Abstract
This study investigates the feasibility of applying Gene Ontology (GO)-derived semantic similarity methods to the biological pathway analysis. The results derived from the analysis of human metabolic and regulatory pathways are consistent with the network biology. It suggests that the semantic similarity measurement may be used to help the pathway modeling.
Introduction
In the post-genomic era, a systems biology approach is critical for the understanding of human health. We have to take a global view of the entire biological network at many levels of abstraction to manage complex biological states such as disease. However, network reconstruction requires a nontrivial integration of various functional genomic data and background knowledge. High-throughput genomic data sacrifice specificity for scale, yielding relatively lower quality measurements. Thus, incorporating prior knowledge becomes critical for the pathway modeling and network rebuilding.
Gene Ontology (GO) is a controlled vocabulary of over 17,000 terms used to describe molecular function, biological process and cellular location of genes and gene products in a generic cell. One strategy to exploit the information encoded consists of processing GO to measure the semantic similarity between gene products. The more information two terms share, the more similar they are. The shared information is indicated by the information content of the terms that subsume them in the directed acyclic graphs (DAGs). The notion of information content p(t) is defined as the frequency of each term, or any of its children occurring within the corpus. Less frequently occurring terms are “more informative”. Since GO allows multiple parents for each term, the similarity score between two terms can be defined as
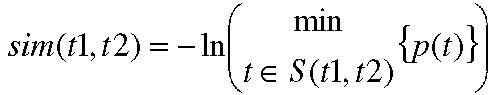 |
where S(t1, t2) is the set of terms that subsume both t1 and t2. Lord et al have verified that this measurement is significantly correlated with sequence similarity [1]. In the current study, we investigated the feasibility of applying the semantic similarity methods to the biological pathway analysis and reconstruction.
Methods
The semantic similarity measures were implemented by Perl scripting against a local copy of GO database. The information content of each GO term was calculated based on their frequency appearing in UniProt-HUMAN. Human regulatory and metabolic pathways from KEGG were analyzed separately using the implemented semantic similarity measurement. The statistical significance of similarity values was estimated by permutation test.
Results and Discussion
Our results indicate that gene products within pathways have significantly stronger semantic similarity than random pairs of gene products in terms of process and cellular component. The similarity values in terms of function have different characteristics for regulatory and metabolic pathways. In addition, protein pairs belonging to the same complex exhibit stronger similarities than other pairs in all of the GO taxonomies. It suggests that semantic similarity measures could be used for the pathway modeling. One popular paradigm for cellular modeling involves creating a comprehensive scaffold of molecular interactions and then rebuilding signaling, regulatory and metabolic pathways from this scaffold. The mining of scaffold is usually done based on high-throughput experimental data [2]. Putative pathways extracted from the scaffold may be ranked using the semantic similarity between direct and indirect interacting pairs. The integration of prior biological knowledge in the pathway modeling would greatly increase the reliability of constructed pathways.
References
- 1.Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 2.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends in Biotechnology. 2003;21:255–262. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]


