Abstract
Objective
To investigate the indirect alignment of two anatomical ontologies through a reference ontology and to compare it to direct alignment between these two ontologies. The ontologies under investigation are the Adult Mouse Anatomical Dictionary (MA) and the NCI Thesaurus (NCI). The Foundational Model of Anatomy serves as reference ontology.
Methods
The direct alignment employs a combination of lexical and structural similarity. The indirect alignment simply derives mappings from direct alignments to the reference ontology.
Results
The indirect MA-NCI alignment yielded 703 mappings and the direct alignment 715, 654 of which are common to both. The mappings specific to one approach were analyzed.
Conclusions
When a reference ontology exists, indirect alignment of multiple ontologies through a reference represents a valid, cost-effective alternative to pairwise alignment.
INTRODUCTION
Anatomical knowledge is central to biomedical applications. Various representations of anatomy have been developed including anatomical ontologies (e.g., the Foundational Model of Anatomy, Adult Mouse Anatomical Dictionary) and broader ontologies covering anatomy (e.g., GALEN, NCI Thesaurus). Despite differences in modeling principles and representation formalisms, these ontologies are expected to be compatible with each other. Mappings among ontologies constitute an enabling resource for applications such as knowledge sharing and application system communication. In particular, such mappings represent a crucial component of the Semantic Web in which the semantic annotation of resources will inevitably draw on multiple ontologies [1].
Mappings among ontologies can be built pairwise, i.e., an alignment is created between every two ontologies. Alternatively, one ontology can be selected as the reference for mapping. All other ontologies only need to be mapped to this reference ontology and the pairwise mappings can be derived from the mappings to the reference ontology. These two approaches to aligning multiple ontologies are illustrated in Figure 1.
Figure 1.
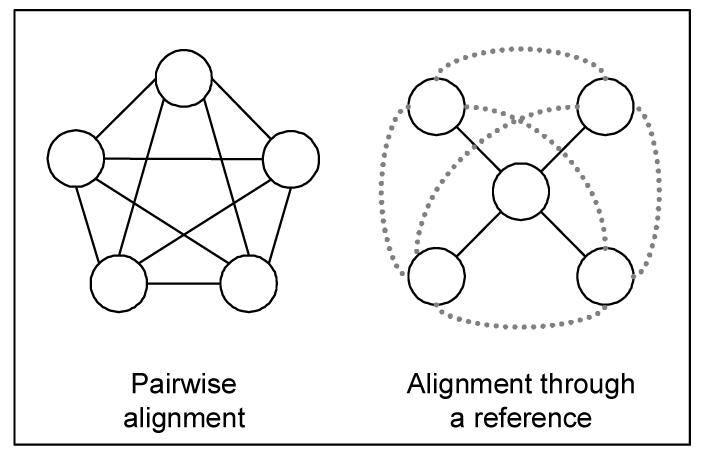
Aligning multiple ontologies
The objective of this study is to compare two approaches to aligning multiple ontologies: pairwise ontology alignment and alignment through a reference ontology. More precisely, we investigate the quality of a mapping between two ontologies (O1–O2) automatically derived from mappings created between these two ontologies and a reference ontology (O1-R and O2-R), compared to the direct mapping between O1 and O2. The three ontologies of anatomy under investigation in this study are: the Foundational Model of Anatomy (FMA)1, the Adult Mouse Anatomical Dictionary (MA)2, and the anatomy subset of the NCI Thesaurus (NCI)3. To our knowledge, this is the first attempt of deriving mappings automatically among anatomical ontologies from the alignment of these ontologies to a reference.
The creation of pairwise mappings among ontologies of anatomy draws on previous work, namely the methodology developed for aligning the Foundational Model of Anatomy and GALEN [2]. For a survey of ontology alignment techniques, see for example [3].
MATERIALS
The Adult Mouse Anatomical Dictionary (MA) is a structured controlled vocabulary describing the anatomical structure of the adult mouse [4]. It comprises 2,404 concepts. Each concept has one name (e.g., Head muscle and Adrenal artery). Additionally, 240 concepts have a total of 259 synonyms (e.g., Limb has synonym Extremity). The ontology is represented as a directed acyclic graph whose edges represent the relationships IS-A and PART-OF. Every concept is connected to others through IS-A or PART-OF relationships. However, about 38% of the concepts do not have any IS-A relationship to others (e.g., Knee PART-OF Hindlimb is the only hierarchical relation available for Knee). On the other hand, nearly 4% of the concepts have more than one IS-A relationship to others (e.g., Hand phalanx is both a kind of Phalanx and Hand digit bone). The version used in this study was downloaded on December 22, 2004 (under the name Mus adult gross anatomy in the Open Biomedical Ontologies4).
The NCI Thesaurus (NCI) provides standard vocabularies for cancer research [5] and its anatomy class describes naturally occurring human biological structures, fluids and substances. The ontology is available in the Ontology Web Language (OWL). There are 4,410 anatomical concepts (accounting for about 12% of all NCI concepts). Every concept has one preferred name (e.g., Abdominal esophagus). Additionally, 1,207 concepts have a total of 2,371 synonyms (e.g., Orbit has synonym Eye socket). Except for the root (Anatomic Structure, System, or Substance), every anatomical concept has at least one IS-A relationship to another concept, and nearly 4% of concepts have more than one IS-A relationship to others (e.g., Radius bone is both a kind of Long bone and Bone of the upper extremity). In addition, anatomical concepts are also connected by a PART-OF relationship (named ANATOMIC STRUCTURE IS PHYSICAL PART OF). The version used in this study is version 04.09a (September 10, 2004).
The Foundational Model of Anatomy (FMA) is an evolving ontology that has been under development at the University of Washington since 1994 [6]. Its objective is to conceptualize the physical objects and spaces that constitute the human body. The underlying data model for FMA is a frame-based structure implemented with Protégé. 71,202 concepts cover the entire range of macroscopic, microscopic and subcel-lular canonical anatomy. In addition to preferred terms (one for each concept), 52,713 synonyms are provided (up to 6 per concept). For example, there is a concept named Uterine tube and its synonym is Oviduct. Because single inheritance is one of the modeling principles used in the FMA, every concept (except for the root) stands in a unique IS-A relation to other concepts. Additionally, concepts are connected by seven kinds of PART-OF relationships (e.g., part of, constitutional part of, regional part of). For the purpose of this study, we considered as only one PART-OF relationship and its inverse HAS-PART the various kinds of partitive relationships present in FMA. The version used in this study was downloaded on December 2, 2004.
METHODS
As illustrated in Figure 2, this study compares the direct alignment between MA and NCI to the indirect alignment automatically generated from mapping both MA and NCI to FMA, the reference ontology. Details about the three phases of our study follow: 1) three direct alignments: MA-NCI, MA-FMA, and NCI-FMA; 2) indirect alignment between MA and NCI through their direct alignments with the FMA; and 3) comparison of the direct alignment MA-NCI to the indirect alignment obtained through the FMA.
Figure 2.
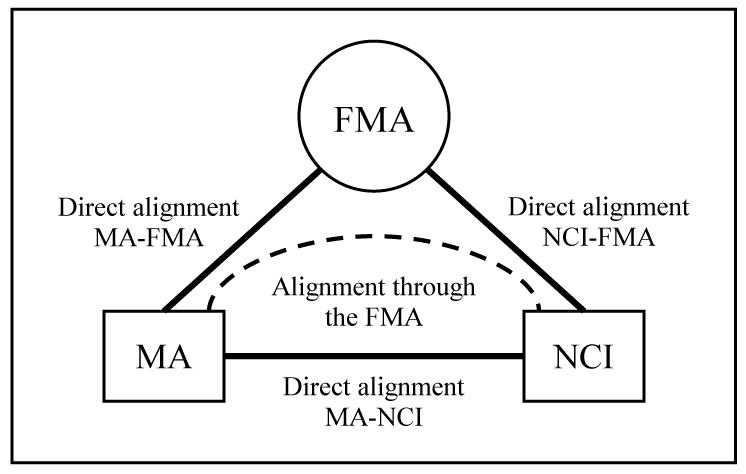
Direct vs. indirect alignment
Direct alignment
Aligning two ontologies directly consists of comparing terms across systems in order to identify one-toone concept matches. This first step constitutes the lexical component of our method. Following is the identification of structural matches. Inter-concept relationships are compared in order to identify similar relations among lexical matches across systems. The interested reader is referred to [7] for additional precisions about our method.
Identifying matches lexically
The lexical alignment compares two systems at the term level, by exact match and after normalization. This process makes the source and target terms potentially compatible by eliminating such inessential differences as inflection, case, hyphen, and word-order variation. Both preferred terms and synonyms in the two systems are used in the alignment process. Moreover, UMLS synonymy is used to identify additional matches. For example, Profunda femoris artery in MA and Deep femoral artery in NCI, although lexically different, are considered as a match because they name the same anatomical concept in UMLS. Our method does not address partial lexical matches.
Identifying matches structurally
In order to facilitate the comparison of relations across systems, the structural alignment first consists of acquiring the inter-concept hierarchical relationships, including IS-A and PART-OF, and their inverses. Missing inverse relations are complemented as necessary. Inference rules are used to generate a partitive relation between a specialized part and the whole or between a part and a more generic whole. Finally, the relations reified in the names of some FMA concepts are represented explicitly (e.g., <Heel, PART-OF, Foot> was augmented from <Heel, IS-A, Subdivision of foot>).
Once all relations are represented consistently, the structural alignment is applied on the matches resulting from the lexical alignment in order to identify similar relations to other matches across systems. For example, match concepts Forelimb in MA and Upper extremity in NCI exhibit similar relations to other matches in the two systems, including Limb (through IS-A), Arm and Hand (through HAS-PART) across systems. Such structural similarity is used as positive evidence for the alignment. Instead of similar relations, one match may exhibit relations to other matches in opposite directions in the two systems. Such relations suggest a structural conflict across systems. For example, in MA Pericardial cavity is in HAS-PART relationship to Pericardium, while in the FMA Pericardial cavity is defined as part of Pericardial sac which is part of Pericardium. These conflicts are used as negative evidence for the alignment, indicating the semantic incompatibility between concepts across systems despite their lexical resemblance.
Indirect alignment MA-NCI using FMA as a reference ontology
The following method was used for automatically deriving a mapping between MA and NCI from the two direct alignments MA-FMA and NCI-FMA. When a FMA concept CF is aligned with both a MA concept ({MA: CM, FMA: CF}) and a NCI concept ({NCI: CN, FMA: CF}), the concepts CM and CN are automatically aligned ({MA: CM, NCI: CN}).
For example, as shown in Figure 3, the direct alignment MA-FMA identifies the match {MA: Forelimb, FMA: Upper limb (synonym: Forelimb)}, which is supported by positive evidence. The direct alignment NCI-FMA identifies the match {NCI: Upper extremity, FMA: Upper limb (synonym: Upper extremity)}, also supported by positive evidence Therefore, the match {MA: Forelimb, NCI: Upper extremity} is derived automatically, through the FMA concept Upper limb, supported by positive structural evidence in both direct alignments.
Figure 3a.
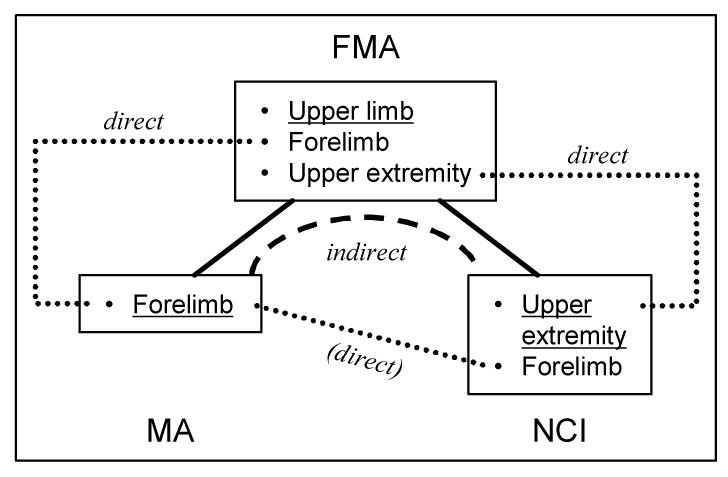
Indirect MA-NCI alignment through FMA
Comparing two alignments between MA and NCI
We compared the matches obtained by direct alignment MA-NCI and by indirect alignment through the FMA. The matches were classified into three groups: matches identified by both alignments; matches specific to the direct alignment MA-NCI; and matches specific to the alignment through the FMA. As shown in Figure 3, the match {MA: Forelimb, NCI: Upper extremity} belongs to the first group. Further analysis of structural evidence for the matches was performed in the three groups.
RESULTS
Three direct alignments
Results for three direct alignments are summarized in Table 1. The alignment NCI-FMA yielded the largest number of matches (2,173) and MA-NCI the smallest (715). A very small number of conflicts was identified in the two direct alignments to FMA; none in the direct MA-NCI alignment. In the three direct alignments, a vast majority of the matches (> 90%) was supported by positive structural evidence. No evidence (positive or negative) was found for 5–9% of the matches in three direct alignments. For example, although Elbow joint has relations to other matches in both MA (e.g., PART-OF Forelimb) and NCI (e.g., PART-OF Skeletal system), none of these relations are shared.
Table 1.
Three direct alignments
| MA - NCI 715 matches | MA - FMA 1,353 matches | NCI - FMA 2,173 matches | |
|---|---|---|---|
| No evidence | 62 (8.7%) | 66 (4.9%) | 205 (9.4%) |
| Positive evidence | 653 (91.3%) | 1,283 (94.8%) | 1,958 (90.1%) |
| Negative evidence | 0 | 4 (0.3%) | 10 (0.5%) |
Indirect alignment MA-NCI through FMA
703 matches between MA and NCI were automatically derived from the 1,353 matches in the direct alignment MA-FMA and the 2,173 matches in NCI-FMA. 649 of them (92%) received positive structural evidence in both direct alignments MA-FMA and NCI-FMA, 8 (1%) received negative evidence in one of the two direct alignments, and 46 (7%) received no evidence in at least one of the two direct alignments.
Comparison between direct and indirect alignment for MA-NCI
Quantitative results
We compared the 715 matches obtained in the direct alignment MA-NCI to the 703 matches resulting from the indirect alignment through the FMA. The results of this comparison are summarized in Figure 3. The most important finding is that 654 matches are shared by both alignments, leaving 61 matches specific to the direct alignment and 49 specific to the indirect alignment through the FMA.
Matches supported by structural evidence
Among the 654 shared matches, 583 (89%) received positive structural evidence in all three direct alignments, e.g., {MA: Forelimb, NCI: Upper extremity}.
Matches without structural evidence
65 (10%) of the 654 shared matches received no structural evidence in at least one of the three direct alignments. Although linked to other matches in MA (e.g., PART-OF Cranium), Chondrocranium has no hierarchical relations to any other matches in NCI and FMA. This is why the matches of Chondrocranium receive no evidence in any of the three direct alignments.
Conflicts
At last, 6 (1%) of the 654 shared matches received negative evidence in one of the three direct alignments. For example, while a concept Pericardial cavity is present in the three ontologies, the corresponding match received negative evidence in the direct MA-FMA alignment, no evidence in MA-NCI, and positive evidence in NCI-FMA. Because of the presence of a conflict in MA-FMA, no match is identified in the indirect alignment through FMA. In this case, the indirect alignment MA-NCI (no match suggested) is consistent with the direct alignment MA-NCI (match not supported by structural evidence). In fact, domain knowledge is required to evaluate the match in these cases.
DISCUSSION
Why are some matches in the alignment through the FMA not identified in the direct alignment?
49 matches in the indirect alignment through the FMA (7%) are not identified in the direct alignment between MA and NCI, most of them being valid (supported by structural evidence). The analysis of these cases reveals two major causes.
Additional synonyms
First, the FMA records a much larger number of synonyms for anatomical entities than the other two ontologies, increasing the chances of finding a lexical match between names in these ontologies and the FMA. For example, the terms Integumental system in MA and Integumentary system in NCI do not match directly, but are listed as synonyms in the FMA.
Additional relations
The FMA also provides a much larger number of relations among anatomical entities, amplified by the inference techniques used in our method. The presence of these additional relations increases the chances of finding similar relations between an ontology and the FMA and maximizes the chances for matches to receive positive structural evidence. It also increases the chances of identifying conflicts, therefore enhancing the overall quality of the mappings.
Why are some matches in the direct alignment not identified in the alignment through the FMA?
61 matches in the direct alignment between MA and NCI (nearly 9%) are not identified in the indirect alignment through the FMA, most of them being valid (supported by structural evidence). Analogously, among the 44 matches identified both directly and indirectly, but not supported by structural evidence in the indirect alignment, 14 received positive evidence in the direct alignment. Differing coverage and representation of anatomy seem to be the cause of these differences.
Differing coverage
Some concepts present in MA and NCI are absent from the FMA. This is the case, for example, of Iliac artery. While Common iliac artery, Internal iliac artery and External iliac artery are present in all three ontologies, Iliac artery is present in MA and NCI, but absent from the FMA. Thus, this concept can be aligned directly, but not through the FMA. We leave anatomists to decide whether and how iliac arteries should be represented. Such discrepancies in coverage are indicative of some potential problem and should be reviewed.
Differing representation
Unlike particular veins and arteries, general concepts such as Blood vessel are defined as a General anatomical term in the FMA and do not form the root of a hierarchy of blood vessels, as it is the case in MA and NCI. Therefore, the relations of particular veins and arteries to Blood vessel present in MA and NCI are not shared with FMA, although the concepts themselves are present. Again, automatic alignment techniques can at best identify such issues. Normalizing the representation generally requires domain experts.
Alignment through a reference ontology vs. pairwise alignment
As suggested in the introduction, mapping through a reference ontology is cost-effective: n ontologies require n(n-1)/2 paiwise mappings, but only n-1 mappings to a reference ontology. As illustrated in Figure 1, for five ontologies – which is a small number by Semantic Web standards – the difference already represents a 60% economy (4 vs. 10).
This study confirms the feasibility and efficiency of indirect alignment through a reference ontology. Of the 715 matches identified by direct alignment, 654 (91%) have been discovered by the indirect alignment. Moreover, the indirect alignment was able to identify matches not discovered by direct alignment. Overall, this study suggests that the performance of the indirect alignments is consistent with – if not better than – that of direct alignments.
The indirect alignment assumes the existence of an ontology that can serve as reference. Desirable characteristics for such an ontology include broad coverage (in terms of both concepts and relations), inclusion of many synonyms and compatibility with standard representation principles. In our experiment, we found the FMA to have many of these characteristics: its large size and comprehensive set of synonyms certainly contributed to the high percentage of mappings discovered (compared to direct alignment) and outweigh its idiosyncrasies.
Current limitations and future work
The MA-NCI alignments, direct and indirect, have identified a total of 764 matches. These only account for about one third of the concepts in MA and NCI anatomical concepts (excluding the 2000 NCI concepts for cell types and subcellular components, not represented in MA). Our alignment approach relies heavily on the lexical similarity and is limited to the identification of one-to-one concept matches. We plan to investigate complementary approaches based on structural similarity, as well as complex matches (one-to-many and many-to-many).
The three ontologies aligned in this study all represent the same domain: vertebrate anatomy. The analysis of fine differences between human and mouse anatomies is beyond the scope of this paper but is addressed in [8].
The absence of validation of the alignment obtained by our fully automatic techniques is another limitation of this study. The manual review of the matches by an expert is labor intensive and costly. While manual validation remains an objective of this project, we believe that the comparison between direct and indirect alignments provides some elements of cross-validation of the results.
CONCLUSIONS
This study suggests that, when a reference ontology exists, indirect alignment of multiple ontologies through a reference represents a valid, cost-effective alternative to pairwise alignment. We believe that reference ontologies will be a key component of semantic integration of biomedical information and interoperability of biomedical applications. Besides anatomy, biochemistry is one of the domains which would benefit most from the development of reference ontologies (e.g., an ontology of small molecules).
Figure 3b.
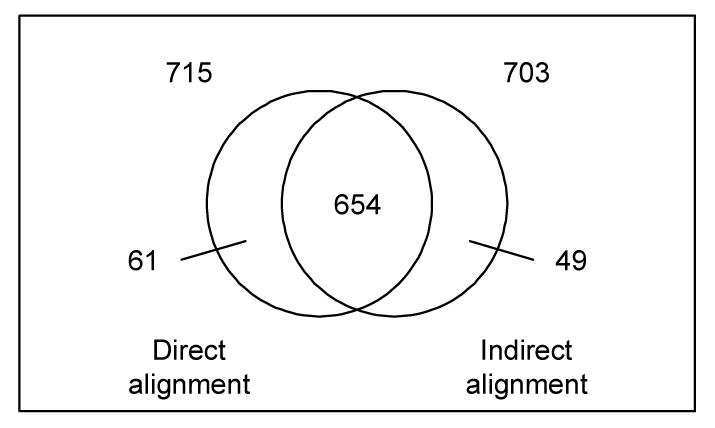
Direct vs. indirect alignment
Acknowledgements
The research was supported in part by an appointment to the National Library of Medicine Research Participation Program administrated by the Oak Ridge Institute of Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine. Thanks for their support to Cornelius Rosse and José Mejino for the Foundational Model of Anatomy, Terry Hayamizu for the Adult Mouse Anatomical Dictionary, and Sherri De Coronado for the NCI Thesaurus.
Footnotes
References
- 1.Uschold M, Gruninger M. Creating semantically integrated communities on the world wide web. Proc. International Workshop on the Semantic Web 2002 http://semanticweb2002.aifb.uni-karlsruhe.de/USCHOLD-Hawaii-InvitedTalk2002.pdf
- 2.Rector AL, Bechhofer S, Goble CA, Horrocks I, Now-lan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9(2):139–71. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 3.Noy NF. Tools for mapping and merging ontologies. In: Staab S, Studer R, editors. Handbook on Ontologies: Springer-Verlag; 2004. p. 365–384
- 4.Hayamizu T, Mangan M, Corradi J, Kadin J, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biology. 2005;6(3):R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: Using Science-based Terminology to Integrate Cancer Research Results. Medinfo. 2004;2004:33–7. [PubMed] [Google Scholar]
- 6.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. AMIA Annu Symp Proc 2003:753–7 [PMC free article] [PubMed]
- 8.Travillian RS, Gennari JH, Shapiro LG. Of mice and men: Design a comparative anatomy information system. AMIA Annu Symp Proc 2005:(in press) [PMC free article] [PubMed]


