Abstract
The task of gathering detailed patient information from narrative text presents a significant barrier to clinical research. A prototype information extraction system was developed to identify concepts and their associated values from narrative echocardiogram reports. The system uses a Unified Medical Language System compatible architecture and takes advantage of canonical language use patterns to identify sentence templates with which concepts and their related values can be identified. The data extracted from this system will be used to enrich an existing database used by clinical researchers in a large university healthcare system to identify potential research candidates fulfilling clinical inclusion criteria. The system was developed and evaluated using ten clinical concepts. Concept-value pairs extracted by the system were compared with findings extracted manually by the author. The system was able to recall 78% [95%CI, 76–80%] of the relevant findings, with a precision of 99% [95%CI, 98–99%].
Background
Collecting specific clinical data from electronic patient records continues to be a major obstacle to taking full advantage of clinical information systems in health care1. One strategy to augment the availability of data has been to train authors of clinical reports to submit structured data. Not surprisingly, for most clinical purposes, this strategy is considered too restrictive and the computer-generated narrative, if any, is frequently altered to fit the author’s needs. In these cases, the narrative contains important clinical data that may not be found elsewhere in the document and the problem of ‘hidden’ data remains.
Information extraction (IE) is the process of extracting user-specified text from a set of documents—the goal is to capture structured information without sacrificing feasibility2. While it requires deeper analysis than simple key word searches, IE tasks are generally easier to implement than general-purpose natural language processing (NLP) systems since complete syntactic characterization and language understanding are not necessary3. While one of the most promising systems developed to extract information from medical narrative is a comprehensive NLP system-- MEDLEE achieved a sensitivity of 81% and a specificity of 98% for six clinical conditions found in chest radiography reports4-- information extraction systems that do not rely on full parsing have also demonstrated promising results, especially when applied to domains that are limited in scope and in which the language displays more regularity5–8.
Documentation of diagnostic procedures, e.g. echocardiogram reports, contains simpler narrative than reports detailing patient care or chronicling the patient’s medical history. With their narrow terminology, little need for outside knowledge and predictable routine, procedural reports lend themselves to a comparatively shallow analysis9. Like the physical exam, the subject of the description is always implicitly the patient, descriptions generally refer only to the present and sentences are generally independent units of description5. These properties permit the desired knowledge to be sufficiently described by a relatively simple and fixed template with slots that can be filled in with material from the text. This intermediate strategy works well when applied to procedural reports where identifying a concept alone is usually insufficient, but in which the relation that exists between the condition of interest and its modifying value is usually explicit. Such values may specify a condition’s absence, its presence or in the case of the latter, the degree of disease (e.g. severe aortic stenosis).
This work describes the implementation and evaluation of an IE system in which sentence templates are generated from named entities recognized as concepts by the MetaMap Transfer (MMTx) program* and by an auxiliary value lexicon. These templates are then used to extract clinical conditions and their related values. This paper demonstrates how this simple method can be applied to echocardiogram reports and yield patient-specific detailed cardiac findings in a precise way.
Methods: Development of Extraction System
Pre-processing
Corpus Overview: 703 echocardiogram reports were analyzed for this study – 483 reports from university hospital 1 (UH1) and 220 reports from university hospital 2 (UH2). These reports were chosen from 19 randomly selected days between June and December 2003. 295 reports from UH1 were used to train the system. The remaining 188 reports from UH1 and all 220 reports from UH2 were used to complete a test set of 408 reports.
Concept Selection: The following ten concepts were used as benchmark conditions to train and test the system: mitral valve insufficiency, aortic valve stenosis, pulmonary hypertension, mitral valve prolapse, valvular vegetations, cardiac shunt, intracardiac thrombus, ejection fraction, pericardial effusion and left ventricular hypertrophy. These ten concepts span a range of conditions that researchers in this university setting have frequently requested, but up to this point, have not been able to obtain easily.
Section Parsing: Regular expressions were used to extract the narrative sections of the test report. Coded fields were excluded from further analysis.
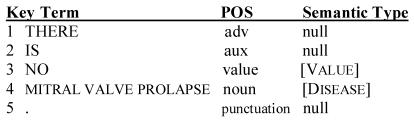
Concept Mapping and Sentence Reconstruction: Both the UMLS Metathesaurus and the strategy used by MMTx have been extensively described and will not be detailed here10. A supplied Java API provides a way to manipulate the input and output to MMTx. In order to simplify the template generation process, only the following UMLS concept categories were permitted to match: disease or syndrome [Disease], body part [BP], anatomical structure [AS], and diagnostic procedure [DP]. Up to three words could be grouped together to generate a ranked list of mappings to concepts in the UMLS. Lexical information and the top scoring mapping for each identified noun phrase are used in this analysis. Each sentence was then reconstructed so that the position of the term in the sentence serves as a key to the details of that term (see Figure A).
Figure A.
Sentence reconstruction involves linking concept and lexical information to each term in the sentence. Each term position or ‘key’ (e.g. “4”) is assigned its associated term (“mitral valve prolapse”), the part-of-speech (“noun”) and the semantic type of that concept ([Disease]). If MMTx could not map a concept to a term, no semantic type was recorded. [Value] assignments are discussed in the following section.
Post-processing
A Perl program was written to accomplish the following three tasks: identification of values, template recovery, and concept-value extraction.
Identification of values: The terms used to characterize disease severity in echocardiogram reports come from a fairly limited domain. However, the semantics behind some of the similar-seeming value terms (e.g. trace vs. mild vs. minimal) are sufficiently ambiguous that a fixed value scheme would likely not be generally applicable. Instead, values were extracted literally; this would permit researchers to specify their own search criteria. While some of these terms map to a quantitative concept in the UMLS (e.g. “moderate”), some terms (e.g. “trace”) do not. Because of this variability, values were mapped in this phase using a separate value lexicon. The following terms were identified from the training set as a value: trace, mild, moderate, severe, insignificant, trivial, small, large, minimal, marked, slight, borderline, significant, modest, critical, substantial, less, very, neither, without, no, not and absent.†
Training & sentence template recovery: Non-null semantic types and conjunctions‡ comprising a sentence formed the concept pattern for that sentence. For example, the concept pattern for:

Those concept patterns matching 3 or more unique sentences and containing at least one of the ten study conditions were added to the system as sentence templates. For example, [Value][Disease] would be added as a sentence template since it matches:
For each sentence template, a rule was defined which associates a value term to a concept. For the example above, this is trivial since there is only one option. A more complicated sentence template looked like this:
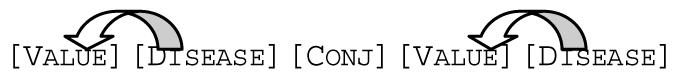
In this way, both diseases and their associated values could be identified and extracted.
Testing and concept-value pair extraction: Once all sentence templates identified in the training phase were added to the system, the concept pattern for each sentence in the test set was identified. If no concept pattern could be identified for a particular sentence, no information could be extracted. If a concept pattern did exist, then it was matched against the possible sentence templates. At this point there were three possibilities-- the concept pattern of a given test sentence could:
Completely match a sentence template,
Partially match a sentence template, or
Not match any of the sentence templates.
In the first and second cases, concept-value pairs were extracted according to what the template dictated. In the third case, if no match existed, the information contained within that sentence was ignored.
Extracted concept-value pairs were output to a Microsoft Access database for further analysis.
Methods: Evaluation
Generation of the Reference Standard
The first author is a board-certified internist and is familiar with the information contained in echocardiogram reports. Because the objective was to quantify how well this system could extract explicit disease findings, the author did not use inference to conclude disease. Only explicitly reported study concepts along with their literal values were input into an Access table. Given the nature of echocardiogram reports, which is to explicitly state the presence, absence, and degree of disease, in most cases, there was little ambiguity or need for inference. In cases where ambiguity was inherent in the sentence (“either a small pericardial effusion or an epicardial fat pad”), the condition (“pericardial effusion”) took on the value that was found associated with it (“small”).
Mapping Rules
A set of mapping rules was created in order to normalize the representation of the ten test conditions in the database. The [Disease] along with a [Body Part] or [Anatomical Structure] characterizes each condition. For example, the condition “mitral valve insufficiency” can be mapped from:
“mitral valve insufficiency [Disease]”,
“mitral valve [body part]” and “insufficiency [Disease]”,
“mitral [anatomical structure]” and “insufficiency [Disease]”. .
The set of mapping rules for each concept was used to group entries by each parent concept. Ultimately, the goal is to use the concept hierarchy built into the UMLS to generate these mapping rules.
Information Extraction
Concept-value pairs related to the ten test conditions were identified from the experimental set using these mapping rules and were compared with the identified pairs in the reference standard.
The following definitions were used:
True positive (TP): Concept-value pair present in both reference standard and experimental set.
False positive (FP): Concept-value pair found only in the experimental set. Since values were matched via an exact string match process, false positive cases occurred when value terms were semantically mismatched (“significant” v. “no significant”), but also when value terms were lexically mismatched (“(mild) 1+/4+” v. “mild) 1+/4+”).
False negative (FN): Concept-value pair found only in the reference standard.
Using these three parameters, recall and precision measures were calculated as follows:
Results
Corpus Description
Table 1 provides a description of the training and the test sets. On average the training set (UH1) had 7–8 unique sentences per report; the entire test set (UH1 and UH2) had an average of 4–5. This disparity is largely due to the differences between the UH1 corpus and the UH2 corpus; the latter had 2–3 unique sentences per report.
Table 1.
Description of corpus.
| Train | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| UH1+UH2 (n=408) | ||||||||
| UH1 (n=295) | UH1 (n=188) | UH2 (n=220) | UH1+UH2 | |||||
| Corpus Characteristic | Total | Single | Total | Single | Total | Single | Total | Single |
| Unique sentences | 2296 | 1720 | 1474 | 1068 | 494 | 242 | 1939 | 1293 |
| Unique words | 1390 | 518 | 1108 | 339 | 576 | 153 | 1350 | 393 |
| Patterns | 1255 | 885 | 863 | 581 | 321 | 146 | 1130 | 698 |
| Unmatched sentences | - | 105 | - | 74 | - | 35 | - | 109 |
p-value compares "total" numbers for each parameter in the TRAIN set and the complete TEST sets (compares both columns in greyed out areas)
Patterns
Table 2 shows the number of total patterns isolated and the number of templates selected in the training phase. The discrepancy exists because only those patterns mapping to one of the ten study conditions were entered into the system as templates. For example, only 10 patterns matched ≥50 sentences each. Of these 10 patterns, only 4 mapped to sentences containing information on at least one of the 10 test conditions. These 4 patterns matched 903 sentences in the test set demonstrating the ability of a few templates to extract information from a large number of sentences. A total of 55 templates were used to extract concept-value pairs from 1390 sentences in the test set.
Table 2.
Pattern analysis
| Matched Sentences/Pattern* | # patterns total | # templates used | # sentences |
|---|---|---|---|
| ≥50 | 10 | 4 | 903 |
| 10–49 | 62 | 14 | 317 |
| 5–9 | 59 | 13 | 89 |
| 3–4 | 91 | 24 | 81 |
| Total | 160 | 55 | 1390 |
Each pattern was categorized according to the number of unique sentences it can map and was grouped as indicated in the first column. The number of sentences mapped by the templates in that category is shown in the last column.
Information Extraction
Using the largest test set (all 55 patterns used), 1258 concept-value pairs related to the ten study conditions were extracted. Table 3 shows the distribution of extracted pairs over the test conditions. The final column is a measure illustrating the variable effectiveness of this method across conditions. This method works best for extracting “Ejection Fraction” and worst for “cardiac shunts.” This variability in performance across conditions suggests how language use may vary from disease to disease
Table 3.
Comparison of extraction efficiency among test conditions.
| n | ||||
|---|---|---|---|---|
| Condition | Expected (E) | Observed (O) | FP | (O-FP)/E |
| Mitral Valve Insufficiency | 389 | 326 | 2 | 0.83 |
| Aortic Valve Stenosis | 115 | 89 | 0 | 0.77 |
| Ejection Fraction | 450 | 415 | 10 | 0.90 |
| Mitral Valve Prolapse | 50 | 36 | 2 | 0.68 |
| Pulmonary Hypertension | 45 | 31 | 0 | 0.69 |
| Left Ventricular Hypertrophy | 139 | 18 | 0 | 0.13 |
| Pericardial Effusion | 330 | 323 | 0 | 0.98 |
| Cardiac shunt (excluding patent foramen ovale) | 36 | 2 | 0 | 0.06 |
| Patent Foramen Ovale, alone | 19 | 8 | 0 | 0.42 |
| Valvular Vegetations | 14 | 4 | 0 | 0.29 |
| Intracardiac Thrombus | 9 | 6 | 0 | 0.67 |
| 1596 | 1258 | 14 | ||
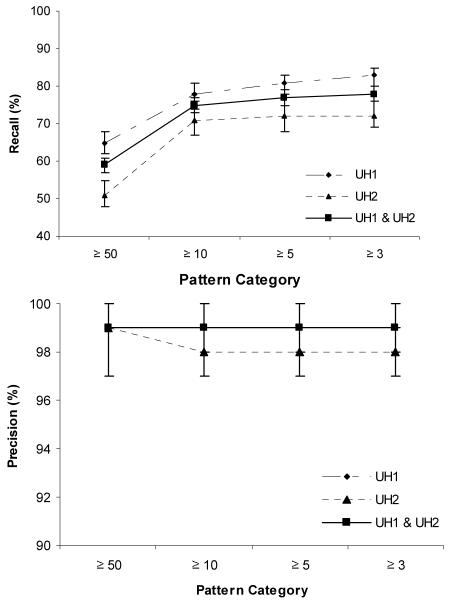
Figure B displays the recall and precision estimates from the entire test set, as well as each hospital. As expected, performance on echocardiograms from UH1 outperformed those from UH2, though not significantly—especially when fewer patterns were used. Sequentially smaller template bases were analyzed in order to demonstrate how recall and precision could be maintained even when small numbers of templates are used.
Figure B.
Recall and precision performance in test set. The precision of the combined result and that of UH1 are overlaid.
By the most conservative estimate-- the system using all patterns was able to recall 78% of the relevant findings (95% CI, 76% to 80%), with a precision of 99% (95% CI, 98%–99%).
Discussion
Clinical information resides in a wide range of formats. While some exist in a completely structured format, others exist as 'free-text'. In the middle of this spectrum, however, there lies some amalgamation of the two, of which echocardiogram reports are an oft-cited example. Echocardiogram narrative invariably contains some degree of coded data, some computer-generated text and some human-generated text. The number of unique sentences occurring as singletons and the mismatch between patterns and unique sentences suggests that there is clearly opportunity for authors to revise and that this opportunity is variably taken. The discrepancy in the number of unique sentences at each institution in the corpus description suggests that this practice is likely institutional.
There were two goals for this project. First, we sought an implementation that could reliably capture explicit concept-value pairs from this more ‘regularized’ narrative. The methods described here, while not trivial to implement nor necessarily novel, do leverage publicly available tools and clearly demonstrates how important clinical data buried within limited types of narrative can be accessed without implementing a full NLP system. Second, because this system will be used to populate a research data registry, we emphasized precision. Our limited evaluation demonstrates high precision at reasonable levels of recall. Importantly, this high level of precision was maintained even when fewer patterns were used suggesting the potential to extract a significant amount of information with minimal review of the most frequently occurring patterns. Despite the reported disparities in complexity between the two hospital records, the usefulness of the templates appears to be preserved across echocardiogram reporting customs.
Limitations
While this method shows promise, there are several key limitations of this system and of its analyses that should be addressed:
Shallow-parsing: Because of the shallow parsing, most IE systems can only extract what is explicit. Handling the most common phenomena gets you to 60% relatively quickly—getting to 100% requires handling increasingly rare phenomena. To get the rest of this information requires deeper analysis and inference2. Because the system was not robust enough to account for atypical values, certainty or inference, much data was not captured. The limited ability to deal with atypical expressions of negation also accounted for some of the loss in recall and precision. For echocardiogram reports where positive as well as negative findings are generally explicitly noted and canonically expressed, our findings suggest that this level of analysis is sufficient—especially if the goal is to preserve precision. This fact, however, does limit the generalizability of these results to other more complex narrative—even in the procedural domain.
Generation of the reference standard: Hripscak et al. found that one to two raters were needed to achieve a reliability of 0.70, and six raters, on average were required to achieve a reliability of 0.95 in information extraction tasks4. In this preliminary evaluation, only the first author generated the gold standard. This was likely sufficient for two reasons: 1.) Findings and their associated values are almost always explicitly stated in echocardiogram reports, 2.) The purpose of this system was to extract only explicit relations between a concept and its value. No attempt was made by either the system or in the construction of the reference standard to draw inference from findings suggestive of a condition. While this restricted analysis likely has the effect of overestimating the recall of the system, we believe this effect to be small given the previously noted characteristics of the echocardiogram narrative. Future evaluation of the system, however, will adapt these recommendations.
Conclusions
String matching, concept pattern matching and predefined tagging methods have all been used successfully to locate information in narrative records. This paper offers promising evidence of the utility of concept-based templates in extracting disease details from a subset of clinical narrative and shows how public tools can be leveraged to facilitate the development process.
Acknowledgements
This research was supported in part by the NLM fellowship training grant number 2T15LM07092-11.
Footnotes
MetaMap Transfer (MMTx) is a program developed at the National Library of Medicine to map biomedical text to concepts in the Unified Medical Language System (UMLS) Metathesaurus.
The adverb form of each [Value] (e.g. “moderately”) was also permitted. Value phrases were identified when [Value] terms occurred in tandem (e.g. “moderately severe”), or when a value range was described (e.g. “trace to mild”). Values identified singly or as a value phrase were identified as a single [Value] term.
Even though conjunctions are not a semantic type, they were included in the construction of sentence templates because they play a significant role in determining distribution of value terms over concepts—in future implementations, the role of conjunctions in this context will be further specified.
References
- 1.McDonald CJ. The barriers to electronic medical record systems and how to overcome them. JAMIA. 1997;4(3) doi: 10.1136/jamia.1997.0040213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobbs JR. Information extraction from biomedical text. J Biomed Inform. 2002;35(4) doi: 10.1016/s1532-0464(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 3.Jurafsky D, Martin JH. Speech and Language Processing: An introduction to natural language processing, computational linguistics, and speech recognition. 1 ed. Upper Saddle River, New Jersey: Prentice Hall; 2000.
- 4.Hripsack G, Kuperman GJ, Friedman C, Heitjan DF. A reliability study for evaluating information extraction from radiology reports. JAMIA. 1999;6(2):143–150. doi: 10.1136/jamia.1999.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin R, Lenert L, Middleton B, Shiffman S. A free-text processing system to capture physical findings: Canonical phrase identification system (CAPIS). In: 15th Annual SCAMC; 1991; Washington, DC: McGraw-Hill, Inc.; 1991. p. 843–847. [PMC free article] [PubMed]
- 6.Mikkelsen G, Aasly J. Manual semantic tagging to improve access to information in narrative electronic medical records. Int J Med Inform. 2002;65(1) doi: 10.1016/s1386-5056(01)00227-1. [DOI] [PubMed] [Google Scholar]
- 7.Brown PJB, Sonksen P. Evaluation of the quality of information retrieval of clinical findings from a computerized patient databse using a semantic terminological model. JAMIA. 2000;7:392–403. doi: 10.1136/jamia.2000.0070392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrows RC, Busuioc M, Friedman C. Limited parsing of notational text visit notes: ad-hoc vs. NLP approaches. In: J. Marc Overhage MP, editor. Proc. AMIA; 2000; Los Angeles, CA: Hanley & Belfus, Inc.; 2000. p. 51–55. [PMC free article] [PubMed]
- 9.Johnson SB, Friedman C. Integrating data from natural language processing into a clinical information system. Proc AMIA 1996. [PMC free article] [PubMed]
- 10.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In: Suzanne Bakken RD, editor. Proc. AMIA; 2001; Washington, DC: Hanley & Belfus, Inc.; 2001. p. 17–21. [PMC free article] [PubMed]