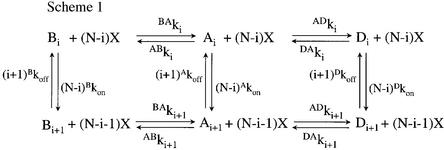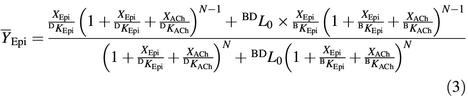Abstract
The molecular mechanisms of nicotinic receptor activation are still largely unknown. The crystallographic structure of the acetylcholine binding protein (AChBP) reveals a single H-bond between two different acetylcholine binding loops. Within these homologous loops we systematically introduced α4 residues into the α7/5HT3 chimeric receptor and found that the single point mutations G152K (loop B) and P193I (loop C) displayed a non-additive increase of equilibrium binding affinity for several agonists compared with the double mutant G152K/P193I. In whole-cell patch–clamp recordings, G152K, P193I and G152K/P193I mutants displayed an increase up to 5-fold in acetylcholine potency with a large decrease of the apparent Hill coefficients (significantly smaller than one). Concomitantly, the G152K/P193I mutant showed a dramatic loss of high-affinity α-bungarotoxin binding (100-fold decrease), thus pinpointing a new contact area for the toxin. Fitting the data with an allosteric–kinetic model, together with molecular dynamic simulations, suggests that the presence of the inter-backbone H-bond between positions 152 and 193, revealed in α4 and in α7 double mutant but not in α7, coincides with a large stabilization of both open and desensitized states of nicotinic receptors.
Keywords: activation mechanism/α-bungarotoxin binding/H-bond/myasthenic mutant/neuronal nicotinic receptor
Introduction
The precise molecular mechanisms underlying ligand-gated ion channel (LGIC) activation and desensitization remain largely unresolved. In the Torpedo nicotinic acetylcholine receptor (nAChR), these allosteric transitions involve a global conformational reorganization of the pentameric transmembrane proteins. In the presence of acetylcholine (ACh) these structural changes (known as gating) manifest themselves in a concerted manner in the ACh binding sites carried by the extracellular domain (ECD) and in the ion channel within the transmembrane domain. At the amino acid level, mutations in several domains of LGICs have been shown to alter these allosteric processes profoundly: (i) within the four transmembrane segments (M1–M4) (reviewed by Corringer et al., 2000; Karlin, 2002); (ii) within the loops connecting the M1–M2 (Wilson and Karlin, 1998) and M2–M3 (Grosman et al., 2000) segments; and (iii) at the bottom of the ECD (Kash et al., 2003). Few mutagenesis data have been reported so far on ECD, and little is known overall about the precise mechanisms at the atomic level of allosteric transitions of this domain. The crystal structure of the ACh binding protein (AChBP), which is homologous to nicotinic ECD, offers new insights into the molecular mechanisms that contribute to nAChR activation and desensitization, at least in the ACh binding area (Brejc et al., 2001).
In a previous work on α4/α7 chimeric receptors, we identified in the ECD some key residues involved in the pharmacological diversity of nAChRs (Corringer et al., 1998). Among them, the mutation α7G152K in the vicinity of the ACh binding site was found to alter the allosteric transitions of both activation and desensitization, resulting in a dramatic increase of the apparent binding affinity for nicotine. At a homologous position, a naturally occurring point mutation in muscle nAChR α1G153S also causes a ‘gain of function’ phenotype associated with a congenital myasthenic syndrome (Sine et al., 1995). The mutation produces an increase in agonist binding affinity, causing the receptor to open repeatedly during ACh occupancy, and in addition enhances the level of desensitization. Together, these data emphasize the functional role of this position in both activation and desensitization of nAChR.
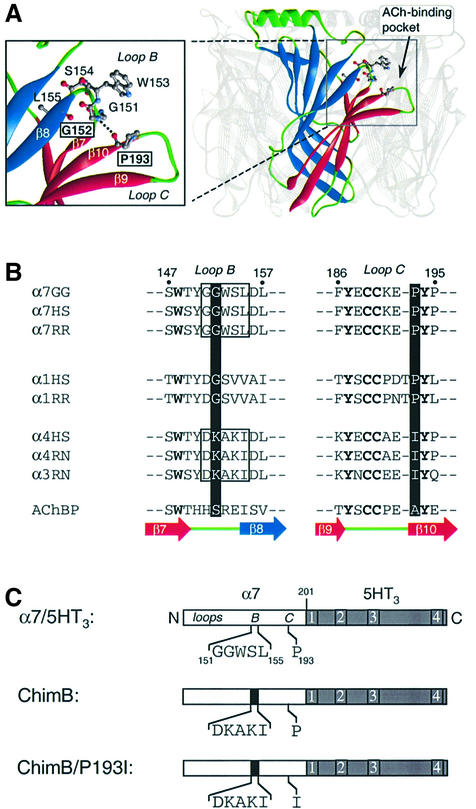
In order to obtain insights into the precise molecular mechanisms underlying the gain of function phenotype of α1G153S mutant, we visualized the homologous residue S147 on the atomic structure of AChBP. We observed that S147 interacts through an inter-backbone H-bond with A191. Comparative modeling of the three main ECDs of the nAChRs indicated the possibility of an H-bond at the homologous position (e.g. α7 in Figure 1A) (Le Novère et al., 2002). Yet, because these nAChR models are assigned to be as close as possible to the template, i.e. AChBP, we wondered whether this H-bond was actually present in nAChRs.
Fig. 1. (A) Three-dimensional model of the extracellular domain of α7 receptor based on the crystal structure of AChBP (Brejc et al., 2001) by comparative modeling (Le Novère et al., 2002). For clarity, one subunit of the pentamer is highlighted. Each subunit is folded in a twisted β-sandwich motif composed of the inner (blue) and outer (red) β-sheets. Nomenclature as described (Unwin et al., 2002). In the enlarged view, a plausible H-bond between G152 and P193 (dotted line) links loop B (fragment 151–155) and the β-hairpin of loop C (fragment 186–195), thus connecting the inner and outer β-sheets at the level of the ACh binding site. (B) Aligned sequences of loops B and C of neuronal (α7, α4 and α3) and muscle (α1) nAChRs: GG, Gallus gallus; HS, Homo sapiens; RR, Rattus norvegicus; AChBP, acetylcholine binding protein. The corresponding strands from the AChBP structure are also indicated under the sequence alignment (Brejc et al., 2001). (C) Schematic representation of the chick α7 ECD (white box) of the chimera α7/5HT3. In this chimera, amino acids beyond residue 201 (gray box) correspond to those of the 5HT3 receptor sequence (Eiselé et al., 1993). The five amino acids of loop B (segment 151–155) and P193 of the wild-type α7/5HT3 are indicated. The microchimera ChimB corresponds to the homologous substitution of rat α4 sequence (black box) loop B, whereas ChimB/P193I corresponds to the same microchimera plus the single mutant P193I in loop C.
Several lines of evidence suggest that this putative H-bond in nAChR might be involved in a critical region. (i) The positions are carried by loops B and C, respectively, which contain amino acids directly contributing to the ACh binding site (α7W148, α7Y188 and α7Y194). Photoaffinity labeling experiments performed on Torpedo receptor showed that in the time course of desensitization, labeling of loop B increases with respect to the loop C region (Galzi et al., 1991), indicating relative motions of these loops. (ii) The residues of these loops are not conserved between two nAChR subtypes that exhibit functional differences in ACh potency, i.e. the low-affinity (α7) neuronal and muscle nAChRs versus the high-affinity (α4) neuronal nAChRs (Figure 1B). (iii) AChBP forms a homopentamer where each subunit is folded into a twisted β-sandwich composed of two main β-sheets rotated against each other: the ‘inner’ and ‘outer’ β-sheet fragments. These two β-sheets are connected at the underside of the protein (near the membrane) by the disulfide bond C123–C136, while at the equatorial position they are connected by the S14:A191 H-bond. Thus, we hypothesized that this H-bond plausibly stabilizes a particular fold of AChBP and consequently would control specific pharmacological and functional features in nAChRs, such as the activation and/or desensitization transition. The present study is aimed at identifying the specific role of loops B and C, and especially this H-bond, in the allosteric properties of nAChRs.
Here we report a combined experimental and molecular modeling study of loops B and C performed by transferring α4 residues into the corresponding loops in the ECD of α7/5HT3 receptor. We find that the single point mutations G152K and P193I (homologous to S147 and A191 in AChBP) display a non-additive increase of the affinity for agonists and ACh potency compared with the double mutant G152K/P193I. Rather unexpectedly, a loss of high-affinity α-bungarotoxin binding (∼100-fold) took place concomitantly, thus pinpointing a new contact area for the toxin. Dynamic molecular simulations reveal that the glycine and proline residues prevent the formation of the H-bond in α7 receptor, whereas introducing a side chain at position 152 restores the inter-backbone H-bond as observed in α4 subunits. These data support the notion that interaction of loops B and C contributes to the activation mechanisms of nicotinic receptors, and provide for the first time a rationale for a congenital myasthenic syndrome (α1G153S) at the atomic level.
Results
Exploration of α7/5HT3 chimeras using [3H]epibatidine and [125I]α-BgTx binding
Alignment of α7 and α4 neuronal nAChR subunits showed that they are not conserved in segment 151–155 (loop B) and in position 193 (loop C) (Figure 1B). Hence, our strategy was to create chimeras by introducing α4 residues into the corresponding regions in the α7/5HT3 receptor (ChimB and ChimB/P193I) (Figure 1C), and to express the mutant chimeras in HEK-293 cells. We used the α7/5HT3 chimera as a template, which is efficiently transiently expressed in HEK-293 cells whereas α7 is not (Eiselé et al., 1993).
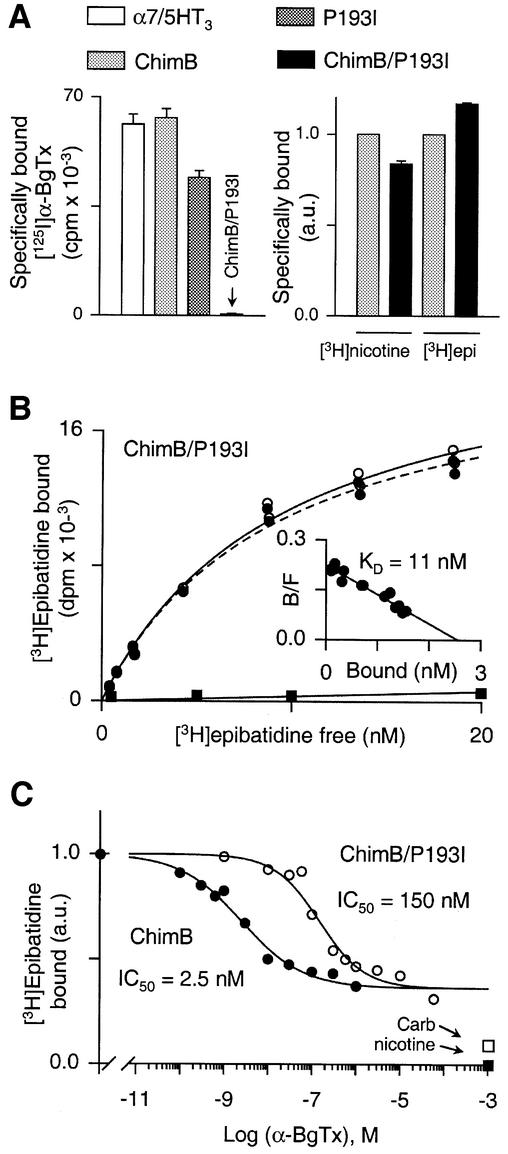
Before testing the functionality of the mutant proteins by electrophysiology, we checked their expression by [125I]α-BgTx binding since the toxin binds to the α7/5HT3 receptor in the nanomolar range. [125I]α-BgTx bound to the ChimB chimera and the P193I mutant, at a level comparable to the α7/5HT3 receptor (Figure 2A, left panel). Unexpectedly, the radioactive toxin did not bind the double mutant protein ChimB/P193I (Figure 2A, left panel). Conversely, [3H]nicotine and [3H]epibatidine bound in a specific manner to both ChimB and ChimB/P193I microchimeras (Figure 2A, right panel), whereas these radioligands did not exhibit a high-affinity binding for the α7/5HT3 receptor (not shown). These results show that (i) [3H]epibatidine bound to ChimB-containing mutant, confirming that segment 151–155 contributes to high-affinity binding (Corringer et al., 1998), and (ii) the loss of [125I]α-BgTx binding was not due to a low level of receptor expression. Hence, two apparent opposite effects were detected for the ChimB/P193I microchimera: a dramatic loss of the competitive antagonist [125I]α-BgTx binding, and an increase of the agonists [3H]nicotine and [3H]epibatidine binding. This led us to investigate further the molecular mechanisms that produced these two phenotypes. In the following, we describe first a detailed analysis of the loss of toxin binding and, secondly, the precise molecular reasons for the increase in agonist binding.
Fig. 2. (A) Left panel: representative experiment of [125I]α-BgTx binding to α7/5HT3 receptor (white bar), ChimB (light gray bar), P193I (dark gray bar) and ChimB/P193I (black bar). Membrane fragments of each construction were incubated with 5 nM [125I]α-BgTx for 2 h and samples were filtered and counted. Values indicated represent specific [125I]α-BgTx binding (non-specific binding was determined in the presence of 1 mM nicotine). Data are means ± SE of triplicates. Right panel: representative experiment of specific (non-specific binding was determined in the presence of 1 mM nicotine) [3H]nicotine and [3H]epibatidine binding on ChimB (light gray bar) and ChimB/P193I (black bar). Membrane fragments were incubated with 100 nM [3H]nicotine or 10 nM [3H]epibatidine until equilibrium. Samples were filtered and counted. Data were normalized (a.u., arbitrary units) to ChimB values and are means ± SE of triplicates. (B) Representative experiment of [3H]epibatidine binding to the double mutant ChimB/P193I. Membrane fragments were incubated with various concentrations of [3H]epibatidine (0.5–20 nM) in the presence (filled squares) or absence (open circles) of 1 mM nicotine until equilibrium and samples were filtered and counted. Specific [3H]epibatidine is also indicated (filled circles). The inset shows a Scatchard plot of specific binding of [3H]epibatidine. Data were fitted to non-linear and linear least-squares analysis for the saturation and Scatchard plots, respectively, according to a single-site model. In this experiment, KD = 11 ± 1 nM and Bmax = 2.56 ± 0.07 nM. (C) Representative competition for [3H]epibatidine binding sites by α-BgTx for ChimB (filled circles) or ChimB/P193I (open circles) microchimeras. Membrane fragments were first incubated with various concentrations of α-BgTx (as indicated) until equilibrium, 10 nM [3H]epibatidine was then added for 1 h and samples were filtered and counted. Data were normalized to values determined in the absence of α-BgTx. Data points are means of duplicates of a representative experiment. Specific [3H]epibatidine binding was determined in the presence of 1 mM nicotine (filled square). Also indicated is [3H]epibatidine binding in the presence of 1 mM carbamylcholine (open square) for both mutants. Data were fitted to non-linear least-squares analysis according to a single-site inhibition model. Note that in this representative experiment, α-BgTx displaced at most 60% of bound [3H]epibatidine, whereas nicotine or carbamycholine displayed almost 100 and 91% of bound [3H]epibatidine, respectively.
G152K/P193I mutant is sufficient to inhibit toxin binding specifically
As [125I]α-BgTx cannot be used to investigate the ChimB/P193I microchimera, we used [3H]epibatidine binding to quantify α-BgTx binding by competitive displacement to all chimeras. Saturation experiments at equilibrium showed that [3H]epibatidine bound to the double mutant ChimB/P193I protein with a high affinity in the nanomolar range (Figure 2B). The linearity of the Scatchard plot suggests that [3H]epibatidine titrated a homogeneous population of sites (Figure 2B, insert). Similar results were obtained with ChimB (not shown).
In competition experiments, we set up a new binding protocol in which the toxin was first incubated with the receptor until equilibrium and then [3H]epibatidine was added for 1 h (see Materials and methods). Considering the slow association and dissociation constants of α-BgTx, it is likely that a period of 1 h for [3H]epibatidine incubation was not sufficient for equilibrium to be reached. Indeed, we observed that equilibrium was not reached for ChimB (data not shown). Nevertheless, the α-BgTx concentration that yielded half-inhibition of [3H]epibatidine binding (IC50) for ChimB (5.3 ± 1.3 nM; Table I) was close to, if not identical with, the apparent affinity determined by following initial velocity binding of [125I]α-BgTx (Kp = 4.6 ± 1.1 nM; Figure 3A). Therefore, we assumed that the different IC50 values measured in our binding assays (Ki values cannot be used) closely reflected the actual dissociation constant of the toxin (Kp or KD).
Table I. Binding properties of α7/5HT3 mutants for [3H]epibatidine and α-bungarotoxin.
| Construction | [3H]epibatidine binding |
α-Bgtx binding |
N | ||
|---|---|---|---|---|---|
| KD (nM) | nH | IC50 (nM) | nH | ||
| α7/5HT3 | NQ | NQ | 5.7 ± 1.4a | 1.05 ± 0.16 | 3 |
| α4β2 | 1.0 ± 0.1 | 0.87 ± 0.06 | >100 000 | – | 1 |
| ChimB | 11.2 ± 3.5 | 0.93 ± 0.08 | 5.3 ± 1.3 | 0.61 ± 0.05 | 4 |
| 4.6 ± 1.1a | 1.49 ± 0.23 | 2 | |||
| P193I | 5.6 ± 0.3 | 0.93 ± 0.07 | 22.5 ± 4.4 | 1.14 ± 0.01 | 2 |
| G152S | 14.3 ± 4.2 | 0.97 ± 0.05 | 38 ± 25 | 0.71 ± 0.05 | 1 |
| G152D | 4.5 ± 1.5 | 1.08 ± 0.10 | 25 ± 9 | 0.89 ± 0.67 | 2 |
| G152E | 76 ± 31 | 0.98 ± 0.03 | 53 ± 30 | 0.98 ± 0.12 | 3 |
| G152K | 10.7 ± 0.5 | 1.00 ± 0.01 | 39 ± 12 | 0.83 ± 0.01 | 2 |
| ChimB/P193I | 9.3 ± 2.1 | 0.98 ± 0.02 | 202 ± 65 | 1.21 ± 0.63 | 2 |
| G151D/P193I | 4.7 ± 0.4 | 1.01 ± 0.01 | 7.9 ± 0.3 | 0.63 ± 0.08 | 2 |
| G152K/P193I | 6.4 ± 1.0 | 0.97 ± 0.02 | 600 ± 132* | 1.03 ± 0.06 | 4 |
| W153A/P193I | 7.1 ± 1.9 | 0.99 ± 0.01 | 30 ± 1 | 1.02 ± 0.88 | 2 |
| L155I/P193I | 9.4 ± 2.7 | 1.00 ± 0.02 | 19.3 ± 4.8 | 0.92 ± 0.19 | 2 |
| G152S/P193I | 6.7 ± 1.7 | 0.96 ± 0.05 | 40 ± 9 | 0.85 ± 0.13 | 1 |
| G152D/P193I | 27 ± 11 | 0.98 ± 0.01 | 109 ± 14 | 0.97 ± 0.21 | 2 |
| G152N/P193I | 50 ± 20 | 0.97 ± 0.04 | >100 000 | – | 2 |
| G152N/S154A/P193Ib | 21 ± 6 | 0.99 ± 0.03 | 502 ± 128 | 0.92 ± 0.12 | 2 |
| G152E/P193I | 13.1 ± 4.2 | 1.01 ± 0.02 | 184 ± 67 | 0.84 ± 0.12 | 2 |
IC50, concentration of α-BgTx that yields 50% inhibition of [3H]epibatidine binding (mean ± SE); KD, dissociation constant (mean ± SE); nH, Hill coefficient (mean ± SE); N, number of independent experiments, each performed at least in duplicate; NQ, not quantifiable.
*Statistically different from α7/5HT3 using an unpaired Student’s t-test (p < 0.025).
aProtection constant Kp determined from initial velocities of [125I]α-BgTx binding (mean ± SE) (Corringer et al., 1995).
bMutant in which the putative N-glycosylation site NWS (created by G152N mutation) was suppressed by introducing the mutation S154A (sequence NWA).
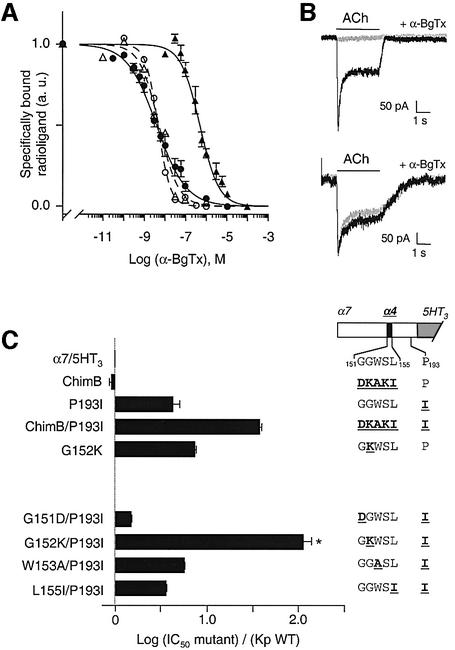
Fig. 3. (A) Competition binding curves for either [125I]α-BgTx (broken curves; open symbols) or [3H]epibatidine (solid curves; filled symbols) binding sites by α-BgTx to α7/5HT3 (open triangles), ChimB (filled and open circles) and G152K/P193I (filled triangles). For [125I]α-BgTx binding, membrane fragments of α7/5HT3 or ChimB were incubated with various concentrations of α-BgTx for 2 h, 2.5 nM [125I]α-BgTx was added for 5 min (initial velocity conditions) and samples were filtered and counted. For [3H]epibatidine binding, the protocol given in the caption to Figure 2C was used. Data were normalized to values determined in the absence of α-BgTx. Non-specific binding was determined in the presence of 100 µM α-BgTx. Data are means ± SE of at least three independent experiments performed in duplicate. For clarity, standard errors were omitted for [125I]α-BgTx binding for ChimB and α7/5HT3 data. (B) Representative whole-cell patch–clamp traces of inhibition of ACh-induced currents by α-BgTx. Upper panel: application of 1 mM ACh for 3 s to a cell expressing α7/5HT3 receptors induced an inward current (dark trace). Preincubation of the same cell with 20 nM α-BgTx for 10 min inhibited 1 mM ACh-induced current strongly (light trace). Lower panel: same protocol as in the upper panel but with the G152K/P193I double mutant. In this representative experiment, preincubation with 100 nM α-BgTx for 10 min did not inhibit 1 mM ACh-induced current. (C) Mutational scanning of α-BgTx affinity binding for the loop B region. The drawing on the right is a schematic representation of the double mutant series (see caption to Figure 1 for explanation). The α4 sequence (black box in the schematic representation) is shown in bold underlined type, while α7 is represented in light-face type. Data are means ± SE of at least two experiments (see Table I). The asterisk represents values that are statistically different from α7/5HT3 using an unpaired Student’s t-test (p < 0.025).
High α-BgTx concentrations displaced at most ∼60% of the bound [3H]epibatidine, while nicotine or carbamylcholine almost completely displaced bound [3H]epibatidine for each mutant tested (Figure 2C). Similar results were observed with both intact cells and membrane preparations, ruling out cell-dependent internalization mechanisms. Hence a fraction of [3H]epibatidine sites, which also bound nicotine and carbamylcholine, was insensitive to α-BgTx. We did not perform further experiments to investigate this issue, but one explanation may be a clustering of α7/5HT3 receptors (and the mutants described in this article) by an unknown mechanism which prevents access of the large toxin to a fraction of binding sites.
Binding data were normalized to the maximum displacement of α-BgTx (Figure 3A). As a control, we checked that α-BgTx affinity for α7/5HT3 (Kp = 5.7 ± 1.4 nM; Table I) was consistent with previous data (Corringer et al., 1995) and that the α4β2 receptor protein was not sensitive to α-BgTx up to 100 µM (Table I). Compared with α7/5HT3, ChimB and P193I mutant proteins displayed a slight increase in the KD for α-BgTx (4-fold for P193I; Table I). On the other hand, the double mutant protein ChimB/P193I displayed a markedly reduced apparent affinity for α-BgTx (∼40-fold; Figure 2C; Table I), as observed previously following [125I]α-BgTx binding (Figure 2A). These results suggest that the double mutant receptor ChimB/P193I displays a large decrease in α-BgTx binding affinity.
In order to identify the residues of ChimB that caused the loss of α-BgTx binding affinity, we introduced the P193I mutation into the α7/5HT3 receptor together with mutation of each single residue of segment 151–155. Four of the mutant proteins displayed [3H]epibatidine binding (Table I). S154K/P193I did not bind [3H]epibatidine, probably because of a lack of expression, in agreement with a previous observation (Corringer et al., 1998). Clearly, G152K/P193I displayed a significant loss of the apparent α-BgTx high affinity (∼100-fold affinity decrease; Figure 3A and C). Moreover, neither G152K nor P193I showed a significant loss of α-BgTx binding affinity (Figure 3C). Furthermore, the loss of α-BgTx affinity was also found through functional analysis of the G152K/P193I mutant in whole-cell patch–clamp recordings (Figure 3B). While application of 20 nM α-BgTx for 10 min strongly inhibited (almost 100%) ACh-evoked currents in α7/5HT3 receptor, no inhibition of ACh-evoked currents was observed for cells expressing G152K/P193I mutant with application of up to 100 nM α-BgTx (Figure 3B). Together, these data demonstrate that the loss of α-BgTx high-affinity binding is due to the combination of mutations at positions G152 and P193.
Steric hindrance governs toxin binding at position 152
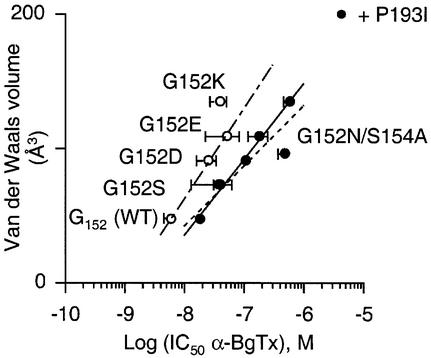
To investigate the mechanism of the loss of α-BgTx binding, a variety of amino acid side chains were introduced at position 152 in the P193I mutant background. Introducing amino acids with an increasing volume (serine, aspartate, glutamate and lysine) at position 152 reduced the apparent α-BgTx binding affinity (Figure 4). A linear correlation (R = 0.996) was noted between the gradual loss of α-BgTx affinity and the van der Waals volumes of the residues introduced at position 152 (Figure 4, filled circles). A similar linear correlation (R = 0.758) was also observed when the amino acid substitutions were performed at the same position 152 without the P193I background mutation (Figure 4, open circles). The fact that correlations are almost parallel suggests that the two positions 152 and 193 contribute independently, in a simple additive manner, to α-BgTx binding.
Fig. 4. van der Waals volume contribution to α-BgTx binding explored by mutation at position 152 with (filled circles) or without (open circles) mutant P193I. Each point is the mean ± SE from receptors containing the indicated mutation at position 152. Note that suppression of the putative N-gycosylation signal NWS by mutating S154A in G152N/P193I mutant (sequence NWA) increased apparent affinity binding for α-BgTx (see Table I for values). Data were fitted to the linear regression y = a log[x] + b. Filled circles, R = 0.996 for solid line (a = 56 ± 3 Å3 l/mol) and R = 0.902 for broken line (a = 45 ± 11 Å3 l/mol); open circles, R = 0.758 for chain line (a = 67 ± 33 Å3 l/mol). For the solid line, G152N/S154A/P193I mutant data (sequence NWA) were not included in the linear regression fit.
In addition, we created the G152N/P193I mutant, which introduced a consensus putative N-glycosylation site (sequence NWS) and noted a complete loss of toxin binding [no displacement of [3H]epibatidine up to 100 µM α-BgTx (Table I)], still with high-affinity [3H]epibatidine sites (Table I). Moreover, suppression of the N-glycosylation signal of NWS by mutating S154A (sequence NWA) resulted in an increase in toxin binding (502 ± 128 nM) close to that found with the G152D mutation (109 ± 14 nM). Including G152N/S154A/P193I mutant data in the linear correlation still gave a linear relationship (R = 0.902). Hence, the isosteric mutation of aspartate to asparagine did not significantly affect the toxin binding despite the removal of the negative charge (Table I). Together, these results clearly support the proposal that steric hindrance governs α-BgTx binding at position 152.
G152K and P193I mutants increase the affinity of agonists
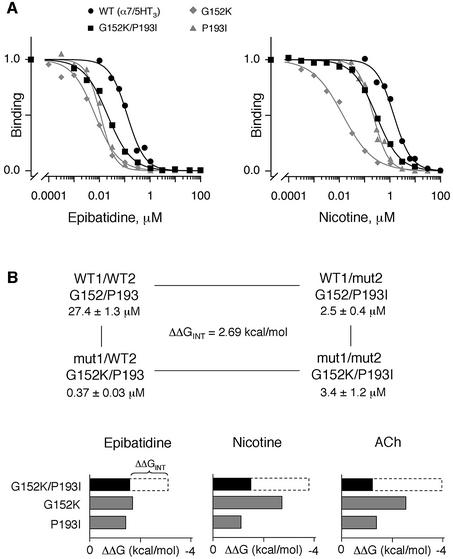
The second phenotype we observed for ChimB/P193I chimera was an increase of the binding affinity for [3H]epibatidine when compared with the wild-type α7/5HT3 receptor (Table I). It has been shown previously that the G152K mutant displayed the most critical effect within segments 151–155 of loop B (Corringer et al., 1998). Here, we have shown that, within fragment 192–195 of loop C, the proline in position 193 was responsible for the high-affinity phenotype, since the mutation of the other non-conserved proline residue within this loop (the P195L mutation) did not increase, but rather decreased, ACh, nicotine and epibatidine binding affinity (Table II).
Table II. Affinity constants and pairwise interaction energies of α7/5HT3 mutants for ACh, nicotine and epibatidine.
| K (µM) | nH,Y | ΔK | ΔΔG (kcal/mol) | Ω | ΔΔGINT (kcal/mol) | |
|---|---|---|---|---|---|---|
| ACh |
|
|
|
|
|
|
| α7/5HT3 | 27.4 ± 1.3a | 1.57 ± 0.13 | 1.0 | – | – | – |
| G152K | 0.37 ± 0.03a | 1.07 ± 0.08 | 74 | –2.51 | – | – |
| P193I | 2.5 ± 0.4a | 2.00 ± 0.10 | 11 | –1.40 | – | – |
| G152K/P193I | 3.4 ± 1.2b | 0.94 ± 0.06 | 8.1 | –1.22 | 101 | 2.69 |
| P195L |
74.2 ± 11.6a |
2.09 ± 0.67 |
0.37 |
– |
– |
– |
| Nicotine |
|
|
|
|
|
|
| α7/5HT3 | 1.43 ± 0.10a | 1.25 ± 0.10 | 1.0 | – | – | – |
| G152K | 0.014 ± 0.002a | 0.72 ± 0.04 | 102 | –2.70 | – | – |
| P193I | 0.22 ± 0.02a | 1.45 ± 0.15 | 7.0 | –1.09 | – | – |
| G152K/P193I | 0.113 ± 0.009b | 0.94 ± 0.05 | 13 | –1.48 | 52 | 2.31 |
| P195L |
6.93 ± 0.95a |
1.30 ± 0.22 |
0.21 |
– |
– |
– |
| Epibatidine |
|
|
|
|
|
|
| α7/5HT3 | 0.126 ± 0.011a | 1.25 ± 0.11 | 1.0 | – | – | – |
| G152K | 0.0068 ± 0.0033a | 0.95 ± 0.15 | 19 | –1.70 | – | – |
| P193I | 0.0111 ± 0.0030a | 1.53 ± 0.29 | 11 | –1.42 | – | – |
| G152K/P193I | 0.0082 ± 0.0016b | 0.92 ± 0.05 | 15 | –1.59 | 13 | 1.53 |
| P195L | 0.603 ± 0.135a | 0.91 ± 0.18 | 0.21 | – | – | – |
aProtection constants Kp (mean ± SE) were obtained from at least two different independent experiments performed in duplicate and were determined from initial velocities of [125I]α-BgTx binding (Corringer et al., 1995).
bInhibition constants Ki (mean ± SE) were obtained from at least two different independent experiments performed in duplicate and were calculated using the measured IC50 at equilibrium from [3H]epibatidine binding (Cheng and Prusoff, 1973).
ΔK = Kp(α7/5HT3)/Kp(mut) or Kp(α7/5HT3)/Ki(G152K/P193I).
ΔΔG = –RT ln(ΔK) where R = 1.99 cal mol–1K–1 and T = 293 K.
ΔΔGINT = RT lnΩ where Ω = K(mut1/mut2)K(WT1/WT2)/K(mut1/WT2)K(WT1/mut2) and K is Kp or Ki, mut1 is G152K, mut2 is P193I, WT1 is G152 and WT2 is P193.
G152K and P193I mutations performed in the same receptor molecule also increased the binding affinity for agonists. In control experiments, we checked that the affinities of ACh (Kp = 27.4 ± 1.3 µM), nicotine (Kp = 1.43 ± 0.10 µM) and epibatidine (Kp = 0.126 ± 0.011 µM) binding to the wild-type receptor were very close to those published in the literature (Corringer et al., 1998; Quiram and Sine, 1998) (Figures 5A and 6B). Following mutation, the Kp of ACh, nicotine and epibatidine decreased for G152K, P193I and G152K/P193I mutants (Figures 5A and 6B). More precisely, the increase in binding affinity was similar (ΔK ∼ 10–15; Table II) for all three agonists for the P193I and G152K/P193I mutants. For the single G152K mutation, the affinity was more pronounced for nicotine (ΔKp = 102) and ACh (ΔKp = 88) than for epibatidine (ΔKp = 19) (Table II). Hence, our binding results not only confirm that the G152K mutation suffices to confer a high affinity for nicotine and ACh, as identified previously (Corringer et al., 1998), but also demonstrate that P193I mutation confers a high affinity for those ligands.
Fig. 5. (A) Competition binding curves by epibatidine (left panel) or nicotine (right panel) to [125I]α-BgTx binding sites for α7/5HT3 wild-type receptor (filled circles), G152K mutant (diamonds) and P193I mutant (triangles) or to [3H]epibatidine binding sites for G152K/P193I mutant (squares). For [125I]α-BgTx binding, membrane fragments of mutant and wild-type receptors were incubated with various concentrations of the indicated ligands until equilibrium, 2.5 nM [125I]α-BgTx was added for 5 min (initial velocity conditions) and samples were filtered and counted. For [3H]epibatidine binding, membrane fragments of G152K/P193I mutant were incubated with various concentrations of the indicated ligands, 10 nM [3H]epibatidine was added until equilibrium and samples were filtered and counted. Data normalized to values determined in the absence of ligand are the means of at least two independent experiments performed in duplicate. For clarity, standard errors are omitted. Data were fitted by the empirical Hill equation. (B) Interaction energies of G152K and P193I mutations. Upper panel: mutant cycle analysis for ACh binding between G152K and P193I. Lower panel: histograms indicating changes in the binding free energy ΔΔG upon mutations of G152K, P193I (gray boxes) and G152K/P193I (black boxes) for epibatidine, nicotine and ACh. The white dashed segments of the bars represent interaction free energies ΔΔGINT between mutants (see Table II for values).
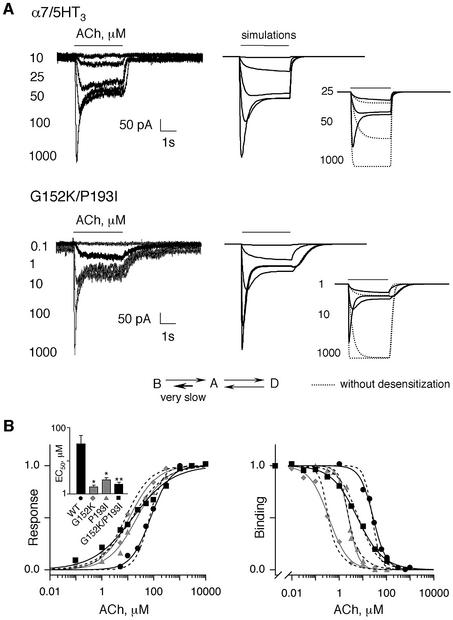
Fig. 6. (A) Typical whole-cell patch–clamp traces evoked by the indicated concentration (µM) of ACh for 3 s in cells transfected with wild-type α7/5HT3 (upper panel) or the double mutant G152K/P193I (lower panel). To the right of the recordings, the solid lines correspond to the model-generated currents evoked by the indicated concentrations of ACh (same concentrations as experimental recordings) using the minimal three-state allosteric– kinetic model. The inset shows model-generated currents evoked by the indicated concentrations (µM) of ACh using the three-state (solid lines) or two-state (without desensitization, dotted lines) model. The kinetic pathways between the basal (B), open (A) and desensitized (D) states are also indicated. The short bold arrow indicates the affected kinetic pathway in the G152K/P193I mutant. (B) Left panel: ACh dose–response relationship of α7/5HT3 wild-type receptor (filled circles) and G152K (diamonds), P193I (triangles) and G152K/P193I (squares) mutant receptors. Peak responses were normalized to maximum currents of each cell. For clarity, standard errors are omitted. Data were fitted by the empirical Hill equation (solid curves). The inset shows mean EC50 ± SE from curves obtained from different cells. Statistical differences from from wild type are indicated as follows (for values see Table III): *p < 0.005); **p < 0.001. Right panel: competition binding curves by ACh either to [125I]α-BgTx binding sites for α7/5HT3 wild-type receptor (filled circles), G152K mutant (diamonds), P193I mutant (triangles) or to [3H]epibatidine binding sites for G152K/P193I mutant (squares). The protocol is the same as described in the legend to Figure 5. Data are normalized to binding determined in the absence of ACh and are the means of at least two independent experiments performed in duplicate. For clarity, standard errors are omitted. Data were fitted by the empirical Hill equation (solid curves), while dashed curves are the best fit using the three-state allosteric model. Note that model-generated dose–response curves are identical for P193I and G152K/P193I mutants. For G152K/P193I mutant, binding data were fitted using equation (3) (see Materials and methods) with the parameters indicated in Table IV and with XEpi = 10 nM, BKEpi = 4 nM, DKEpi = 1 µM and BDL0 = 42.9. For G152K mutant data, the best fit was obtained with BKD = 33.2 µM, AKD = 1.2 µM and DKD = 0.2 µM, keeping all other parameters in Table IV unchanged.
However, at variance with α-BgTx binding data, non-additive effects were noted between G152K and P193I. We quantified this observation by the pairwise interaction method using the thermodynamic mutant cycle analysis (Figure 5B, upper panel). This technique has been used extensively to calculate coupling energies between amino acid residues involved in close interactions (Carter et al., 1984; Molles et al., 2002). We found interaction energies ΔΔGINT for ACh (2.69 kcal/mol), nicotine (2.31 kcal/mol) and epibatidine (1.53 kcal/mol) that were smaller than the sum of the ΔΔG values of the two single mutations (Figure 5B, lower panel). Therefore, the interaction between the two mutated residues K152 and I193 contributed to the high-affinity phenotype for agonists.
G152K and P193I mutants increase ACh potency
Since the two positions 152 and 193 conferred high-affinity binding for agonists, it became important to determine whether a similar increase of agonist potency (gain of function) could be recorded with cells expressing the α7/5HT3 constructs, using whole-cell patch–clamp. Dose–response curves in cells expressing wild-type α7/5HT3 showed a mean EC50 for ACh of 75.7 ± 12.1 µM and an apparent Hill coefficient slightly above unity (nH,A = 1.16 ± 0.07) (Figure 6B, left panel). These values were close to those found with α7/5HT3 expressed in oocytes (Corringer et al., 1998), although with a lower apparent Hill coefficient. Conversely, the dose–response curves established with either the single mutants G152K or P193I, or the double mutant G152K/P193I, were significantly left-shifted (5-fold; Figure 6B, left panel; Table III). These data further confirm that mutations promote a high-affinity phenotype for nicotinic agonists.
Table III. Functional and binding properties of ACh for the different α7/5HT3 mutants.
| Construction | Patch–clamp recording dataa |
Binding dataa |
|||
|---|---|---|---|---|---|
| EC50 (µM) | nH,A | No. of cells | Kp or Ki (µM) | nH,Y | |
| α7/5HT3 | 75.7 ± 12.1 [68.4] | 1.16 ± 0.07 [1.64] | 6 | 27.4 ± 1.3 [25.4] | 1.57 ± 0.13 [2.55] |
| G152K | 11.4 ± 2.6** [8.2] | 0.83 ± 0.07* [0.91] | 5 | 0.37 ± 0.03 [0.38] | 1.07 ± 0.08 [1.83] |
| P193I | 22.6 ± 1.9** [14.5] | 0.79 ± 0.04** [0.69] | 5 | 2.5 ± 0.4 [2.8] | 2.00 ± 0.10 [1.82] |
| G152K/P193I | 15.6 ± 2.2*** [14.5] | 0.57 ± 0.03*** [0.69] | 11 | 3.4 ± 1.2 [2.6] | 0.94 ± 0.06 [1.02] |
EC50, concentration of ACh that yields 50% of the response (mean ± SE); nH,A, Hill coefficient of dose–response curves (mean ± SE); nH,Y, Hill coefficient of binding curves (mean ± SE).
aTheoretical values derived from the allosteric–kinetic model are indicated in brackets.
*Significantly different from wild-type α7/5HT3 receptor using an unpaired Student’s t-test (p < 0.01).
**Significantly different from wild-type α7/5HT3 receptor using an unpaired Student’s t-test (p < 0.005).
***Significantly different from wild-type α7/5HT3 receptor using an unpaired Student’s t-test (p < 0.001).
Surprisingly, low values for the apparent Hill coefficients (nH,A between 0.57 ± 0.03 and 0.83 ± 0.07) were observed with the mutants compared with wild-type α7/5HT3 receptor (1.16 ± 0.07; Table III). Hill coefficients below unity usually indicate a heterogeneous population of functional receptors. However, in the equilibrium binding experiments, the Hill coefficients nH,Y were close to unity (0.94 ± 0.06 for ACh, 0.94 ± 0.05 for nicotine and 0.92 ± 0.05 for epibatidine; Table II). Thus the apparent decrease in nH,A did not seem to result from an obvious heterogeneity of functional receptors.
Mutations affect selectively the activation transition of nAChR
Mutations that produce an increase in the apparent binding affinity and potency for an agonist can be interpreted by modifications of the intrinsic binding parameters (K-phenotype) and/or by modifications of the conformational transitions of the receptor protein (L-phenotype) (see Materials and methods). In order to discriminate between these two possibilities, we applied the kinetic model (Edelstein et al., 1996) derived from the Monod–Wyman–Changeux (1965) model, which includes a closed but activatable state (B state), an active state in which the ion channel is open (A state) and a desensitized closed non-activatable state (D state) according to a linear cascade scheme (see Scheme 1).
Scheme 1.
First, our strategy was to define a set of 12 parameters for the wild-type α7/5HT3 receptor using ACh, assumed to bind to five equivalent sites per molecule (Palma et al., 1996), and to elicit activation and desensitization with the kinetic constants established with α7/5HT3 receptor (Galzi et al., 1996; Corringer et al., 1998). Theoretical traces were fitted to experimental data and adequately described the rising peak current, the steady-state current and the deactivation current of the wild-type receptor (Figure 6A, upper panel). Model-generated dose–response and binding curves gave an apparent EC50 and binding affinity Kp very close to the experimental values (Figure 6B; Table III). On the other hand, a systematic discrepancy in theoretical Hill coefficients for dose–response curves (nH,A) and binding curves (nH,Y) was present when compared with wild-type values (Table III) which we pursued with kinetic modeling.
Secondly, we varied the parameters of the wild-type receptor in order to fit the data for the double mutant G152K/P193I. Changing only the kinetic constant of the pathway A→B, by decreasing ABk0 from 105 to 102 s–1 (Table IV) and adjusting all other parameters by <13-fold, resulted in an adequate fit of the in vivo current recordings (Figure 6A, lower panel). Model-generated dose–response and binding curves adequately matched the experimental data (Figure 6B). Thus, the double mutant was interpreted as an L-phenotype. More importantly, the values of EC50 (15 µM) and nH,A (0.69) obtained from the simulation were close to, if not identical with, the experimental values (EC50 = 15.6 µM and nH,A = 0.57) (Table III). According to these parameters, the apparent low value of the Hill coefficient nH,A resulted from an underestimation of the actual peak currents at high agonist concentrations because desensitization intervenes prior to reaching full response amplitude (Figure 6A, inset to lower panel). It should be noted that a similar, although less pronounced, effect is also predicted for the wild-type α7/5HT3 receptor (Figure 6A, inset to upper panel), thereby explaining its rather low Hill coefficient (nH,A ∼ 1.2).
Table IV. Parameter values for the three-state allosteric kinetic mechanism for the α7/5HT3 receptor and mutants with five equivalent ACh binding sites per molecule.
| α7/5HT3 receptor parameters |
|
|
|
| State parameters |
B state |
A state |
D state |
| ACh on rates (M–1 s–1) | Bkon = 107 | Akon = 3.33 × 106 | Dkon = 106 |
| ACh off rates (s–1) | Bkoff = 880 | Akoff = 7.5 | Dkoff = 1.6 |
| Equilibrium constants (µM) |
BKD = 88 |
AKD = 2.25 |
DKD = 1.6 |
| Interconversion parameters |
B↔A |
A↔D |
|
| Transition state positional parameters | BAp = 0.75 | ADp = 0.99 | |
| Interconversion rates (s–1) | BAk5 = 22.8 | ADk5 = 2.54 | |
| ABk5 = 0.107 | DAk5 = 1.10 | ||
| BAk0 = 0.233 | ADk0 = 2.5 | ||
| ABk0 = 105 | DAk0 = 6.0 | ||
| Allosteric constants | BAL0 = 4.3 × 105 | ADL0 = 2.4 | |
| |
BAL5 = 4.7 × 10–3 |
ADL5 = 0.43 |
|
| G152Ka, P193I or G152K/P193I mutant parameters |
|
|
|
| State parameters |
B state |
A state |
D state |
| ACh on rates (M–1 s–1) | Bkon = 107 | Akon = 3.33 × 106 | Dkon = 106 |
| ACh off rates (s–1) | Bkoff = 1173.33 | Akoff = 6.66 | Dkoff = 1.42 |
| Equilibrium constants (µM) |
BKD = 117 |
AKD = 2.0 |
DKD = 1.42 |
| Interconversion parameters |
B↔A |
A↔D |
|
| Transition state positional parameters | BAp = 0.85 | ADp = 0.99 | |
| Interconversion rates (s–1) | BAk5 = 63.5 | ADk5 = 4.07 | |
| ABk5 = 3.06 × 10–6 | DAk5 = 0.92 | ||
| BAk0 = 3.0 | ADk0 = 4.0 | ||
| ABk0 = 100 | DAk0 = 5.0 | ||
| Allosteric constants | BAL0 = 33 | ADL0 = 1.30 | |
| BAL5 = 4.82 × 10–8 | ADL5 = 0.226 |
aFor the G152K mutant, BKD = 33.2 µM, AKD = 1.2 µM and DKD = 0.2 µM.
Finally, the same set of kinetic parameters adequately described P193I and G152K single mutant data (Figure 6B), except that a better fit for the G152K mutant also required a change of the affinity constants for all three states (K-phenotype; Table IV). Altogether, the modeling data suggest that mutations G152K, P193I and G152K/P193I affect primarily the conformational transition of activation between the closed- and open-channel states.
The presence of the H-bond correlates with a high-affinity activation
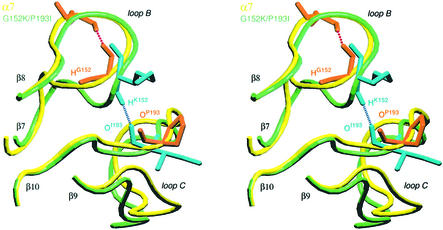
In all the structural models of the ECDs generated previously, by comparative modeling, on the basis of the experimental crystallographic structure of AChBP (Le Novère et al., 2002), an H-bond was present between positions 152 and 193. However, when the initial model of (α7)5 ECD was subjected to an equilibration by a progressive increase in temperature from zero to 300 K, the constraints on the resemblance with AChBP were relaxed and the H-bond disappeared. The presence of a glycine at position 152 allowed greater flexibility of loop B because of the absence of a side chain. Another conformation became possible, with G152 sometimes forming an H-bond with L155 instead of P193 (Figure 7). Interestingly, the mutation in muscle nAChR α1V156M, a position homologous to α7L155, also causes a congenital myasthenic syndrome (Croxen et al., 1997). Similarly to α7 ECD, the H-bond disappeared during the equilibration of α1β1γδ (not shown). Conversely, the formation of the H-bond was still possible after equilibration of α4β2 ECD and α7 ECD double mutant G152K/P193I (Figure 7). During a 10 ps simulation at 300 K following the equilibration, the H-bond was present in ∼40% of the conformers of α4β2 ECD and 20% of the conformers of the double mutant (see Table V and the video available at http://www.pasteur.fr/recherche/unites/neubiomol/SOFTWARES/a7ggG152K_P193I.mpg).
Fig. 7. Stereo view of loops B and C of α7 ECD model (yellow) and G152K/P193I mutant (green) after equilibration and dynamic simulation for 10 ps. The side chains of residues 152 and 193 are orange for α7 and cyan for G152K/P193I mutant. The figure was constructed using VMD (Humphrey et al., 1996) and generated with RASTER3D (Merritt and Bacon, 1997).
Table V. Correlation between the presence of the H-bond between loops B and C and the potency in wild-type and mutant receptors.
| |
% H-bonda |
Potency |
|---|---|---|
| α4β2 | 42 | High |
| α7 | 0 | Low |
| α7 G152K | 14 | High |
| α7 P193I | 0 | High |
| α7 G152K/P193I | 20 | High |
| α1β1γδ | 0 | Low |
| α1β1γδ α1G153S | 39 | High |
aDetermined as the number of recorded conformers that displayed the H-bond relative to the total number of recorded conformers.
The situation was more complicated in the single mutants. In the P193I mutant, loop B displayed a structure similar to that of α7 wild type, while the structure of loop C was similar to the double mutant. The interloop H-bond was never present in a period of 10 ps after equilibration, whereas an intraloop H-bond in the β-hairpin structure of loop C was observed between Y194 and S185. In G152K, loop B displayed a structure similar to that of the double mutant, while loop C was similar to that of α7 wild type (absence of the intraloop H-bond). The interloop H-bond represented 14% of the conformers that were less frequent than the double mutant (Table V). Therefore, the absence of a glycine in position 152 allowed the formation of the interloop H-bond, which was otherwise impossible. Thus, its presence impaired the formation of the interloop H-bond. However, even in the case where the glycine at position 152 was replaced by another residue, the interloop H-bond was formed less frequently because of the structure of loop C, which was constrained in an unfavorable conformation by the proline at position 193 (Table V). The same results were obtained in α1β1γδ nAChR with the congenital myasthenic mutant α1G153S, where the interloop H-bond was present as often as in α4β2 ECD (Table V).
Discussion
In the present study we report that mutations of residue G152 (loop B) and P193 (loop C) of the ACh binding pocket of neuronal α7 nicotinic receptor increase both the binding affinity for agonists and potency for ACh. We suggest that G152K, P193I and G152K/P193I mutants affect primarily the isomerization constant between the closed- and open-channel states. Unexpectedly, G152K/P193I mutant alters α-BgTx binding dramatically and thus displays two apparent opposite effects: (i) an increase in the apparent affinity for agonists; and (ii) a decrease in the apparent affinity for the competitive antagonist α-BgTx. In the following two sections we discuss the implications of the two phenotypes in order to reveal a plausible common mechanism.
Interaction between loops B and C contributes to the activation mechanism
The increase in both ACh binding affinity and potency caused by either single mutants G152K and P193I or the double mutant G152K/P193I is interpreted on the basis of a minimal three-state allosteric–kinetic model (see Scheme 1). Several lines of evidence argue that these mutations primarily affect the allosteric constant of isomerization (L-phenotype). (i) A 10- to 15-fold increase in the apparent binding affinity of the agonists, regardless of their chemical structure, was observed for G152K/P193I and P193I mutants. (ii) Fitting the data with the model revealed that a reduction of the kinetic constant ABk0 of the pathway A→B in G152K, P193I and G152K/P193I mutants resulted in reduced ACh EC50 and nH,A values, unaltered desensitization and slowed deactivation (Figure 6). However, it should be noted that the fitting procedure reflected only one interpretation of the data since multiple parameters (12) were used (Table IV). (iii) Previous studies provided evidence for a major effect of the G152K mutation and its homologues in the myasthenic mutant α1G153S on the allosteric transitions of activation and desensitization (Sine et al., 1995; Corringer et al., 1998). Moreover, the model shows that a better fit for the G152K mutant is obtained, in addition to an L-phenotype, by slightly enhancing the ACh microscopic binding affinity for all three states (K-phenotype) (Figure 6B, right panel; Table III). This K-phenotype interpretation agrees with nicotine and epibatidine binding data, since for the G152K mutant their apparent affinities differ from those for the P193I and G152K/P193I mutants, depending on the nature of the ligand (Table II). Interestingly, the P193I mutant cancels the K-phenotype of G152K in the G152K/P193I mutant. Altogether, our allosteric–kinetic model suggests that G152K, P193I or both G152K/P193I mutations cause a ‘gain of function’ phenotype primarily associated with a modification of the activation properties of α7 nAChR.
The energetically coupled interaction between G152K and P193I monitored by epibatidine, nicotine and ACh binding further suggests a close contact of the two mutant residues in the three-dimensional space of the α7 receptor. This result is consistent with the α7 ECD model derived from AChBP by comparative modeling (Le Novère et al., 2002) in which the two residues are sufficiently close to form an H-bond. However, molecular dynamics of α7 ECD causes this H-bond between the glycine and proline residues to disappear despite the fact that the two residues are spatially close (Figure 7). Conversely, dynamic simulations of the G152K/P193I mutation performed in silico restored the H-bond between the lysine and isoleucine residues (Figure 7), as observed initially for AChBP between S147 and A191 (Brejc et al., 2001). Hence we conclude that the presence of the critical glycine residue, and to a lesser extent the proline residue, prevents the formation of an optimal H-bond between loops B and C in α7 receptor.
Interestingly, 10 ps simulations of α4β2 ECD showed an inter-backbone H-bond between the lysine and isoleucine residues, as observed for the mutant G152K/P193I in α7 ECD. Thus, the presence of this non-covalent interaction correlates with a high-affinity activation process (Table V). The correlation is not perfect, since the interloop H-bond was not present in the P193I mutant (Table V). The high-affinity phenotype observed for this mutant may result from an alternative mechanism such as an enhanced stabilization of loop C structure caused by the P193I mutation (formation of an intraloop H-bond). Overall, we propose that the inter-backbone H-bond between loops B and C observed for the G152K/P193I mutant stabilizes a tertiary folding of each α subunit corresponding to a high-affinity state, therefore explaining the reduction of ABK0 in the kinetic activation pathway. Furthermore, AChBP displays a relatively high-affinity phenotype for most nicotinic ligands compared with the α7 receptor (Smit et al., 2001; Hansen et al., 2002). Hence, we conclude that the crystal structure of AChBP and other related models that display such an H-bond would represent a tertiary fold structure very close to the high-affinity states of nAChRs, i.e. open-channel (A) and/or desensitized (D) states.
Consistent with these views, a recent electron microscopy study revealed that AChBP structure fits into the electron density maps corresponding to the active high-affinity state of Torpedo nAChR, although images were resolved at only 9 Å (Unwin et al., 2002). The authors also suggested that activation causes tertiary structural changes of each α subunit by changing the relative orientations of the inner and outer sheets near strands β8 and β10. Interestingly, this region is located near the region that we identified in this study. Hence, these structural data further emphasize the notion that the molecular switch of nAChR activation requires concerted motions of loops B and C.
Finally, we provide a rationale for understanding at the atomic level the naturally occurring ‘gain of function’ mutant α1G153S, which produces a congenital myasthenic syndrome (Sine et al., 1995). According to our dynamic simulations (Table V), the presence of the serine residue in this mutant restores an H-bond interaction with the proline, thus stabilizing a high-affinity state of α1 subunits and consequently producing the myasthenic syndrome. Two ‘gain of function’ myasthenic mutants, α1G153S (Sine et al., 1995) and α1V156M (Croxen et al., 1997), are located in the loop B region. As this loop bridges the inner and outer β-sheets of each α subunit and contributes through W148 to the ACh binding site, we propose that, in addition to the functional importance of the interaction, the loop region represents some kind of pivot that controls the tertiary fold of the protein necessary for activation of LGICs.
Residues G152 and P193 directly contact α-bungarotoxin
The second striking result obtained with the G152K/P193I mutant is the unanticipated loss of α-BgTx binding. We wondered whether modification of the allosteric transition in the mutant would be sufficient to explain the loss of α-BgTx affinity.
A systematic analysis of snake venom α-toxin binding to nAChR has revealed multiple interaction points for this bulky ligand that overlap the ACh binding pocket and its two ‘principal’ and ‘complementary’ components (Osaka et al., 1999, 2000; Servent and Ménez, 2001). Here, we show that mutations at new positions within the B (G152) and C (P193) loops of the ‘principal’ component cause a dramatic loss of α-BgTx binding. Steric hindrance governs α-BgTx binding at position 152 for the following reasons: (i) the linear relationship between the van der Waals volumes of the introduced amino acids and the α-BgTx binding (Figure 4); (ii) the lack of effect on toxin binding of the isosteric mutation aspartate to asparagine despite the removal of the negative charge; and (iii) the significant increase of toxin affinity following the elimination of the consensual N-glycosylation site. Together, these data demonstrate an exclusion mechanism of α-BgTx binding by steric hindrance in the downstream region of loop B and therefore pinpoint a new contact area for the toxin.
Recently, several groups have proposed docking models of α-toxins and nAChRs. Based on the analysis of specific pairwise interactions, a docking model of α-CbTx was proposed for α7 nAChR (Fruchart-Gaillard et al., 2002). Models based on X-ray (Harel et al., 2001) and NMR (Moise et al., 2002; Samson et al., 2002) studies were also reported for the docking of α-BgTx as well as for a short neurotoxin (Teixeira-Clerc et al., 2002). All these studies suggested that α-toxins bind to the external face of the pentameric structure at the subunit interfaces, thus occluding the agonist binding site.
Since we have identified a new contact area for α-BgTx, our data offer the opportunity to refine current molecular models of the toxin–receptor interaction. Accordingly, we docked α-BgTx to α7 ECD in silico using the model established for α-CbTx and α7 ECD as a template (Fruchart-Gaillard et al., 2002). We found that finger I of α-BgTx is closer to the nAChR region that we identified by mutagenesis, i.e. G152 and P193 (arrow in Figure 8), than the homologous positions of α-CbTx. Supporting this hypothesis, binding data on G152K/P193I mutant reveal a loss of specific binding more pronounced for α-BgTx (100-fold) than for α-CbTx (∼15-fold, data not shown). In our model, the toxin cleft framed by fingers I and II wraps the β-hairpin structure of loop C, as described previously, but additionally a contact zone is disclosed between toxin finger I and G152 of the α7 receptor (arrow in Figure 8). Our data thus dictate a model of α-BgTx docked on nAChRs that excludes alternative positions which, for instance, tilt the toxin (∼45°) in the pentameric structure of α7 in the direction of the membrane surface (Moise et al., 2002). Hence, substituting larger residues in position 152 would reduce the capacity of toxin binding most probably by steric hindrance with finger I of the toxin.
Fig. 8. Stereo view of the interaction of α-BgTx (blue) and the principal (orange) and complementary (green) α7 subunits. The arrow pinpoints the close contact between the toxin and G152 and P193 nAChR. The figure was constructed using VMD (Humphrey et al., 1996) and generated with RASTER3D (Merritt and Bacon, 1997).
In conclusion, we discovered two mutations, G152K from loop B and P193I from loop C of the ACh binding pocket of neuronal α7 nicotinic receptor, which control both α-BgTx binding by steric hindrance and the allosteric transition between the closed and open states. The effects of the double mutation on the activation transition can be explained by the formation of an inter-backbone H-bond between loops B and C which would stabilize both the open and desensitized states. This molecular mechanism also sheds light on a congenital myasthenic syndrome.
Materials and methods
Equilibrium binding
Equilibrium binding experiments using [125I]α-BgTx as radioligand were performed as described previously at room temperature on membrane fragments (Corringer et al., 1995). Briefly, mutant cDNAs were transiently transfected into HEK-293 cells by calcium phosphate precipitation, and cells were collected 48 h after transfection. Membrane fragments were prepared as described previously (Corringer et al., 1995). Ligand apparent affinity (Kp value) was determined as the concentration reducing 50% of the initial velocity of 2.5 nM [125I]α-BgTx binding in the same buffer (ES) as the electrophysiological experiments (140 mM NaCl, 2.8 mM KCl, 500 mM EGTA, 10 mM glucose, 10 mM HEPES–NaOH pH 7.3) supplemented with protease inhibitor cocktail tablets (Roche). In the case of the [125I]α-BgTx assay, membrane fragments were incubated with 5 nM [125I]α-BgTx for 2 h at 37°C. Samples were diluted in 5 ml of cold ES buffer, filtered through a GF-C filter (Whatman), rinsed with 5 ml of cold ES buffer and counted. Non-specific binding was determined in the presence of 1 mM l-nicotine.
Equilibrium saturation experiments using [3H]epibatidine were performed as follows. After transfection, receptor membranes were incubated with various concentrations of [3H]epibatidine (0.5–20 nM) until equilibrium was reached in a 100 µl ES-buffered solution. Samples were filtered as described above and counted. Non-specific binding was determined in the presence of 1 mM l-nicotine. For all concentrations tested, we verified by either dilution of membranes or measurement of the free ligand concentration by centrifugation that no depletion of ligand occurred. In all cases, the Bmax values of the mutants were at least 0.3-fold less than the measured KD values, which exclude ligand depletion. We also verified that [3H]nicotine binding was prevented by epibatidine.
Equilibrium binding experiments using [3H]epibatidine as radiotracer were performed as follows. Receptor membranes were incubated in the presence of ligand (ACh, nicotine, epibatidine or α-BgTx with the concentrations indicated in the text) until equilibrium was reached (>2 h at room temperature for α-BgTx) to 100 µl ES-buffered solution. Then, [3H]epibatidine was added for 1 h (in the case of α-BgTx) and samples were filtered as described above. Non-specific binding was determined in the presence of 1 mM l-nicotine.
Electrophysiological recordings
ACh-evoked currents were monitored from transfected cells in the whole-cell patch–clamp configuration (Hamill et al., 1981) using an EPC-9 amplifier (HEKA Electronics, Liambrecht, Germany) at a holding potential of –60 mV. Since macroscopic currents mediated by 5-HT3 channels are suppressed by external Ca2+ and Mg2+ (Maricq et al., 1991; Eiselé et al., 1993), cells were perfused with an external solution containing both ions only until the whole-cell configuration was obtained, and afterwards with a Ca2+- and Mg2+-free solution of similar composition but from which both ions were omitted and 1 mM EGTA was added. The external solutions (ES) contained 140 mM NaCl, 2.8 mM KCl, 10 mM HEPES–NaOH pH 7.3 supplemented with 2 mM CaCl2, 2 mM MgCl2 or 1 mM EGTA. Patch pipettes were filled with a solution containing 140 mM CsCl2, 2 mM MgATP, 10 mM HEPES–CsOH and 10 mM BAPTA pH 7.3.
ACh and control solutions were delivered with the aid of a computer-driven fast-step perfusion system (SF 77A Perfusion Fast-Step; Warner, USA) at a rate of 0.7 ml/min through three parallel square tubes (0.6 × 0.6 mm). The first tube, placed immediately above the recorded cell, delivered the control solution. The two other tubes delivered the agonist-containing solutions. Rapid displacement of the tubes ensured a 10–90% solution exchange in ∼10 ms, as measured by open electrode controls (external solution and 1:10 external solution in water). For dose–response curves, eight different concentrations of the agonist were connected to the same application tube through a manifold and valves. An interval of 1 min was observed between applications.
To investigate the antagonism of ACh-induced responses by α-BgTx, test concentrations of 1 mM ACh were applied until stable responses were obtained (when present, run-down occurred within the first 5–10 min of recording). Perfusion was then stopped, α-BgTx (concentrated twice, 10–200 nM final) was added directly to the 1 ml perfusion chamber and the cells were allowed to incubate for 10 min before starting the perfusion and ACh test applications again. We also verified that after application of α-BgTx to α7/5HT3 receptor-expressing cells, no recovery of the response could be detected after a 10 min wash, in agreement with the pseudo-irreversible blockade of α-BgTx in α7 receptor expressed in oocytes (Johnson et al., 1995).
Functional and binding data modeling
Binding and functional data were interpreted in terms of a minimal three-state model derived from the Monod–Wyman–Changeux scheme which postulates that the protein is in equilibrium between different discrete states (Edelstein et al., 1996), i.e. the basal (B), active (A) and desensitized (D) states. In the absence of effectors, the B state is predominant while the presence of an agonist (X) will select the following linear pathways according to the lowest transition state barrier B→A→D (Edelstein et al., 1996):
In Scheme 1, each column corresponds to a series of agonist binding events at five identical sites per receptor (N = 5) with the constants specified along the vertical arrows by the state-specific intrinsic ‘on’ and ‘off’ rates corrected for statistical factors. Each row corresponds to a series of transitions between states governed by rate constants that vary with the number of molecules bound, the initial and final states in the superscript, and the number of ligands in the subscript (0 ≤ i ≤ 5). Intrinsic affinity for a specified state is given by:
KD = koff/kon(1)
A K-phenotype is defined by mutations affecting koff and/or kon (Galzi et al., 1996). The allosteric constant governing spontaneous equilibrium between two states, for example between the B and A state, is given by:
BAL0 = ABk0/BAk0(2)
An L-phenotype is defined by mutations affecting the allosteric constant L (Galzi et al., 1996). The model includes linkage relationship involving ligand binding, rates of interconversion between allosteric states and transition state properties (Edelstein et al., 1996). We used the simulation program STOIC (Simulations of Transient Openings in Ionotropic-receptor Channels), which runs within the MATLAB environment (Edelstein et al., 1996). Model-generated current and binding data were performed by assigning values for the basic 12-parameter set, formulating the appropriate differential equations and solving the system at a selected concentration of agonist. The set of parameters was changed until simulated currents reproduced electrophysiological recordings adequately.
When [125I]α-BgTx was used as the radioligand at initial velocity conditions, model-generated binding data were considered as reflecting experimental data. On the other hand, when [3H]epibatidine was used as radioligand at equilibrium, an underestimation of the real affinity is observed (Cheng and Prusoff, 1973). Hence, we used the following equation, which integrates in terms of the allosteric model the presence of the radioligand [3H]epibatidine:
where XEpi and XACh are the actual concentrations of [3H]epibatidine and ACh, respectively, BKEpi, DKEpi and BKACh, DKACh are the intrinsic dissociation constants of [3H]epibatidine and ACh for the basal and desensitized states, respectively, and N represents the number of binding sites (N = 5).
Molecular modeling
Comparative models of (α7)5 receptor from chick, (α4)2(β2)3 receptor from rat and (α1)2β1γδ receptor from Torpedo have been designed based on the experimental structure of AChBP (Protein Data Bank code 1I9B) (Le Novère et al., 2002). In silico mutagenesis of these initial models was performed within Deep-View (Guex and Peitsch, 1997). Molecular dynamics experiments were run with program Sander of the AMBER7 package (Case et al., 2002). After energy minimization (100 cycles), the residues corresponding to loops B, F and C (α7 146–196) of the ‘principal component’, together with the residues of loops D, E and F (α7 52–55, 102–109, 114–118 and 166–168) of the ‘complementary component’ were equilibrated from 0 to 300 K for 10 ps. Molecular dynamic simulations were then run for 10 ps. Visualization of the simulation results, including analysis of H-bond formation, was performed with VMD (Humphrey et al., 1996) by recording one conformer every 0.04 ps and generated using RASTER3D (Merritt and Bacon, 1997). A video presenting the molecular dynamic simulations of wild-type α7 together with the double mutant G152K/P193I is avail able at http://www.pasteur.fr/recherche/unites/neubiomol/SOFTWARES/a7ggG152K_P193I.mpg
Acknowledgments
Acknowledgements
HEK-293 cells were kindly provided by Dr Y.Paas. The authors thank M.Soudan for her technical support, Y.Archambeau for excellent help with the electrophysiological set-up and N.Duclert for her help with AMBER. This work was supported by the Collège de France, the Commission of the European Communities (CEC), the Association pour la Recherche sur le Cancer and the Association Française contre les Myopathies. T.G. is supported by CEC contract 2000-00318.
References
- Brejc K., van Dijk,W.J., Klaassen,R., Schuurmans,M., van der Oost,J., Smit,A.B. and Sixma,T.K. (2001) Crystal structure of AChBP reveals the ligand-binding domain of nicotinic receptors. Nature, 411, 269–276. [DOI] [PubMed] [Google Scholar]
- Carter P.J., Winter,G., Wilkinson,A.J. and Fersht,A.R. (1984) The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell, 38, 835–840. [DOI] [PubMed] [Google Scholar]
- Case D.A. et al. (2002) AMBER 7. University of California, San Francisco, CA.
- Cheng Y.C. and Prusoff,W.H. (1973) Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol., 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- Corringer P.J., Galzi,J.-L., Eisele,J.-L., Bertrand,S., Changeux,J.-P. and Bertrand,D. (1995) Identification of a new component of the agonist binding site of the nicotinic α7 homooligomeric receptor. J. Biol. Chem., 270, 11749–11752. [DOI] [PubMed] [Google Scholar]
- Corringer P.J., Bertrand,S., Bohler,S., Edelstein,S.J., Changeux,J.-P. and Bertrand,D. (1998) Critical elements determining diversity in agonist binding and desensitization of neuronal nicotinic acetylcholine receptors. J. Neurosci., 18, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corringer P.J., Le Novère,N. and Changeux,J.-P. (2000) Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol., 40, 431–458. [DOI] [PubMed] [Google Scholar]
- Croxen R., Newland,C., Beeson,D., Oosterhuis,H., Chauplannaz,G., Vincent,A. and Newsom-Davis,J. (1997) Mutations in different functional domains of the human muscle acetylcholine receptor α subunit in patients with the slow-channel congenital myasthenic syndrome. Hum. Mol. Genet., 6, 767–774. [DOI] [PubMed] [Google Scholar]
- Edelstein S.J., Schaad,O., Henry,E., Bertrand,D. and Changeux,J.-P. (1996) A kinetic mechanism for nicotinic acetylcholine receptors based on multiple allosteric transitions. Biol. Cybern., 75, 361–379. [DOI] [PubMed] [Google Scholar]
- Eiselé J.-L., Bertrand,S., Galzi,J.-L., Devillers-Thiéry,A., Changeux,J.-P. and Bertrand,D. (1993) Chimaeric nicotinic–serotonergic receptor combines distinct ligand binding and channel specificities. Nature, 366, 479–483. [DOI] [PubMed] [Google Scholar]
- Fruchart-Gaillard C., Gilquin,B., Antil-Delbeke,S., Le Novère,N., Tamiya,T., Corringer,P.J., Changeux,J.-P., Ménez,A. and Servent,D. (2002) Experimentally based model of a complex between a snake toxin and the α7 nicotinic receptor. Proc. Natl Acad. Sci. USA, 99, 3216–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galzi J.-L., Revah,F., Bouet,F., Menez,A., Goeldner,M., Hirth,C. and Changeux,J.-P. (1991) Allosteric transitions of the acetylcholine receptor probed at the amino acid level with a photolabile cholinergic ligand. Proc. Natl Acad. Sci. USA, 88, 5051–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galzi J.-L., Edelstein,S.J. and Changeux,J.-P. (1996) The multiple phenotypes of allosteric receptor mutants. Proc. Natl Acad. Sci. USA, 93, 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosman C., Salamone,F.N., Sine,S.M. and Auerbach,A. (2000) The extracellular linker of muscle acetylcholine receptor channels is a gating control element. J. Gen. Physiol., 116, 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hamill O.P., Marty,A., Neher,E., Sakmann,B. and Sigworth,F.J. (1981) Improved patch–clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch., 391, 85–100. [DOI] [PubMed] [Google Scholar]
- Hansen S.B., Radic,Z., Talley,T.T., Molles,B.E., Deerinck,T., Tsigelney,I. and Taylor,P. (2002) Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J. Biol. Chem., 277, 41299–41302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel M. et al. (2001) The binding site of acetylcholine receptor as visualized in the X-ray structure of a complex between α-bungarotoxin and a mimotope peptide. Neuron, 32, 265–275. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke,A. and Shulten,K. (1996) VMD-Visual molecular dynamics. J. Mol. Graph., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Johnson D.S., Martinez,J., Elgoyhen,A.B., Heinemann,S.F. and McIntosh,J.M. (1995) α-Conotoxin ImI exhibits subtype-specific nicotinic acetylcholine receptor blockade: preferential inhibition of homomeric α7 and α9 receptors. Mol. Pharmacol., 48, 194–199. [PubMed] [Google Scholar]
- Karlin A. (2002) Emerging structure of the nicotinic acetylcholine receptors. Nat. Rev. Neurosci., 3, 102–114. [DOI] [PubMed] [Google Scholar]
- Kash T.L., Jenkins,A., Kelley,J.C., Trudell,J.R. and Harrison,N.L. (2003) Coupling of agonist binding to channel gating in the GABAA receptor. Nature, 421, 272–275. [DOI] [PubMed] [Google Scholar]
- Le Novère N., Grutter,T. and Changeux,J.-P. (2002) Models of the extracellular domain of the nicotinic receptors and of the agonist- and Ca2+-binding sites. Proc. Natl Acad. Sci. USA, 99, 3210–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq A.V., Peterson,A.S., Brake,A.J., Myers,R.M. and Julius,D. (1991) Primary structure and functional expression of the 5HT3 receptor, a serotonin-gated ion channel. Science, 254, 432–437. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) RASTER3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Moise L., Piserchio,A., Basus,V.J. and Hawrot,E. (2002) NMR structural analysis of α-bungarotoxin and its complex with the principal α-neurotoxin-binding sequence on the α7 subunit of a neuronal nicotinic acetylcholine receptor. J. Biol. Chem., 277, 12406–12417. [DOI] [PubMed] [Google Scholar]
- Molles B.E., Tsigelny,I., Nguyen,P.D., Gao,S.X., Sine,S.M. and Taylor,P. (2002) Residues in the ε subunit of the nicotinic acetylcholine receptor interact to confer selectivity of Waglerin-1 for the α–ε subunit interface site. Biochemistry, 41, 7895–7906. [DOI] [PubMed] [Google Scholar]
- Osaka H., Malany,S., Kanter,J.R., Sine,S.M. and Taylor,P. (1999) Subunit interface selectivity of the α-neurotoxins for the nicotinic acetylcholine receptor. J. Biol. Chem., 274, 9581–9586. [DOI] [PubMed] [Google Scholar]
- Osaka H., Malany,S., Molles,B.E., Sine,S.M. and Taylor,P. (2000) Pairwise electrostatic interactions between α-neurotoxins and γ, δ and ε subunits of the nicotinic acetylcholine receptor. J. Biol. Chem., 275, 5478–5484. [DOI] [PubMed] [Google Scholar]
- Palma E., Bertrand,S., Binzoni,T. and Bertrand,D. (1996) Neuronal nicotinic α7 receptor expressed in Xenopus oocytes presents five putative binding sites for methyllycaconitine. J. Physiol., 491, 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiram P.A. and Sine,S.M. (1998) Identification of residues in the neuronal α7 acetylcholine receptor that confer selectivity for conotoxin ImI. J. Biol. Chem., 273, 11001–11006. [DOI] [PubMed] [Google Scholar]
- Samson A., Scherf,T., Eisenstein,M., Chill,J. and Anglister,J. (2002) The mechanism for acetylcholine receptor inhibition by α-neurotoxins and species-specific resistance to α-bungarotoxin revealed by NMR. Neuron, 35, 319–332. [DOI] [PubMed] [Google Scholar]
- Servent D. and Ménez,A. (2001) Snake neurotoxins that interact with nicotinic acetylcholine receptors. In Massaro,E.J. (ed.), Handbook of Neurotoxicology, Vol. 1. Humana Press, Totowa, NJ, pp. 385–425.
- Sine M.S., Ohno,K., Bouzat,C., Auerbach,A., Milone,M., Pruit,J.N. and Engel,A.G. (1995) Mutation of the acetylcholine receptor α subunit causes a slow-channel myasthenic syndrome by enhancing agonist binding affinity. Neuron, 15, 229–239. [DOI] [PubMed] [Google Scholar]
- Smit A.B. et al. (2001) A glia-derived acetylcholine binding protein that modulates synaptic transmission. Nature, 411, 261–268. [DOI] [PubMed] [Google Scholar]
- Teixeira-Clerc F., Ménez,A. and Kessler,P. (2002) How do short neurotoxins bind to a muscular-type nicotinic acetylcholine receptor? J. Biol. Chem., 277, 25741–25747. [DOI] [PubMed] [Google Scholar]
- Unwin N., Miyazawa,A., Li,J. and Fujiyoshi,Y. (2002) Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the α subunits. J. Mol. Biol., 319, 1165–1176. [DOI] [PubMed] [Google Scholar]
- Wilson G.G. and Karlin,A. (1998) The location of the gate in the acetylcholine receptor channel. Neuron, 20, 1269–1281. [DOI] [PubMed] [Google Scholar]