blood’, following the instructions of the manufacturer, also including an incubation with RNase A. The DNA of X.maculatus was isolated from the brain of the fish (anesthetized in ice water) using the QIAamp DNA Mini Kit ‘Tissue’, following the instructions of the manufacturer, again including an RNase A incubation step.
Quality and concentration of all DNA samples were determined by recording a UV absorption spectrum between 220 and 320 nm with a U3000 UV-VIS spectrophotometer (Hitachi/Colora, Lorch, Germany) using a Suprasil-Quarzglas microcuvette (Helma, Müllheim/Baden, Germany). A DNA solution with one A260, corrected for the baseline absorbance at 320 nm (A320), was taken to have a concentration of 50 µg ml–1 dsDNA. The purity was regarded as acceptable when the ratio A260:A280 was between 1.8 and 1.9. The shape of the spectrum was used to confirm the absence of light scattering particles and organic compounds which might impair the A260 value. One A260 unit was assumed to correspond to a DNA concentration of 50 µg ml–1.
Primers and probes
Primers were from MWG Biotech (Ebersberg, Germany). Hybridization probes were from TIB Molbiol (Berlin, Germany). The sequences of primers and probes are shown in Table 1.
Table 1. Sequences of primers and probes.
| Target | Name | Positiona | Length (nt) | Sequence 5′→3′ |
|---|---|---|---|---|
| rps3b | RPS3-F1 | 1285 | 19 | CGCTGACGGTGTCTTCTAC |
| RPS3-F2 | 1376 | 20 | CCAACCAAGACCGAAGTTAT | |
| RPS3-R1 | 1646 | 20 | CGGAAACAACAACTTCACAA | |
| RPS3-R2 | 1531 | 16 | GACAGCGGACAAACCA | |
| xp53c | xp53-F1 | 1010 | 19 | CGGTGCTTCGAGGTCCGTG |
| xp53-F2 | 1198 | 19 | GAGCGCTCCTGCTCCAGAT | |
| xp53-R1 | 1377 | 19 | CACAGATAACGATTACGGC | |
| xp53-R2 | 1267 | 19 | GAGAGTGTAAATCTCCTTG | |
| p53d | p53-F1 | 14513 | 21 | CGGCGCACAGAGGAAGAGAAT |
| p53-F2 | 14674 | 19 | TTCCTAGCACTGCCCAACA | |
| p53-R1 | 14834 | 21 | CAAATGCCCCAATTGCAGGTA | |
| p53-R2 | 14779 | 20 | GACTGGAAACTTTCCACTTG | |
| p53-FAM | 14707 | 34 | CCCCAGCCAAAGAAGAAACCACTGGATGGAGAAT-FAMe | |
| p53-RED | 14742 | 35 | RED-TTTCACCCTTCAGGTACTAAGTCTTGGGACCTCTT-pf |
aFirst nucleotide position of the primer sequence in the GenBank entry (see below).
bSaccharomyces cerevisiae ribosomal protein S3 gene (rps3), GenBank accession no. U34347.
cXiphophorus maculatus p53 gene, GenBank accession no. U34751.
dHomo sapiens sapiens p53 tumor suppressor gene, GenBank accession no. X54156.
eFAM, 6-carboxyfluorescein.
fRED, RED-640; p, 5′ phosphate group.
Synthesis and preparation of standard DNA
PCR products specific for rps3 of yeast, xp53 of X.maculatus and human p53 were generated by conventional PCR using the ‘outer’ primer pairs RPS3-F1/R1, XP53-F1/R1 and p53-F1/R1 (see Table 1), respectively, in a GeneAmp 2400 PCR system (Applied Biosystems, Weiterstadt, Germany) with the isolated genomic DNA as template. The generated PCR products contain the binding sites for the ‘inner’ primers of the specific targets used in the real-time PCR. The reaction mixtures contained 0.5 µM each forward and reverse primer (F1/R1, see Table 1), 200 µM each deoxynucleotide (dATP, dCTP, dGTP and dTTP), 0.5 U Taq DNA polymerase, and 20–50 ng genomic template DNA in a total volume of 25 µl 1× reaction buffer (10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.3 at 25°C). All reagents were purchased from Roche Diagnostics (Mannheim, Germany). The PCR protocol consisted of an initial denaturation step of 95°C for 5 min, followed by 35 amplification cycles with 30 s at 95°C, 30 s at 55°C and 1 min at 72°C. The absence of non-specific PCR products for all targets was confirmed by electrophoretic separation of the products in 12% polyacrylamide gels and ethidium bromide staining. The PCR products were purified using the QIAquick PCR purification kit (Qiagen), following the instructions of the manufacturer. The elution was carried out with water. Quality and concentration of all purified standard DNA samples were determined by UV spectroscopy as described above.
The molar concentrations of the PCR products (RPS3-F1/R1, XP53-F1/R1 and p53-F1/R1) that served as standard DNA in the LightCycler experiments (‘standard’) were calculated by UV absorption spectroscopy as described above.
Real-time PCR
Real-time PCR was used to quantify the amount of a target sequence in a genomic DNA sample. The target sequence was amplified with the ‘inner’ primer pairs PRS3-F2/R2 (S.cerevisiae), XP53-F2/R2 (X.maculatus) and P53-F2/R2 (H.sapiens sapiens). The accumulation of PCR product was monitored in real time once per cycle during the amplification process by measuring specific fluorescence signals, whose intensities are proportional to the amount of PCR product. The PCR process was followed using either the dsDNA-specific fluorescent dye SYBR Green I (measured after the extension phase) or sequence-specific pairs of fluorophor-labeled oligonucleotides (hybridization probes, measured after the annealing phase).
The signal curves of standards and unknowns, measured in the same run, are used for quantification, which is done automatically by the software. In brief, the (fractional) cycle number, CT, which indicates where the signal curve crosses an arbitrary threshold intersecting the signal curves in their exponential phases, are determined. The CT values are proportional to the logarithms of the initial target concentrations. A calibration curve of the CT values of the standard dilution series versus the concentrations is calculated and used to determine the concentrations of the unknowns, based on their CT values.
Quantitative real-time PCR experiments were performed in a LightCycler instrument from Roche Diagnostics (18–20). This instrument allows the simultaneous measurement of the fluorescence of SYBR Green I and fluorescein in channel 1 (maximum emission wavelength at 520 nm) and of RED-640 in channel 2 (maximum emission wavelength at 640 nm). The 10 µl PCRs contained 2 µl template DNA, 0.5 µM each forward and reverse primer (F2/R2, see Table 1), 200 µM each deoxynucleotide (dATP, dCTP, dGTP and dTTP), 0.5 g/l bovine serum albumin, 0.5 U Taq DNA polymerase, and either SYBR Green I in a 1:30 000-fold dilution of the commercial stock solution or 0.2 µM each hybridization probe (p53-FAM and p53-RED), and 6 mM MgCl2 in 1× reaction buffer (10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.3 at 25°C). The template DNA was either PCR generated DNA (standard) or genomic DNA (sample). Each run contained a series of standards (with 102, 103, …, 108 copies of standard DNA as template) and the unknowns (5-fold replicates with genomic DNA as template) for one target (rps3, xp53 or p53). All reaction components, except the oligonucleotides, were purchased from Roche Diagnostics. The cycling protocol was as follows: initial denaturation for 2 min at 95°C followed by 50 cycles with 0 s at 95°C, 5 s at 55°C and 10 s at 72°C. The temperature gradient was always 20 K × s–1. Fluorescence was measured either after the extension phase at 72°C for SYBR Green I detection (rps3 and xp53) or after the annealing phase at 55°C for hybridization probe detection (p53). When SYBR Green I was used for detection, a melting curve analysis with a temperature gradient of 0.2 K × s–1 from 72 to 95°C was performed following the PCR amplification, to confirm that only specific product was amplified.
The fluorescence data were evaluated with the software SoFAR (21), using the preset adjustments (background correction, sigmoidal amplification function and automatic threshold determination) to calculate the absolute initial concentrations of the template copies of the unknowns for each reaction. The data recorded in channel 1 were used to evaluate experiments with SYBR Green I detection, whereas in the case of the hybridization probe detection format, the ratio of the fluorescence intensities measured was used. This ratio corresponds to the ratio of acceptor (RED-640) to donor (FAM) fluorescence. Therefore, any effect altering the signal of both fluorophors proportionally (e.g. photobleaching, formation of excimers or temperature-dependent quenching) is eliminated from the signal curves. Therefore, the use of the donor-to-acceptor signal ratio increases the precision of the CT values obtained (22).
Calculation of the genome sizes
The amount of DNA which corresponds to the size of one haploid genome (C value) can be derived from the ratio of the mass of template DNA (m, determined by UV absorbance) and the copy number of the target sequence (N, determined by real-time PCR): C = m × N–1. The genome size, i.e. the number of base pairs per genome, is given by Γ = C × NA × MBp–1, where NA is Avogadro’s number (6.022 × 1023 mol–1) and MBp is the mean molar mass of a base pair (660 g mol–1).
RESULTS AND DISCUSSION
The real-time PCR-based method for the estimation of C values and Γ requires the absolute quantification of a single-copy gene. For this purpose, a DNA solution with known concentration of the amplicon sequence (‘standard’) is required. This standard is generated by conventional PCR from the genomic sample using the ‘outer’ primers; the amplicon includes the binding sites of the ‘inner’ primers which are subsequently used in the real-time PCR. Because the lengths of these PCR products are known, their concentration, again determined by UV absorbance, can be calculated as copies per microliter. For the quantification of the target gene sample, the PCR amplifications of the target gene from a dilution series of the standard and from the sample are monitored in real time and evaluated by an appropriate software. Because the concentration of the standard is known as copies per microliter, the absolute number of target genes in the sample and, thus, the genome size can be calculated.
The results of the quantitative real-time PCR and the calculated genome sizes for the samples analyzed are shown in Table 2.
Table 2. Estimated haploid genome sizes.
| Target | Sample concentration | Product length (bp) | Calibration curvea y = mx + b (R2) | Target copies (copies µl–1) (mean ± CV)b | Γ (bp) | C (pg) | |
|---|---|---|---|---|---|---|---|
| (ng µl–1) | Standard | Real-time PCR | |||||
| rps3 (S.cerevisiae) | 17.9 | 382 | 172 | –4.0x + 38.6 (0.9993) | 1.35 × 106 ± 6% | 12.1 × 106 | 0.013 |
| xp53 (X.maculatus) | 16.8 | 387 | 89 | –3.9x + 43.0 (0.9998) | 2.78 × 104 ± 8% | 5.5 × 108 | 0.603 |
| p53 (H.sapiens sapiens) | 15.0 | 342 | 125 | –3.71x + 38.3 (0.9978) | 4.69 × 103 ± 5% | 2.9 × 109 | 3.178 |
aCurve from CT versus log copies. m, slope; b, intercept; R2, coefficient of correlation.
bCV (coefficient of variation) = 100% × (standard deviation) × (mean)–1 (n = 5).
For the determination of the C value, the mass of the pure genomic DNA sample and the concentration of a single copy gene must be known. For all examples shown, the generated standard DNA was longer than the products amplified in the real-time PCR. This kind of ‘nested PCR’ has two advantages for exact quantification: first, the error in the determination of the product concentration by UV spectroscopy is smaller for longer products; second, tail effects for the hybridization of the primers will be nearly the same for genomic DNA and standard DNA. The tail effects may be caused by G:C-rich sequences or complex secondary structures, which may disturb the hybridization of the primers (23). Due to these effects, the amplification efficiency may be reduced in the first PCR cycles, compared to the amplification efficiency of a template which does not cover the flanking regions outside the amplified sequence. Therefore, to ensure that the conditions for the amplification are identical for both standard and genomic template DNA, the standard DNA templates should have the same flanking sequences, like the genomic templates.
However, an absolute quantification by real-time PCR is only possible when (i) the absolute copy number of the target sequence in the standard is known and (ii) the absolute mass concentration of genomic DNA in the sample can be determined accurately. For both requirements, the UV absorption at 260 nm (A260) of the DNA solution was used in this study. The accurate determination of A260 is a crucial point in this assay: it must not be influenced by factors other than by the DNA, something that cannot be excluded when only A260 is determined. Most frequently, the A260 is increased by light scattering and the presence of contaminating proteins. For pure DNA, the ratio of A260 to A280 is between 1.8 and 1.9. Because proteins have an absorbance maximum at 270–290 nm, the A260:A280 ratio is decreased when the DNA solution is contaminated with proteins. The influence of light scattering cannot be estimated from the A260:A280 ratio, because the effect decreases exponentially with wavelength. Therefore, a complete absorbance spectrum between 220 and 320 nm should be recorded to obtain reliable information when the A260 value is used to calculate the accurate DNA concentration. Note that RNA contamination was excluded in this study by an exhaustive RNase digestion step during the DNA purification.
The sequenced strain of S.cerevisiae was chosen as a ‘gold standard’ for validation of this technique, because the complete genomic sequence of this organism and, therefore, its exact length are known. The genome size measured here for S.cerevisiae (12.1 Mb) is identical within 1% to the size derived from the published sequence (12.069 Mb) (24). This demonstrates the accuracy which can be achieved using this technique.
The platyfish, X.maculatus, is not only a popular fish for aquarists, but also an important animal model for behavioral studies and, in particular, for the study of tumorigenesis (25,26). Therefore, the genome size of this species has been determined frequently by several techniques. The values of Γ given in the literature vary between 410 and 550 Mb. The value of Γ determined with the c0t assay was 530 Mb, whereas the absorption cytometric analysis after Feulgen staining yielded a slightly higher value of 540 Mb (27,28). The value of 550 Mb as determined by the real-time PCR-based method used here is in excellent agreement with the latter two values.
The Γ value of H.sapiens sapiens, as reported in common textbooks, varies between 2.8 and 3.2 Gb (29–31). Morton (32) reported in 1991 a value of 3.286 Gb, which is still cited by The International Human Genome Mapping Consortium in 2001 (33). Estimations for the genome size based on the sequencing results of the human genome project predict a Γ value between 2.69 and 3.0 Gb: The International Human Genome Sequencing Consortium (34) reported 2.6929 Gb of sequenced bases for the draft genome sequence, excluding gaps (i.e. an underestimated value of Γ), while the total length of the scaffold, including gaps, is given as 2.916 Gb, a value nearly identical to that found by Venter et al. (35) who generated a 2.907 Gb consensus sequence of the human genome by the whole-genome shotgun sequencing method. The value of 2.9 Gb determined here is identical to this sequence-based data.
Whereas the dsDNA-specific dye SYBR Green I was used in the real-time PCR with the sequences from S.cerevisiae and X.maculatus, sequence-specific hybridization probes were used for the human sequence. Both detection formats yielded comparable accuracies under the assay conditions. For exact quantification it is important that no non-specific products are amplified by PCR to measurable amounts before the signal curves reach the threshold. The accumulation of primer dimers can be seen in non-template controls only when SYBR Green I is used for detection: amplification curves of standards and samples with CT values similar to those of the non-template controls must not be used for quantification or cannot be quantified, respectively. The amplification of other non-specific PCR products during PCR due to the existence of false binding sites for the primers can be detected neither by SYBR Green I nor by sequence-specific probes. This has to be checked after the PCR by a melting curve analysis (36), which, however, is only possible when SYBR Green is used (Fig. 1), or by gel electrophoresis, which can distinguish different PCR products (data not shown). The melting curve analysis can be carried out directly after the PCR in the same reaction tube, which reduces the risk of carry-over contamination of PCR products. In conclusion, SYBR Green I seems to be the better choice for real-time quantification than sequence-specific probes, because (i) the accuracies achieved are similar, (ii) it is less expensive, (iii) it is easier to handle, (iv) the potential accumulation of primer dimers can be analyzed in real time and (v) the absence of other non-specific PCR products can be confirmed by melting curve analyses in a closed-tube format.
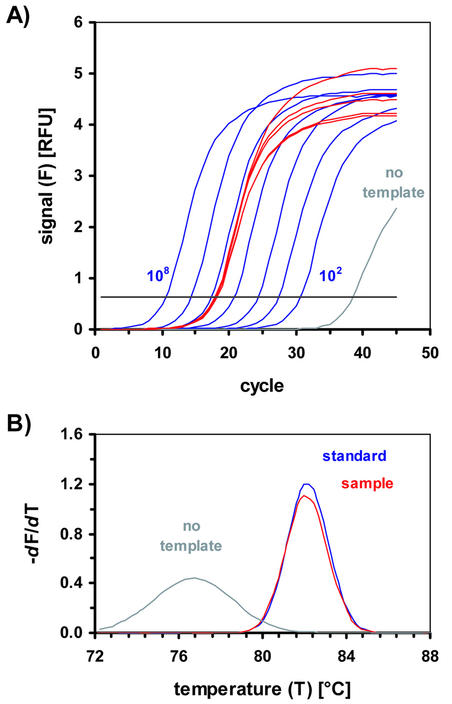
Figure 1.
Signal curves obtained in real-time PCR. (A) Amplification curves used for quantification of the rsp3 sequence from S.cerevisiae. Fluorescence signals from SYBR Green I measured in channel 1 (520 nm). The curves obtained for the standards with (from left to right) 108, 107, …, 102 copies of the standard PCR product as template are shown in dark blue. The curves for the genomic sample, measured five times, are shown in red. The gray curve represents the amplification profile of the non-template control. The threshold for the determination of the CT values is indicated by the horizontal black line. RFU, relative fluorescence units. (B) Melting curve analysis after amplification with the rps3 specific primers [amplification curves are shown in (A)]. Melting curve analysis was carried out directly after the PCR by slowly increasing the temperature at 0.2 K × s–1 from 72 to 90°C while the signal was recorded continuously. The cooperative melting process of the dsDNA causes a steep decrease in the fluorescence signal around the melting temperature of the PCR product. The signal decrease occurs as a clear peak in the negative derivative (–dF/dT) of the melting curves. The temperature where the highest value is reached is defined as the melting temperature: the specific PCR product obtained from the amplification of the standard PCR product (‘standard’, blue) melts at 82°C. The same melting temperature is determined for the PCR product of the genomic sample DNA (‘sample’, red). The PCR product amplified in the non-template control (‘no template’, gray) has a significantly lower melting temperature of 77°C. T, temperature in °C; F, fluorescence signal measured in channel 1 (520 nm).
In conclusion, the Γ values obtained for the three species analyzed here demonstrate the high accuracy of the method for genome sizes between several Mb and Gb. This method is considerably faster to perform than others described in the literature (1–10). Any genomic DNA, even of low quality, may be used, provided it is free of RNA and UV-absorbing contaminants. The preparation of whole cells with intact nuclei is not required. In contrast to colorimetry-based methods, no nuclear reference standards are needed and the quantitative results are absolute. The accuracy of this method strongly depends on the specificity of the PCR and the precision of the UV spectrophotometric determination of the DNA concentrations. The amplification of non-specific PCR products during the generation of the standard DNA leads to an underestimation of Γ, whereas errors from the UV absorption measurements may cause both under- as well as overestimations of Γ. Therefore, a complete UV absorption spectrum should be used to quantify the DNA concentration and to estimate DNA quality (purity) rather than a two-point measurement (at 260 and 320 nm).
The real-time PCR-based method for the estimation of genome sizes will be a useful tool for the analyses of large numbers of species, individuals and tissues to investigate the changes in genome size during phylogenesis. The method may also provide a new tool to identify species. It is also applicable for samples that cannot be analyzed by flow cytometry, and is more accurate and easier to perform than c0t analyses.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Dr J. Hegemann for supplying the yeast strain and Drs A. Anders and J. Michel for the fishes, discussions and support. This work was supported by the Deutsche Forschungsgemeinschaft (Graduiertenkolleg Molekulare Biologie und Pharmakologie) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Zhang J.Z and Fan,M.Y. (2002) Determination of genome size and restriction fragment length polymorphism of four Chinese rickettsial isolates by pulsed-field gel electrophoresis. Acta Virol., 46, 25–30. [PubMed] [Google Scholar]
- 2.Sun L.V., Foster,J.M., Tzertzinis,G., Ono,M., Bandi,C., Slatko,B.E. and O’Neill,S.L. (2001) Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J. Bacteriol., 183, 2219–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rydkina E., Roux,V. and Raoult,D. (1999) Determination of the genome size of Ehrlichia spp., using pulsed field gel electrophoresis. FEMS Microbiol. Lett., 176, 73–78. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro H.S. (1970) Nucleic Acids. In Sober,H.A. (ed.), CRC Handbook of Biochemistry. Selected Data for Molecular Biology. CRC Press, Cleveland, OH, pp. H-113.
- 5.Britten R.J. and Kohne,D.E. (1986) Repeated sequences in DNA. Hundreds and thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science, 161, 529–540. [DOI] [PubMed] [Google Scholar]
- 6.Couch J.A., Zintel,H.A. and Fritz,P.J. (1993) The genome of the tropical tree Theobroma cacao L. Mol. Gen. Genet., 237, 123–128. [DOI] [PubMed] [Google Scholar]
- 7.De Vita R., Cavallo,D., Eleuteri,P. and Dell’Omo,G. (1994) Evaluation of interspecific DNA content variations and sex identification in Falconiformes and Stringiformes by flow cytometric analysis. Cytometry, 16, 346–350. [DOI] [PubMed] [Google Scholar]
- 8.Vinogradov A.E. (1994) Measurement by flow cytometry of genomic AT/GC ratio and genome size. Cytometry, 16, 34–40. [DOI] [PubMed] [Google Scholar]
- 9.Lamatsch D.K., Steinlein,C., Schmid,M. and Schartl,M. (2000) Noninvasive determination of genome size and ploidy level in fishes by flow cytometry: detection of triploid Poecilia formosa. Cytometry, 39, 91–95. [DOI] [PubMed] [Google Scholar]
- 10.Barow M. and Meister,A. (2001) Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry, 47, 1–7. [DOI] [PubMed] [Google Scholar]
- 11.Siroky J., Lysak,M.A., Dolezel,J., Kejnovsky,E. and Vyskot,B. (2001) Heterogeneity of rDNA distribution and genome size in Silene spp. Chromosome Res., 9, 387–393. [DOI] [PubMed] [Google Scholar]
- 12.Brainerd E.L., Slutz,S.S., Hall,E.K. and Phillis,R.W. (2001) Patterns of genome size evolution in tetraodontiform fishes. Evolution, 55, 2363–2368. [DOI] [PubMed] [Google Scholar]
- 13.Rabinovitch P.S., O’Brien,K., Simpson,M., Callis,J.B. and Hoehn,H. (1981) Flow-cytogenetics II. High-resolution ploidy measurements in human fibroblast cultures. Cytogenet. Cell Genet., 29, 65–76. [DOI] [PubMed] [Google Scholar]
- 14.Voglmayr H. and Greilhuber,J. (1998) Genome size determination in peronosporales (Oomycota) by Feulgen image analysis. Fungal Genet. Biol., 25, 181–195. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 16.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 17.Nazarenko I.A., Bhatnagar,S.K. and Hohman,R.J. (1997) A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res., 25, 2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittwer C.T., Fillmore,G.C. and Hilliard,D.R. (1989) Automated polymerase chain reaction in capillary tubes with hot air. Nucleic Acids Res., 17, 4353–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–131, 134–138. [DOI] [PubMed] [Google Scholar]
- 20.Wittwer C.T., Ririe,K.M., Andrew,R.V., David,D.A., Gundry,R.A. and Balis,U.J. (1997) The LightCycler(TM): a microvolume multisample fluorimeter with rapid temperature control. Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 21.Wilhelm J., Pingoud,A. and Hahn,M. (2003) SoFAR: software for fully automatic and accurate evaluation of real-time PCR data. Biotechniques, 34, 324–332. [DOI] [PubMed] [Google Scholar]
- 22.Wilhelm J., Pingoud,A. and Hahn,M. (2001) Comparison between Taq DNA polymerase and its Stoffel fragment for quantitative real-time PCR with hybridization probes. Biotechniques, 30, 1052–1056, 1058, 1060. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelm J., Hahn,M. and Pingoud,A. (2000) Influence of target DNA melting behavior on real-time PCR quantification. Clin. Chem., 46, 1738–1743. [PubMed] [Google Scholar]
- 24.Mewes H.W., Albermann,K., Bahr,M., Frishman,D., Gleissner,A., Hani,J., Heumann,K., Kleine,K., Maierl,A., Oliver,S.G., Pfeiffer,F. and Zollner,A. (1997) Overview of the yeast genome. Nature, 387 (suppl.), 7–65. [DOI] [PubMed] [Google Scholar]
- 25.Anders F., Schartl,M., Barnekow,A. and Anders,A. (1984) Xiphophorus as an in vivo model for studies on oncogenes. Adv. Cancer Res., 42, 191–275. [DOI] [PubMed] [Google Scholar]
- 26.Anders A., Zechel,C., Schlatterer,B., Gröger,H., Schmidt,D., Smith,A. and Anders,F. (1991) Genetic and molecular approach for breeding and use of laboratory fish for the detection of agents with carcinogenic and/or promoting activity. Bull. Cancer, 78, 415–433. [PubMed] [Google Scholar]
- 27.Schwab M., Vielkind,J. and Anders,F. (1979) Genetic basis of susceptibility for neuroblastoma following treatment with N-methyl-N-nitrosourea and X-rays in Xiphophorus. Cancer Res., 39, 519–526. [PubMed] [Google Scholar]
- 28.Tiersch T.R., Chandler,R.W., Kallman,K.D. and Wachtel,S.S. (1989) Estimation of nuclear DNA content by flow cytometry in fishes of the genus Xiphophorus. Comp. Biochem. Physiol. B, 94, 465–468. [DOI] [PubMed] [Google Scholar]
- 29.Brown T.A. (1991) Molecular Biology. LabFax, Bios Scientific Publishers, Oxford, UK.
- 30.Alberts B., Bray,D., Lewis,J., Raff,M., Roberts,K. and Watson,J.D. (1994) Molecular Biology of the Cell, 3rd Edn. Garland Publishing Group, New York, NY.
- 31.Knippers R. (2001) Molekulare Genetik, 8th Edn. Georg Thieme Verlag, Stuttgart, Germany.
- 32.Morton N.E. (1991) Parameters of the human genome. Proc. Natl Acad. Sci. USA, 88, 7474–7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The International Human Genome Mapping Consortium (2001) A physical map of the human genome. Nature, 409, 934–941. [DOI] [PubMed] [Google Scholar]
- 34.The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 35.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 36.Ririe K.M., Rasmussen,R.P. and Wittwer,C.T. (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem., 245, 154–160. [DOI] [PubMed] [Google Scholar]