METADATA ONTOLOGY
Biologists and clinical researchers are increasingly using ontologies to annotate experiment data, to index biomedical literature, and to integrate heterogeneous data sources [1]. With the ever increasing amount of competing knowledge available for computation, today’s biomedical researcher has to figure out how to use these resources to solve his problem. In this context, to find the right ontology for the purpose, it is usual for a user to ask amongst other things, How well does the Ontology or part of ontology covers his domain of interests? What is the maturity of ontology content? How is the content related to standard biomedical ontologies such as GO, UMLS? What are user’s experiences with the ontology? The problem in providing an answer to these questions is that most of these metadata information is not present in the ontology. The problem could be attributed to the limitations of the underlying knowledge representation to specify such support information and lack of tool support to associate metadata to ontology.
To enable the creation and retrieval of ontology these metadata, we have designed an ontology of metadata elements and have implemented a prototype tool (figure 1) that supports the creation of metadata for ontology resources based on the Metadata ontology. For each resource, the Metadata ontology provides two types of information: (1) source metadata, which include the metadata provided by ontology authors and generated by ontology-development tools; and (2) third-party metadata, which are provided by ontology users and that include peer reviews of ontologies, usage and experience information, and ratings. Some of the metadata categories included in the ontology are:
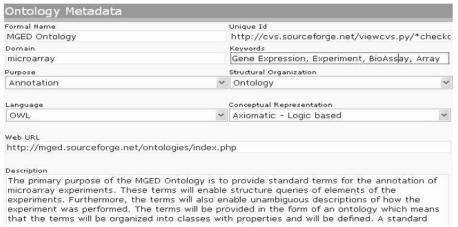
Figure 1.
Form to annotate an ontology resource. The interface shows the metadata information captured for the MGED Ontology.
▪ Domain of the ontology (using controlled terminology, when possible); informal description of the content; intended use of the ontology;
▪ Version number; contact and author information; supporting institutions; availability and licenses; citations and references;
▪ Verification tools used and development methodology;
▪ Naming policy; policy for extensions; reliance on other ontologies
▪ Peer reviews; experience reports; usage data; ratings along different axes, such as coverage; degree of formality.
EVALUATION
The metadata ontology was created using the Protégé ontology authoring environment. We have successfully used the tool to capture source metadata associated with the Gene Ontology , MGED and MeSH. In this evaluation, we observed that the metadata ontology was comprehensive enough to capture all the source information. We are currently, working on evaluating the other part of our ontology that captures third-party annotations such as peer-reviews. We are in the process of analyzing recent publications that have critically evaluated biomedical ontologies. We aim to capture this information as properties for instances of the metadata ontology.
DISCUSSION
To search for ontologies, the biomedical community, currently relies on ontology libraries such as the DAML Ontology Library1 and the OBO2 which are a mere listing of knowledge resources [2]. We intend to utilize the ontology metadata such as keywords that describe the topic of ontology content, to build a ontology library where users can submit metadata about their ontologies, search for existing ontologies against his/her requirements, and view their interrelationships.
Footnotes
References
- 1.Noy NF, Rubin DL, Musen MA. Making Ontologies and Ontology Libraries Work. IEEE Intelligent Systems. 2005;19(6):78–81. [Google Scholar]
- 2.Ding Y, Fensel D. Ontology library systems: The key to successful ontology reuse. The First Semantic Web Working Symposium, Stanford, USA, 2001