Abstract
A common method for generating mice with subtle genetic manipulations uses homologous recombination (HR) in embryonic stem (ES) cells to replace a wild-type gene with a slightly modified one. Generally, a drug resistance gene is inserted with the modified gene to select correctly targeted clones. Often, however, the presence of this drug resistance gene interferes with the normal locus and creates a null or hypomorphic allele. Flanking of the selectable marker by loxP sites followed by Cre-mediated deletion after drug selection can overcome this problem. The simplest method used to remove a loxP-flanked selectable marker is to breed an animal carrying a loxP-flanked drug resistance gene to an animal that expresses Cre recombinase in the germline. To date only outbred transgenic mice are available for this purpose. This can be problematic for phenotypic analysis in many organ systems, including the brain, and for the analysis of behavior. While attempting to make 129S6/SvEvTac inbred background (isogenic to our ES cells) mice that express Cre under the control of several tissue-specific promoters, we serendipitously generated a line that excises loxP-flanked drug resistance genes in all tissues, including the germline. This reagent allows deletion of loxP-flanked sequences while maintaining the mutation on an inbred background.
INTRODUCTION
A common method for generating mice with subtle genetic manipulations, for example inserting point mutations rather than creating a null allele, uses homologous recombination (HR) in embryonic stem (ES) cells to replace a wild-type gene with a modified one. In most cases a neomycin resistance gene (NeoR) or other drug resistance gene is inserted into an intron or in the 3′-untranslated region (UTR) near the desired alteration to allow for selection of correctly targeted clones. The goal of this use of HR and insertion of NeoR is to make precise changes upstream of the NeoR insertion rather than to generate a null allele. However, in many cases the presence of the drug resistance gene interferes with the normal locus, resulting in a null or hypomorphic allele (1,2). To overcome this problem, the drug resistance gene, flanked by loxP sites, can be removed by exposure to Cre recombinase (Cre).
Cre can be introduced by several methods. First, transfection of a Cre-expressing plasmid into ES cells (1). Second, injection of a Cre-expressing plasmid into fertilized eggs (3,4). Finally, mating of the mouse mutant created using ES cell technology to a transgenic mouse that expresses Cre in the germline (1,5). The latter method is the simplest. To date only outbred mice are available at Jackson Laboratories for this purpose. The use of outbred mice can be problematic for phenotypic analysis in many organ systems, including the brain, and for the analysis of behavior (6). Another option is to use a recently described inbred mouse line in which the Cre recombinase expression cassette was inserted into the X-linked hypoxanthine guanine phosphoribosyltransferase gene (Hprt). The advantage of this mouse is that the researcher knows where the Cre is inserted in the genome (7). However, this insertion creates a null allele at the locus and produces a complete knockout of Hprt in the homozygous animal. Although this knockout does not produce a phenotype, it may lead to a synthetic phenotype in its offspring when it is bred to another transgenic mouse with a floxed allele. In order to maintain the mutation in an inbred background, transfection into ES cells or pronuclear injection of a Cre-expressing plasmid is required. Both of these latter methods are labor intensive. We generated a transgenic mouse line in an inbred 129S6/SvEvTac (129) strain that expresses Cre and demonstrate that deletion of a loxP-flanked NeoR gene (loxP– NeoR–loxP) occurs in all tissues, including the germline. This reagent can be used to delete loxP-flanked sequences in all tissues while maintaining a background isogenic to that of commonly used 129 ES cells.
MATERIALS AND METHODS
Transgene construct
The green fluorescent protein (GFP) gene was removed from pEGFP-C1 (Clontech) using Eco47111/XhoI. The GFP sequence was isolated by gel purification and inserted into EcoRV/XhoI-digested pBSKS (Strategene) to generate pBSKSGFP. The 1.1 kb Cre recombinase gene was amplified using the plasmid pCre306C as a template (courtesy of Steve O’Gorman) using the 5′ primer 5′-GCT AGC TCC GGA GAG CAA AAG CTG-3′ and the 3′ primer 5′-CTC TAG CTC GAG CTA ATC GCC ATC-3′, which included a 5′ BspeI site and a 3′ XhoI site for use in generating an in-frame fusion of Cre to GFP. The product was gel purified and inserted into pBSKSGFP at the BspeI/XhoI site, to generate pBSKSGFPCre. The final transgenic vector was constructed by inserting the purified 1.86 kb NotI blunted GFPCre fusion cDNA into blunted KpnI and NotI sites in the plasmid hgPrnp (8,9) downstream of a modified prion promoter. The Prion.GFPCre plasmid (Fig. 1A) was purified using CsCl ultracentrifugation twice according to published methods (10). The purified plasmid was digested using MluI and SalI to release the backbone. The 5.7 kb Prion.GFPCre fragment was isolated after separation on a 1% agarose gel overnight at 4°C using Gene Clean (Qbiogene, Carlsbad, CA).
Figure 1.
Transgene construct. A 5.5 kb minimal promoter and the first intron of the prion gene were ligated to a fusion protein of GFP (Met1–Lys239) and the Cre protein. A rat growth hormone poly(A)+ tail was used to ensure appropriate processing.
Production of transgenic 129 mice
All animal procedures were performed according to protocols approved by both The Salk Institute for Biological Studies and the National Institutes of Health Animal Care and Use Committees. The procedures for superovulation, embryo recovery, microinjection and transfer were similar to those described (10). Female 129 mice at 4 weeks of age were used as embryo donors after superovulation with 2.5 U pregnant mare’s serum (PMS) or gonadotropin–PMS (catalog no. 367222; Calbiochem) and 2.5 U chorionic gonadotropin (catalog no. 0052-0315-10; Organon) 46 h apart. Stud males were 129 mice. Recipient females (CBy6F1, 8 weeks or older) were placed with CB6F1 vasectomized males. Micro injections were performed at 200× magnification using a Zeiss Axiovert 135M, Eppendorf micromanipulator 5171 and automatic Eppendorf microinjector 5242. Embryos were injected in M2 (catalog no. MR-015-D; Speciality Media) and then incubated in BMOC2 (catalog no. MR-013-D; Speciality Media) at 37°C in 5% CO2 overnight. The two-cell embryos were separated from the one-cell embryos the following morning and surgically transferred to primed recipients.
Determining where Cre-mediated excision occurs
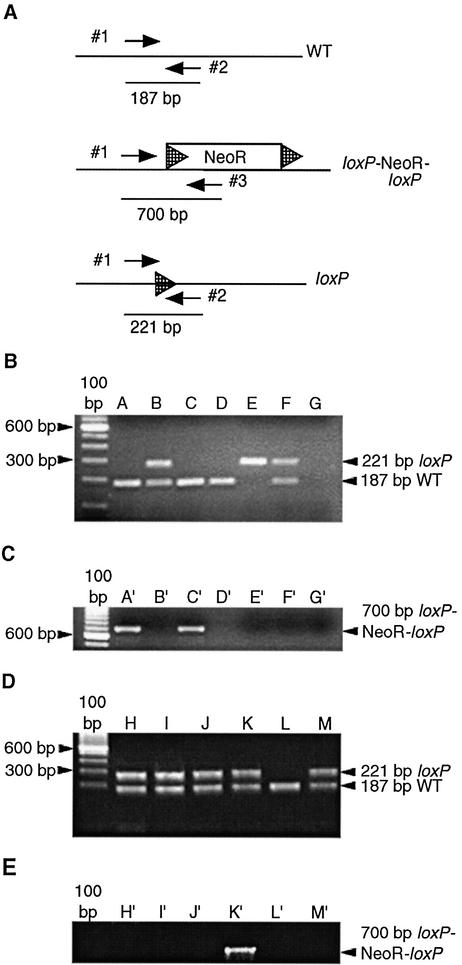
Genotypes were determined by PCR using DNA prepared from tail biopsies or several brain regions, kidney, testes and liver using Phase-Lock Gel reagent (Eppendorf) after overnight lysis (11). Cre(+) mice were identified using the 5′ Cre-specific primer 5′-CCG GGC TGC CAC GAC CAA-3′ and 3′ Cre-specific primer 5′-GGC GCG GCA ACA CCA TTT TT-3′. To determine if Cre-mediated excision occurred, PCR was performed on DNA using primers to detect either a wild-type allele (Fig. 2A, upper panel, primers 1 and 2), the loxP–NeoR–loxP allele (Fig. 2A, middle panel, primers 1 and 3) or an allele where the NeoR gene was deleted leaving a single intact loxP site (Fig. 2A, lower panel, loxP primers 1 and 2). PCRs were run on 0.8% agarose–TBE gels.
Figure 2.
Location of the PCR primers and detection of various alleles. (A) Primers used to detect the wild-type allele (WT) and the deleted loxP-flanked NeoR allele are labeled 1 and 2. These primers amplify a 187 bp wild-type fragment. Primers 1 and 2 will also amplify the deleted NeoR allele (loxP), which has a residual 34 bp loxP site remaining after Cre-mediated excision of the NeoR gene has occurred. In this case, a 221 bp fragment is amplified. This PCR was designed so that the undeleted loxP-flanked NeoR allele would not amplify if present. Primers 1 and 3 amplify a 700 bp fragment when the loxP-flanked NeoR gene (loxP–NeoR–loxP) is present. NeoR indicates neomycin resistance gene and hatched arrowheads indicate loxP sites. (B) PCR results demonstrating amplification of the 187 bp wild-type allele and the 221 bp deleted loxP allele in tail DNA. (C) PCR amplification of the intact 700 bp loxP–NeoR–loxP allele in tail DNA from the same animals shown in (B). Lanes A and A′ represent the female breeder. Lanes B and B′ represent a Cre(+) male offspring of a mating between the animal in lane A and the Cre(+) founder. Lanes C and C′ represent a female carrying a wild-type allele and a loxP–NeoR–loxP allele. The animal in lanes B and B′ was mated to the animal in lanes C and C′ and the genotypes of the offspring are shown in lanes D–F and D′–F′. Lane D represents a homozygous wild-type mouse, lane E represents a mouse carrying two loxP alleles and lane F represents a mouse with both a wild-type and a loxP allele. Lanes G and G′ represent the negative control PCR in the absence of genomic DNA. (D) PCR results demonstrating amplification of the 187 bp wild-type allele and the 221 bp deleted loxP allele in DNA isolated from the liver (lane H), testes (lane I), tail (lane J) and kidney (lane K) of the F1 mouse shown in (B), lane B. (E) PCR amplification of DNA isolated from the liver (lane H′), testes (lane I′), tail (lane J′) and kidneys (lane K′) of the F1 mouse in (B), lane B. Lanes L, L′, M and M′ represent control amplifications of tail DNA isolated from a wild-type mouse and a mouse carrying both the wild-type and deleted loxP alleles, respectively.
Immunoflourescence
Mice were perfused with cold 4% paraformaldehyde transcardially. The kidney, testes, brain, and liver were dissected and submerged in 30% sucrose for 2 days before microtome sectioning (40 µm). Sections were stained with DAPI, analyzed and photographed using confocal microscopy (Bio-Rad).
Generating homozygous Cre(+) mice
A heterozygous 129 Cre(+) male was mated to a heterozygous 129 Cre(+) female. Cre(+) offspring derived from the mating were mated to wild-type animals and the genotypes of the offspring were determined by PCR. The probability of the mice being homozygous was calculated by using the Rule of Multiplication on a standard Punnet Square of a heterozygote mated to a wild type.
RESULTS AND DISCUSSION
Production of transgenic 129 mice
Of 40 superovulated female 129 mice, approximately 200–250 embryos were harvested and 97 embryos at the two-cell stage were recovered after microinjection. These embryos were implanted and 11 pups were born. Screening by PCR identified two Cre(+) founder animals. Both founders were bred to 129 wild-type mice to generate inbred F1 offspring. Only one founder (98030B) produced Cre(+) transgenic offspring.
Cre(+) transgenic line deletes a loxP-flanked gene
The 98030B Prion.GFPCre transgenic founder line was studied to assess where Cre-mediated excision occurred. To generate appropriate mice, the founder male was bred to a Cre(–) female 129 mouse carrying a wild-type allele (Fig. 2A, WT) and a NeoR gene flanked by loxP sites (Fig. 2A, loxP-NeoR-loxP). DNA isolated from tails of Cre(+) offspring was screened for evidence of Cre-mediated deletion of the loxP-flanked NeoR gene. The 187 bp wild-type allele was amplified in both the breeder female (Fig. 2B, lane A, lower band) and the offspring (Fig. 2B, lane B, lower band), as would be expected. An additional 221 bp fragment was amplified in Cre(+) offspring (Fig. 2B, lane B, upper band), consistent with excision of the NeoR gene and retention of a single loxP site. To determine the extent of Cre-mediated excision, PCR was used to detect any intact loxP–NeoR–loxP DNA. The breeder female had an intact loxP–NeoR–loxP allele (Fig. 2C, lane A′), whereas her Cre(+) offspring showed no amplification of the loxP–NeoR–loxP (Fig. 2C, lane B′), consistent with complete deletion of the NeoR gene in the Cre(+) offspring.
To test if the deletion occurred in the germline of the Cre(+) offspring whose tail DNA showed deletion of the NeoR gene, a Cre(+) male offspring (Fig. 2B and C, lanes B and B′) was mated to a Cre(–) female carrying one wild-type allele (Fig. 2B, lane C) and one loxP–NeoR–loxP allele (Fig. 2C, lane C′) and the genotype of three resulting offspring from the mating determined (Fig. 2B and C, lanes D–F and D′–F′). One offspring was Cre(+), but was homozygous for the wild-type allele (Fig. 2B, lane D), and therefore no loxP–NeoR–loxP products were amplified (Fig. 2C, lane D′). One of the offspring was Cre(+) and only showed amplification of the deleted allele (Fig. 2B, lanes E), consistent with inheriting the deleted allele from the male parent and also exhibiting Cre-mediated deletion of the loxP–NeoR–loxP allele (Fig. 2C, lane E′) inherited from the female parent. Additionally, the offspring that was Cre(–) had one copy of the wild-type allele and one copy of the deleted allele (Fig. 2B, lane F) with no amplification of the loxP–NeoR–loxP allele (Fig. 2C, lane F′), demonstrating that paternal germline transmission of the deleted allele occurred. Lanes G and G′ are included as negative controls (Fig. 2B and C).
Excision is complete in virtually all tissues
In order to determine in what other tissues loxP–NeoR–loxP excision occurred in the Cre(+) mice, PCR was performed on DNA isolated from the liver (Fig. 2D and E, lanes H and H′), testes (Fig. 2D and E, lanes I and I′), tail (Fig. 2D and E, lanes J and J′), kidney (Fig. 2D and E, lanes K and K′) and brain (data not shown) of the Cre(+) male animal that showed germline transmission of the deleted allele and no detectable NeoR gene in the tail DNA (Fig. 2C, lane B′). DNA from all tissues amplified both the deleted loxP and the wild-type alleles (Fig. 2D, lanes H–K). Lanes L and M are included as controls (Fig. 2D). A faint intact loxP–NeoR–loxP allele was amplified in the kidney (arrow indicating 700 bp fragment in Fig. 2E, lane K′), indicating that excision of the NeoR gene was incomplete in the kidney, but complete excision occurred in all other tissues studied (Fig. 2E, lanes H′–J′). However, because the intact loxP–NeoR–loxP allele was not passed on to any subsequent offspring, germline excision was complete (data not shown). Similar results were found for multiple other animals (data not shown). Lanes L′ and M′ are included as controls (Fig. 2E).
Cre is not detected in adult tissues
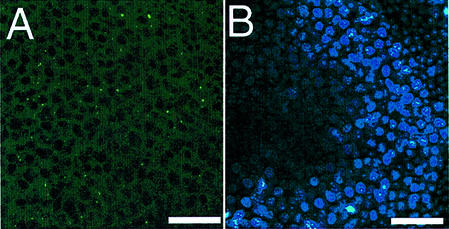
To determine if Cre was expressed in the germline of adult animals, testes from a Cre(+) animal were sectioned and visualized using the GFP marker. No GFP was detected in the testes (Fig. 3A). Therefore, the lack of Cre expression in the adult animal germline suggests that Cre must be expressed during early germ cell or embryo development. However, in spite of attempts to detect Cre in early embryos, we were unable to determine at which stage Cre expression and deletion occurred.
Figure 3.
GFPCre expression is absent in the adult male germline. (A) GFP is not detected in the testes of an adult Cre(+) transgenic mouse. (B) DAPI staining of the testes section serves as a control. Scale bars indicate 5 µm.
Homozygous Cre(+) line is normal and fertile
In order to generate homozygous Cre(+) mice, a heterozygous Cre(+) 129 male was mated to a heterozygous Cre(+) 129 female. To identify homozygous Cre(+) animals derived from the mating, the Cre(+) offspring were mated to wild-type animals and the numbers of Cre(+) mice determined. One female Cre(+) mouse had two litters resulting in five and seven mice, respectively. All of the mice were determined to be Cre(+). Therefore the probability is extremely low (0.00024) that this animal is not homozygous. A similar analysis of a Cre(+) male was performed. The 11 offspring from three litters resulting from mating to a wild-type female (four offspring from the first litter, five from the second and two from the third) were all Cre(+). Therefore, the probability of the male mouse not being homozygous is extremely low (0.000488).
In summary, we generated inbred homozygous Cre-expressing male and female mice that reliably and consistently delete a loxP-flanked gene in the germline. Deletion of the loxP-flanked allele occurs whether the Cre transgene is inherited from the mother or the father. The homozygous mice are both fertile and phenotypically normal. This mouse has been accepted by the Jackson Laboratories for distribution under the name 129S6-TgN(Prnp-GFP/Cre)1Blw (Stock JR3960).
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to acknowledge L. Kitabayashi and C. T. Jacob for assistance with immunohistochemical analysis and genotyping, C. Weissmann and R. Nussbaum for the prion promoter construct and C. Doane for assistance with the manuscript. J.S. is supported by NIH Fellowship F31 NS10860-01, and C.B. by Department of the Army grant number DAMD17-99-1-9561 and the Fredrick B. Rentschler Developmental Chair.
REFERENCES
- 1.Xu X., Wagner,K.U., Larson,D., Weaver,Z., Li,C., Ried,T., Hennighausen,L., Wynshaw-Boris,A. and Deng,C.X. (1999) Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nature Genet., 22, 37–43. [DOI] [PubMed] [Google Scholar]
- 2.Nagy A., Moens,C., Ivanyi,E., Pawling,J., Gertsenstein,M., Hadjantonakis,A.K., Pirity,M. and Rossant,J. (1998) Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr. Biol., 8, 661–664. [DOI] [PubMed] [Google Scholar]
- 3.Araki K., Araki,M., Miyazaki,J. and Vassalli,P. (1995) Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl Acad. Sci. USA, 92, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunaga S., Maki,K., Komagata,Y., Ikuta,K. and Miyazaki,J.I. (1997) Efficient removal of loxP-flanked DNA sequences in a gene-targeted locus by transient expression of Cre recombinase in fertilized eggs. Mol. Reprod. Dev., 46, 109–113. [DOI] [PubMed] [Google Scholar]
- 5.Williams-Simons L. and Westphal,H. (1999) EIIaCre – utility of a general deleter strain. Transgenic Res., 8, 53–54. [DOI] [PubMed] [Google Scholar]
- 6.Crusio W.E. (1996) Gene-targeting studies: new methods, old problems. Trends Neurosci., 19, 186–187; discussion 188–189. [DOI] [PubMed] [Google Scholar]
- 7.Tang S.H., Silva,F.J., Tsark,W.M. and Mann,J.R. (2002) A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis, 32, 199–202. [DOI] [PubMed] [Google Scholar]
- 8.Weissmann C., Fischer,M., Raeber,A., Bueler,H., Sailer,A., Shmerling,D., Rulicke,T., Brandner,S. and Aguzzi,A. (1996) The use of transgenic mice in the investigation of transmissible spongiform encephalopathies Int. J. Exp. Pathol., 77, 283–293. [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer M., Rulicke,T., Raeber,A., Sailer,A., Moser,M., Oesch,B., Brandner,S., Aguzzi,A. and Weissmann,C. (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J., 15, 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 10.Hogan B., Beddington,R., Costantini,F. and Lacy,E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.