Abstract
In Ustilago maydis, pathogenic development is controlled by a heterodimer of the two homeodomain proteins bW and bE. We have identified by RNA fingerprinting a b-regulated gene, kpp6, which encodes an unusual MAP kinase. Kpp6 is similar to a number of other fungal MAP kinases involved in mating and pathogenicity, but contains an additional N-terminal domain unrelated to other proteins. Transcription of the kpp6 gene yields two transcripts differing in length, but encoding proteins of identical mass. One transcript is upregulated by the bW/bE heterodimer, while the other is induced after pheromone stimulation. kpp6 deletion mutants are attenuated in pathogenicity. kpp6T355A,Y357F mutants carrying a non-activatable allele of kpp6 are more severely compromised in pathogenesis. These strains can still form appressoria, but are defective in the subsequent penetration of the plant cuticle. Kpp6 is expressed during all stages of the sexual life cycle except mature spores. We speculate that Kpp6 may respond to a plant signal and regulate the genes necessary for efficient penetration of plant tissue.
Keywords: appressoria/MAP kinase/pathogenic development/phytopathogenic fungus/Ustilago maydis
Introduction
In pathogenic fungi, conserved signaling cascades control distinct stages of the disease process (Xu, 2000). How these cascades are connected and how they link pathogenic development with the new environment in the host is largely unknown. One of the model systems to investigate such processes is the fungus Ustilago maydis, which causes corn smut disease (Kahmann et al., 2000). In this fungus, cell recognition and cell fusion, prerequisites for plant infection, are controlled by the a mating type locus encoding a pheromone–pheromone receptor system (Bölker et al., 1992). The pheromone signal is transduced by a conserved MAP kinase module, which results in the formation of conjugation tubes that can fuse (Mayorga and Gold, 1999; Müller et al., 1999; Andrews et al., 2000).
One of the presumed targets of this MAP kinase module is the transcription factor Prf1 (Hartmann et al., 1996). Prf1 is an HMG domain protein that activates transcription of the bW and bE genes (Hartmann et al., 1996). The b genes encode homeodomain proteins that form a heterodimeric complex after cell fusion if derived from different alleles (Gillissen et al., 1992; Kämper et al., 1995). This complex controls the switch to the filamentous dikaryon and subsequent pathogenic development (Bölker et al., 1995). Maintenance of the dikaryon also requires heterozygosity at the a locus (Banuett and Herskowitz, 1989) to maintain sufficiently high levels of the b gene products (Hartmann et al., 1996). The dikaryotic filaments can easily be visualized on charcoal-containing plates, where they appear white and fuzzy in contrast to the smooth colonies produced by haploid yeast-like strains (Banuett and Herskowitz, 1994a).
On the leaf surface the dikaryon forms appressoria-like structures to allow penetration (Snetselaar and Mims, 1992, 1993). The fungus grows initially through plant cells with intracellular hyphae being separated from the host cytoplasm by the invaginated host plasma membrane. Later in development, fungal hyphae proliferate and accumulate intercellularly (Snetselaar and Mims, 1992), followed by a switch to sporogeneous hyphae that differentiate into the diploid spores (Snetselaar and Mims, 1994). Characteristic symptoms of the disease are tumors in which the spores mature (Banuett and Herskowitz, 1996). Under the infection conditions used in the greenhouse, the host responds with anthocyanin production, a known stress reaction in other plant species. This response occurs prior to tumor formation and is considered to indicate that the fungus has attempted to enter the plant tissue (Banuett and Herskowitz, 1996).
After formation of the dikaryon, the host plant is required for proliferation. To date little is known about genes that control fungal development at this stage. Many b-controlled genes have been identified (Bohlmann et al., 1994; Schauwecker et al., 1995; Bohlmann, 1996; Wösten et al., 1996; Romeis et al., 2000) and, using a system where the active bW/bE heterodimer could be induced, it was estimated that ∼200 transcripts are either up- or downregulated after heterodimer formation (Brachmann et al., 2001). However, none of the genes investigated so far proved crucial for pathogenicity. The cAMP and pheromone signal transduction cascades also play an important role during pathogenic development (Kronstad et al., 1998; D’Souza and Heitman, 2001), partly by transcriptional regulation of the bW and bE genes (Hartmann et al., 1999; Müller et al., 1999). In addition, the integrity of both cascades is required for normal fungal development inside the plant. This suggests crosstalk between the b-controlled regulatory cascade and activities mediated by cAMP and/or the MAP kinase module after the cell fusion step (Sanchez-Martinez and Perez-Martin, 2001).
Here we describe a novel MAP kinase gene, kpp6, as being upregulated by the bW/bE heterodimer. Kpp6 is crucial for penetration of the plant epidermis and might be involved in transmitting plant signals.
Results
Identification of a b-regulated MAP kinase gene
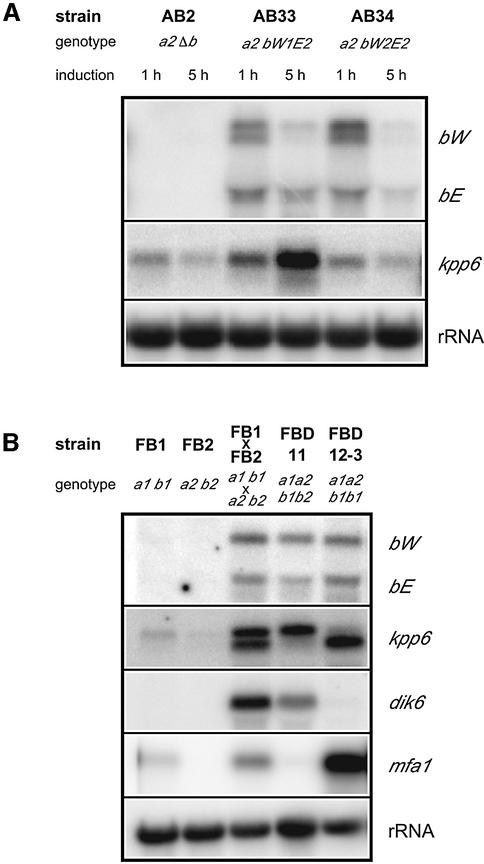
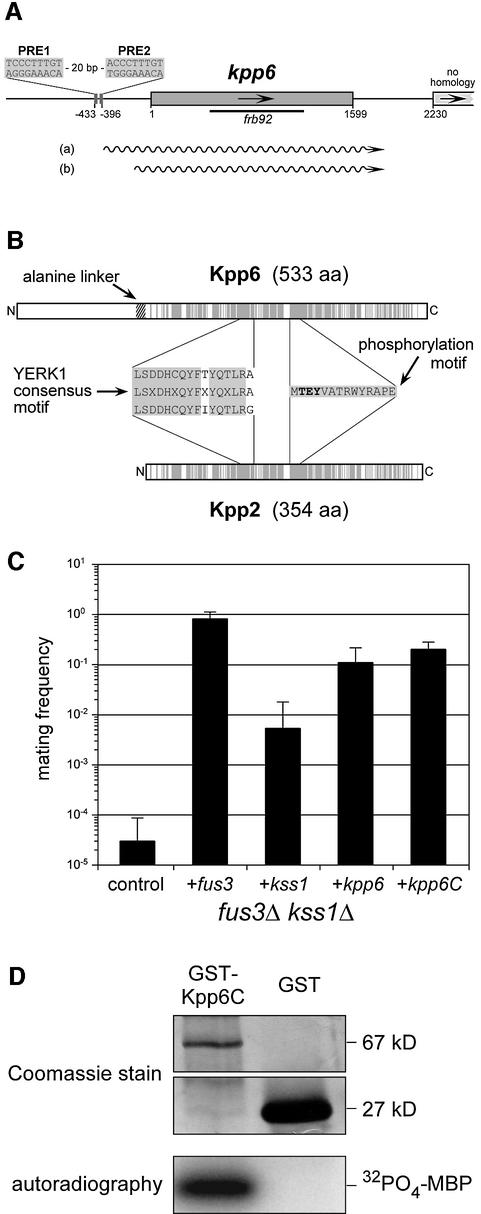
The b-induced amplicon frb92 was isolated by an RNA fingerprinting procedure using the pair of strains AB33 and AB34, in which the combination of either bW2 and bE1 or bW2 and bE2 genes, respectively, is induced by shifting the cells to nitrate-containing medium (Figure 1A) (Brachmann et al., 2001). The corresponding gene was designated kpp6 and the genomic sequence was obtained from strain FB1 (Figure 2A) (DDBJ/EMBL/GenBank accession No. AJ505152). Analysis of cDNA clones did not reveal the presence of introns in the gene. Polyadenylation occured at either position 1818 or position 1821, and the most 5′ end of cDNA clones analyzed was located at position –404 (not shown). The deduced polypeptide shows significant similarity to the YERK1 subfamily of fungal MAP kinases (Figure 2B; Supplementary figure 1 available at The EMBO Journal Online) involved in mating, morphogenesis and pathogenic development in other fungi (Kültz, 1998). Close relatives are PMK1 of Magnaporthe grisea (DDBJ/EMBL/GenBank accession No. MGU70134, 67% identity) involved in appressorium formation (Xu and Hamer, 1996) and Kpp2/Ubc3 of U.maydis (DDBJ/EMBL/GenBank accession No. AF193614, 66% identity) necessary for transmitting the pheromone signal (Mayorga and Gold, 1999; Müller et al., 1999). Similarities are restricted to the C-terminal 367 amino acids of Kpp6. The 154 amino acid N-terminus lacks similarity to database entries. In Kpp6, the two domains are connected by an alanine linker encoded by a trinucleotide repeat domain (GCT)n. The length of this repeat is variable, and alleles coding for 6, 8, 9, 11 and 12 alanine residues have been found in different strains from the ATCC collection (Supplementary table I). Strains FB1 and FB2 used in this study both carry functional alleles (see below) with 12 and 6 alanine residues, respectively. Thus, Kpp6 encoded by strain FB1 is expected to consist of 533 amino acids. The assignment of the translational start to the third in-frame ATG codon in the open reading frame (ORF) was based on the following arguments. Position –81 could be excluded in experiments where C- and N-terminal tagged versions of Kpp6 were generated (not shown). A putative –9 start codon was excluded because this codon is absent in some strains, for example in ATCC22898, 22885 and 22896 (not shown).
Fig. 1. Analysis of kpp6 expression under mating conditions. Parallel northern blots were hybridized to radioactively labeled probes indicated on the right. (A) Strains were grown in AM-Glc to OD600 of 0.5 and induced in NM-Glc for 1 and 5 h prior to total RNA isolation. (B) Strains were grown on charcoal-containing CM solid medium at 28°C for 48 h prior to total RNA isolation.
Fig. 2. The MAP kinase gene kpp6. (A) Schematic representation of the genomic kpp6 locus (DDBJ/EMBL/GenBank accession No. AJ505152). The black bar indicates the relative position of amplicon frb92; wavy lines represent the two different transcripts (a) and (b). (B) Comparison of the two MAP kinases Kpp6 and Kpp2 (Ubc3) in U.maydis. Gray regions in both proteins indicate identical amino acids. (C) Assay of mating frequencies of strain YM107 with plasmids p423ADH (control), p423ADH-Fus3 (fus3), p423ADH-Kss1 (kss1), p423ADH-Kpp6 (kpp6) and p423ADH-Kpp6C (kpp6C). Mating assays were performed against strain 1686. Bars and error bars show means of six experiments and standard deviations, respectively. (D) In vitro kinase assay with a GST-Kpp6C fusion protein. Proteins indicated at the top were purified from E.coli and subjected to kinase assay with MBP as substrate. Upper panels show Coomassie Blue stained SDS–PAGE gels while the lower panel depicts the incorporated radioactive phosphate in MBP.
The novel 154 amino acid extension of the N-terminus of Kpp6 does not display any domain profiles. This prompted us to ask whether Kpp6 exhibits MAP kinase activity. To test this, we attempted to complement a Saccharomyces cerevisiae fus3/kss1 double mutant with either full-length Kpp6 or the C-terminal MAP kinase domain of Kpp6 (Kpp6C, amino acids 167–533) expressed under the control of the constitutive ADH promoter. In both cases mating was restored to ∼20% compared with a strain expressing fus3 under identical conditions (Figure 2C). These results show that kpp6 can functionally replace FUS3 during mating. Surprisingly, the additional N-terminus does not interfere with complementation in yeast. In U.maydis, overexpression of just the N-terminal domain of Kpp6 caused no phenotype (not shown), and Kpp6 lacking the N-terminal domain was able to complement the defect of kpp6 deletion strains (not shown). When this allele, kpp6C, was expressed in Escherichia coli as a GST–Kpp6C fusion protein, the purified protein was able to phosphorylate myelin basic protein in an in vitro kinase assay (Figure 2D). On these grounds we consider Kpp6 to be an active MAP kinase.
Expression pattern of kpp6
kpp6 was isolated as a gene whose expression is upregulated by the bW/bE heterodimer. However, no potential binding sites for the bW//bE heterodimer could be identified within 2.7 kb of the promoter region of kpp6 (not shown). Instead, two PRE boxes which could represent binding sites for Prf1 (Hartmann et al., 1996) were found between positions –433 and –396 (Figure 2A). PRE1 shows a perfect match to the Prf1 consensus binding site (ACAAAGGGA) (Hartmann et al., 1996; Urban et al., 1996), while PRE2 contains one mismatch. To investigate the kpp6 expression pattern during the mating process, transcript levels were compared in haploid strains FB1 (a1 b1) and FB2 (a2 b2), compatible matings of FB1 (a1 b1) and FB2 (a2 b2), and the diploid strains FBD11 (a1a2 b1b2) and FBD12-3 (a1a2 b1b1), which are heterozygous at a and b or only at a, respectively. As markers for pheromone stimulation and b-induced development, mfa1 and dik6 gene expression levels were determined for comparison (Figure 1B).
Northern blot analysis revealed a low basal level of kpp6 expression in the two haploid strains (Figure 1B). Two transcripts differing in size by ∼200 nucleotides were detected in the cross of FB1 and FB2; the larger of them corresponds to the one seen in haploid strains (Figure 1B). The occurrence of two transcripts most likely reflects that only a fraction of the two haploid strains mixed has fused and formed dikaryotic hyphae (Urban et al., 1996). The finding that transcripts of the b-repressed gene mfa1 are still detectable under these conditions (Figure 1B, lane FB1 × FB2) supports this interpretation. In FBD11, which mimics the dikaryon, the larger transcript predominates. In this strain, pheromone signaling is downregulated through the active bW//bE heterodimer (Urban et al., 1996), suggesting that the larger transcript is b-induced. This was corroborated in strain AB33 (a2 Pnar:bW2, bE1), where the larger transcript accumulates upon b-induction by nitrate (Figure 1A). In strain FBD12-3, where pheromone signaling is constitutive and an active b complex is missing, the smaller transcript predominates (Figure 1B). The smaller transcript was also more abundant in mixtures of FB1 (a1 b1) and FB6a (a2 b1), two strains differing at a but identical at b (not shown), a situation where pheromone signaling is activated. To ascertain whether the two transcripts detected with the same probe differ at their 3′ ends, the PCR analysis on cDNA was repeated with RNA from a strain expressing mainly the shorter transcript. No differences in polyadenylation sites were found. Hybridization experiments with internal fragments of kpp6 detected both transcripts in all cases (not shown). To determine the transcriptional start sites of the two transcripts we performed 5′ RACE with RNA isolated from either FBD11 or FBD12-3, which express predominantly the longer or the shorter transcript, respectively (Figure 1B). The 5′ ends of the longer transcripts, designated (a) in Figure 2A, were mapped between positions –432 and –415, which coincides with the location of the two PRE boxes (Supplementary table II). The 5′ ends of the shorter transcript, designated (b) in Figure 2A, were located between positions –205 and –184 (Supplementary table II). To investigate whether differences in transcript length lead to the production of differently sized proteins, a triple c-myc tag was inserted before the stop codon of the kpp6 gene. The construct was inserted by homologous integration in single copy into the ip locus in strains FBD11 and FBD12-3. The resulting strains produce mainly the larger (AB251) or smaller (AB252) transcript of kpp6, respectively (Figure 3, upper panel). In both cases, the most abundant c-myc fusion protein was the same size, and a comparable ladder of presumed degradation products was detected (Figure 3, lower panel). This indicates that the two kpp6 transcripts, which differ in length, code for the same protein.
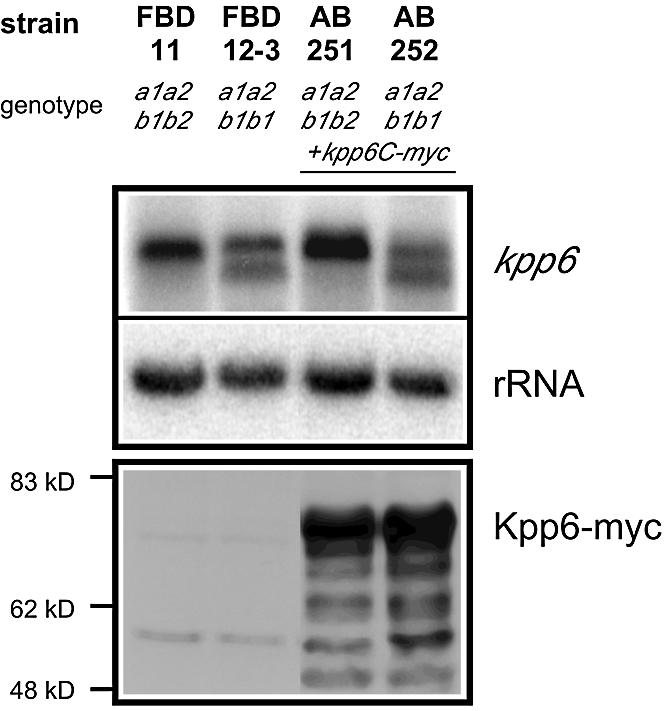
Fig. 3. Expression of Kpp6-myc fusion proteins after pheromone stimulation and b induction. Strains grown were grown on charcoal-containing CM solid medium at 28°C for 24 h prior to RNA isolation and protein extraction. The upper panel shows parallel northern blots hybrid ized to the radioactively labeled probes as indicated on the right. The lower panel shows the western blot. The Kpp6-myc fusion protein is predicted to have a molecular weight of 64 kDa but runs at a higher apparent weight.
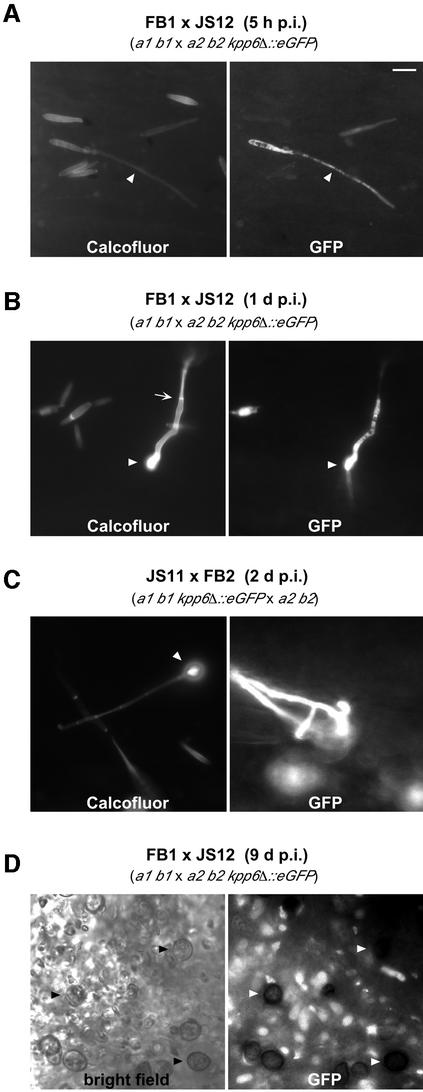
The course of kpp6 expression during development was monitored using strains JS11 (a1 b1 kpp6Δ::eGFP-BleR) and JS12 (a2 b2 kpp6Δ::eGFP-BleR) in which kpp6 is replaced by eGFP. To ensure that the infectious dikaryon carries a functional kpp6 allele, mixtures of JS11 and the compatible wild-type strain FB2, or JS12 and the compatible wild-type strain FB1, were injected into maize plants. On the plant surface, GFP fluorescence was detectable in sporidia that had formed conjugation tubes (Figure 4A) and in dikaryotic hyphae, as well as in appressoria (Figure 4B). GFP expression was also visible in all later stages except mature spores (Figure 4C and D). Since GFP fluorescence is readily detectable in spores (Huber et al., 2002), kpp6 appears to be repressed at this developmental stage.
Fig. 4. Activity of the kpp6 promoter during the life cycle of U.maydis. Crosses between FB1 and JS12 (A, B and D) or FB2 and JS11 (C) were used for plant infection. The fungus was visualized at the times indicated by epifluorescence following calcofluor staining, eGFP fluorescence or bright field microscopy, as indicated in the figure. The bar corresponds to 10 µm in all pictures. (A) Sporidia on the plant surface. One sporidium has formed a conjugation hypha that shows eGFP fluorescence (arrowheads). (B) On the plant surface a septated empty filament (arrow) and an appressorium (arrowheads) can be seen. A filament that emerges from the appressorium into the plant tissue can be visualized by eGFP fluorescence. (C) An appressorium on the plant surface (arrowhead) shows no eGFP fluorescence, presumably because the cytoplasm has moved into the plant tissue. In the right panel the plane of focus is in the leaf tissue. Hyphae exhibit strong eGFP fluorescence. (D) Section through a young tumor in which sporogenous hyphae and spores can be seen. Mature spores show no eGFP fluorescence (arrowheads).
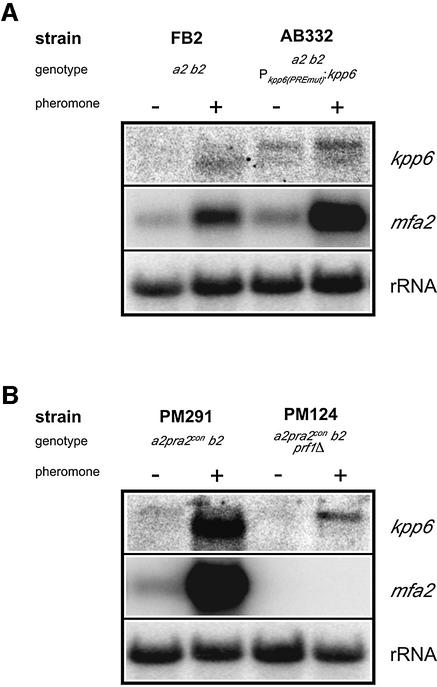
Pheromone-dependent induction of kpp6
To elucidate the contribution of the two Prf1-binding sites (PREs) located in the kpp6 promoter to the observed pheromone responsiveness of this promoter, we have mutated these PREs in FB2. In the resulting strain AB332, the larger transcript was more abundant and its amount increased slightly upon pheromone stimulation, while in the wild-type strain FB2 the smaller transcript was induced (Figure 5A). This shows that deletion of the PREs abolishes pheromone-dependent induction of the shorter transcript. To investigate whether Prf1 is involved we have used strain PM291, which expresses pra2 constitutively, and a derivative of PM291 that lacks prf1 (PM124). Constitutive expression of the pheromone receptor gene pra2 ensured pheromone stimulation since prf1 is essential for transcription of the a locus genes pra and mfa (Hartmann et al., 1996). Upon pheromone addition the smaller transcript was highly induced in PM291 expressing Prf1 (Figure 5B). In the prf1Δ strain PM124, the shorter transcript was absent but a low level of the longer transcript could be detected upon pheromone induction. This shows that expression of the shorter transcript depends on Prf1, while synthesis of the longer transcript does not require this transcription factor.
Fig. 5. The influence of (A) PREs and (B) Prf1 on the expression of kpp6. Strains grown in CM were treated with (+) or without (–) synthetic a1 pheromone for 5 h prior to RNA isolation. Parallel northern blots were hybridized to the radioactively labeled probes as indicated on the right. The mfa2 specific probe served as a positive control for Prf1-mediated pheromone stimulation.
kpp6 deletion mutants are not impaired in cell recognition and cell fusion
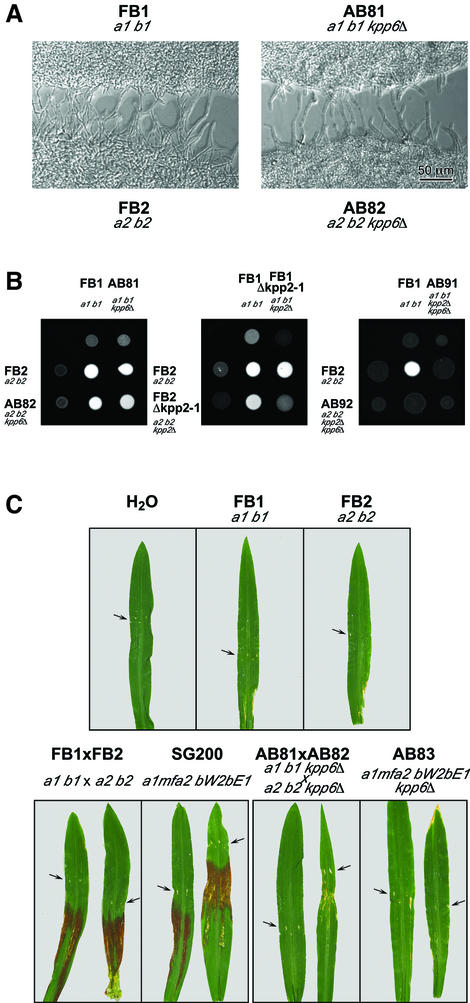
To obtain insight into the function of Kpp6, mutants were generated in strains FB1 (a1 b1), FB2 (a2 b2) and SG200 (a1mfa2 bW2bE1) by replacing the entire kpp6 ORF with a hygromycin resistance cassette (not shown). The respective mutant strains AB81 (a1 b1 kpp6Δ), AB82 (a2 b2 kpp6Δ) and AB83 (a1mfa2 bW2bE1 kpp6Δ) displayed normal growth on minimal media and were unaffected in morphology (not shown). Conjugation tube formation was indistinguishable from wild-type strains (Figure 6A), indicating that both pheromone production and pheromone perception are unaffected by the mutation. When co-spotted on charcoal-containing media to monitor development of dikaryotic hyphae, no difference from wild-type strains was detected (Figure 6B). This shows that Kpp6 is not essential in the a- and b-controlled process of filamentous growth. In addition, when young maize seedlings were infected and fungal structures were visualized prior to penetration, kpp6Δ mutants formed typical filaments that displayed all the characteristics of wild-type hyphae with respect to tip growth, septation and appressorium formation (not shown).
Fig. 6. Influence of kpp6 deletion on mating and pathogenicity. (A) Confrontation assay with compatible wild-type and kpp6 mutant strains. The formation of conjugation hyphae is clearly visible in both cases. (B) Mating assays of kpp6Δ, kpp2Δ, kpp6Δ kpp2Δ mutant and wild-type strains. The appearance of white filaments indicates formation of dikaryotic hyphae. (C) Anthocyanin production on the leaf blades of young maize plants 14 days post inoculation with the strains indicated. Arrows point to the injection punctures.
kpp6 mutants are severely attenuated in pathogenicity
When compatible combinations of kpp6Δ strains (AB81 and AB82) were co-injected into maize plants, only 30% of plants produced tumors compared with 90% in comparable wild-type infections (Table I). A similar reduction in tumor formation was seen after infection with the solopathogenic kpp6Δ strain AB83 (Table I). When kpp6 was deleted in HA103, a solopathogenic strain expressing the bE1 and bW2 genes constitutively, comparable results were obtained (Table I, strain AB84), showing that loss of kpp6 cannot be compensated by an active b heterodimer. When AB81 and AB82 were crossed with the wild-type strains FB2 and FB1, respectively, pathogenicity was restored to the level observed in a cross of two wild-type strains (Table I). This illustrates that the kpp6 alleles present in FB1 and FB2 are both functional (despite differences in length of the alanine linker region in these alleles). Maize plants infected with compatible kpp6Δ mutants were significantly taller and heavier (Supplementary figure 2) than those observed after infection with compatible wild-type strains, and most tumors were considerably smaller (not shown). In the few large tumors that developed after infection with kpp6Δ mutants, spores were produced that were able to germinate and gave rise to haploid sporidia (not shown).
Table I. Pathogenicity assays.
| Inoculum | Genotype | Anthocyanin formationa |
Tumor formation |
||
|---|---|---|---|---|---|
| Totalb | Percentage | Totalb | Percentage | ||
| FB1 × FB2 | a1 b1 × a2 b2 | 264/270 | 98 | 245/270 | 91 |
| FB1 × AB82 | a1 b1 × a2 b2 kpp6Δ | 39/43 | 91 | 30/43 | 70 |
| AB81 × FB2 | a1 b1 kpp6Δ × a2 b2 | 42/45 | 94 | 32/45 | 71 |
| AB81 × AB82 | a1 b1 kpp6Δ × a2 b2 kpp6Δ | 0/262 | 0 | 59/262c | 23 |
| SG200 | a1mfa2 bW2bE1 | 166/175 | 95 | 145/175 | 83 |
| AB83 | a1mfa2 bW2bE1 kpp6Δ | 0/158 | 0 | 70/158c | 44 |
| HA103 | a1 bW2bE1con | 72/96 | 75 | 48/96 | 50 |
| AB84 | a1 bW2bE1con kpp6Δ | 5/181d | 3 | 4/181c | 2 |
| AB91 × AB92 | a1 b1 Δkpp2-1 kpp6Δ × a2 b2 Δkpp2-1 kpp6Δ | 0/97 | 0 | 0/97 | 0 |
| AB91 × FB2 | a1 b1 Δkpp2-1 kpp6Δ × a2 b2 | 84/92 | 91 | 74/92 | 80 |
| FB1 × AB92 | a1 b1 × a2 b2 Δkpp2-1 kpp6Δ | 52/57 | 91 | 47/57 | 82 |
| AB93 | a1mfa2 bW2bE1 Δkpp2-1 kpp6Δ | 0/40 | 0 | 0/40 | 0 |
| AB301 × AB302 | a1 b1 kpp6-CbxR × a2 b2 kpp6-CbxR | 39/39 | 100 | 38/39 | 97 |
| AB303 | a1mfa2 bW2bE1 kpp6-CbxR | 43/44 | 98 | 41/44 | 93 |
| AB311 × AB312 | a1 b1kpp6T355A,Y357F-CbxR × a2 b2 kpp6T355A,Y357F-CbxR | 0/46 | 0 | 1/46e | 2 |
| AB313 | a1mfa2 bW2bE1 kpp6T355A,Y357F-CbxR | 0/46 | 0 | 4/46e | 9 |
aAnthocyanin formation on leaf blades proximal to injection punctures.
bGiven as the number of plants showing disease symptoms compared with the total number of infected plants.
cIn most cases tumor size did not exceed 2 mm in diameter.
dAnthocyanin formation was restricted to small areas of the leaf blades.
ePlants had only a single tumor with a diameter <2 mm.
The most prominent feature seen in all infections with compatible kpp6Δ strains was the lack of anthocyanin induction, usually seen on the leaf blades proximal to the punctures inflicted by the injection procedure (Figure 6C; Table I). Anthocyanin induction on leaf blades is an early symptom elicited by wild-type strains prior to tumor formation. It does not occur when plants are injected with water or haploid strains alone (Figure 6C), demonstrating that this reaction involves recognition of the pathogen.
As the incomplete loss in pathogenicity of kpp6Δ strains could indicate genetic redundancy, we considered the possibility that the closely related MAP kinase Kpp2 (Figure 2B) could partially substitute for Kpp6 during pathogenic development. Therefore, we generated a set of compatible double-mutant strains AB91 (a1 b1 kpp6Δ kpp2Δ) and AB92 (a2 b2 kpp6Δ kpp2Δ). When co-spotted on charcoal plates, these mixtures did not produce dikaryotic hyphae (Figure 6B). In this respect they clearly differed from compatible mixtures of either kpp2Δ or kpp6Δ strains, indicating that Kpp6 can partially substitute for Kpp2 during cell fusion. kpp6Δ kpp2Δ double mutants were unable to induce disease symptoms when co-infected (Table I). This phenotype is not due to a fusion defect as the double mutants co-injected with the respective wild-type strains showed wild-type levels of tumor formation (Table I). Deletion of the kpp6 and kpp2 genes in the solopathogenic strain SG200 (strain AB93) abolished tumor formation (Table I). This indicates that both MAP kinases partially substitute for each other during both mating and pathogenic development.
Kpp6 is essential for penetration
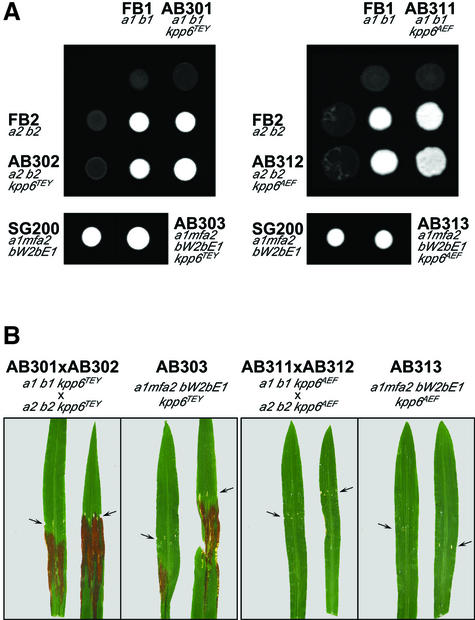
To circumvent a substitution of Kpp6 by Kpp2 in kpp6 deletion strains, the kpp6Δ allele was replaced in strains AB81, AB82 and AB83 by a variant encoding Kpp6T355A,Y357F that cannot be activated due to mutations in the putative phosphorylation motif (Madhani et al., 1997). As control, a set of strains AB301 (a1 b1 kpp6-CbxR), AB302 (a2 b2 kpp6-CbxR) and AB303 (a1mfa2 bW2bE1 kpp6-CbxR) was constructed in which the wild-type kpp6 gene, including a resistance marker, replaced the kpp6Δ allele in strains AB81, AB82 and AB83, respectively. Compatible combinations of these strains developed dikaryotic hyphae when co-spotted on charcoal-containing media, and AB303 grew filamentously when spotted alone (Figure 7A). When tested for tumor formation, the solopathogenic strain AB303 (a1mfa2 bW2bE1 kpp6-CbxR) or crosses of the haploid control strains AB301 (a1 b1 kpp6-CbxR) and AB302 (a2 b2 kpp6-CbxR) behaved indistinguishably from the respective wild-type strains (Table I). Thus, insertion of the carboxin resistance cassette in the 3′ UTR of the kpp6 gene did not affect expression of Kpp6. These results also show that the phenotype caused by deletion of kpp6 is fully complemented by reintroduction of the wild-type allele. However, when mixtures of AB311 (a1 b1 kpp6T355A,Y357F-CbxR) and AB312 (a2 b2 kpp6T355A,Y357F-CbxR) were assayed for pathogenicity, tumor formation was severely reduced compared with kpp6Δ strains, and only one out of 46 infected plants produced a small tumor (Table I). In the solopathogenic derivative AB313 (a1mfa2 bW2bE1 kpp6T355A,Y357F-CbxR), four out of 46 infected plants displayed tumors and all of these remained small (Table I). We did not observe the induction of anthocyanin in any of these infections (Figure 7B).
Fig. 7. Effect of the inactive Kpp6T355A,Y357F allele on mating and pathogenicity. (A) Mating assay and filament formation with wild-type strains (FB1, FB2, SG200), control strains with insertion of the CbxR cassette at the kpp6 locus (AB301, AB302, AB303) and strains with kpp6T355A,Y357F variants (AB311, AB312, AB313). Strains were either spotted alone or in the combinations indicated. (B) Anthocyanin production on the leaf blades 14 d p.i. infected with the strains indicated. Arrows point to the injection punctures.
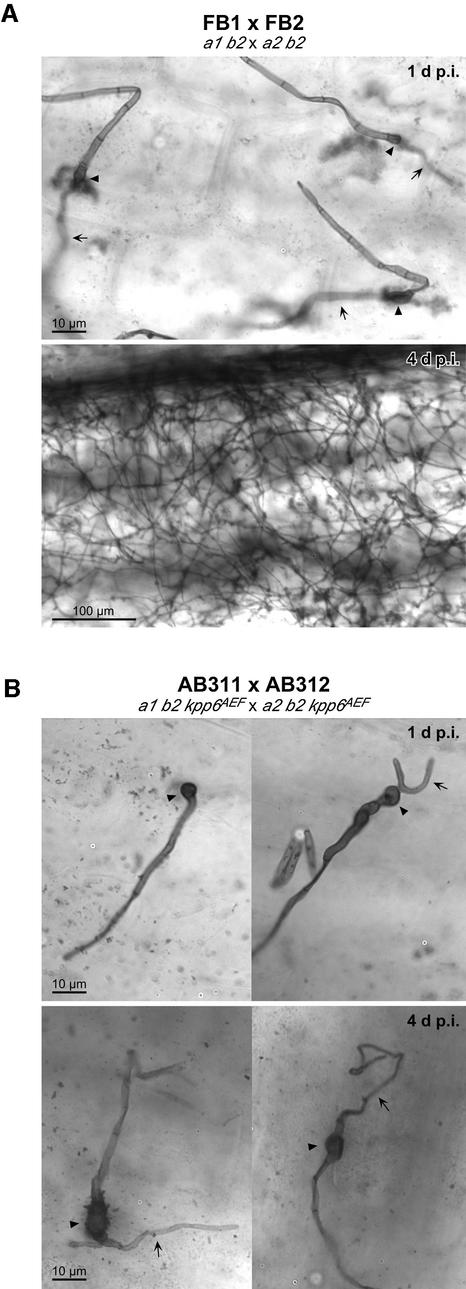
To elucidate which stage during pathogenesis is blocked in these mutants, the infection process was followed. When calcofluor was used to stain appressoria, no differences could be seen between infections by compatible wild-type strains and infections by mixtures of AB311 and AB312, respectively (not shown). To visualize appressoria as well as invading hyphae we adopted a chlorazole black E staining method used extensively to picture arbuscules produced by AM fungi (Brundrett et al., 1996). In crosses of wild-type strains 1 day after inoculation, 92% of all appressoria had penetrated the cuticle and had formed invading hyphae >20 µm long (Figure 8A; Table II). The remainder of appressoria had produced shorter infecting hyphae or had not yet formed an infection peg (Table II). Four days post-inoculation (d p.i.), massive ramification of the fungus was observed in leaf tissue (Figure 8A). It should be noted that most of these fungal structures are collapsed and do not contain cytoplasm, except in the tip cells (Snetselaar and Mims, 1992). In contrast, the majority of appressoria formed after infection with AB311 and AB312 produced only short filaments which failed to enter the plant and either arrested growth or showed limited spread on the plant surface even 4 d p.i. (Figure 8B; Table II). This illustrates that kpp6 has an essential function during the penetration process.
Fig. 8. Early infection structures formed by wild-type and kpp6 mutant strains. Filaments and appressoria were stained with chlorazole black E. (A) Infections with compatible wild-type strains (FB1 and FB2). At 1 d p.i. filaments on the leaf surface form appressoria (arrowheads) from which long filaments emerge into the leaf tissue (arrows). At 4 d p.i. hyphae have ramified through the leaf. (B) Infections with compatible kpp6T355A,Y357F mutant strains (AB311 and AB312). At 1 d p.i. filaments on the leaf surface form appressoria (arrowheads, upper panel). Short thin filaments (arrows), which localize to the plant surface and arrest in growth, emerge from some appressoria. Even at 4 d p.i. the vast majority of hyphae have not invaded the leaf tissue (lower panel).
Table II. Appressoria and emerging filaments.
| Inoculum | Appressoria without filaments or with filaments <20 µm | Appressoria with filaments >20 µm inside the plant |
Appressoria with filaments >20 µm on the plant surface |
|||
|---|---|---|---|---|---|---|
| Total | Percentage | Total | Percentage | Total | Percentage | |
| FB1 × FB2, 1 d p.i. | 17/217 | 8 | 200/217 | 92 | 0/217 | 0 |
| (a1 b1 × a2 b2) | ||||||
| AB311 × AB312, 1 d p.i. | 286/308 | 93 | 8/308 | 3 | 14/308 | 4 |
| (a1 b1kpp6T355A,Y357F-CbxR × a2 b2 kpp6T355A,Y357F-CbxR) | ||||||
| AB311 × AB312, 4 d p.i. | 184/205 | 90 | 1/205 | 0 | 20/205 | 10 |
| (a1 b1kpp6T355A,Y357F-CbxR ×a2 b2 kpp6T355A,Y357F-CbxR) | ||||||
Discussion
In this study we have identified the kpp6 gene as being regulated through both mating type loci. Kpp6 belongs to the YERK1 subfamily of MAP kinases (Supplementary figure 1), but shows an additional unique domain at its N-terminus. Kpp6 is a functional MAP kinase, since both full-length Kpp6 protein and Kpp6 lacking the N-terminal part complemented the mating defect of a S.cerevisiae fus3/kss1 mutant. Attempts to ascribe a role to the novel N-terminal domain failed because neither its deletion nor its overexpression noticeably affected Kpp6 function in U.maydis. The C-terminal part of Kpp6 is closely related to the U.maydis MAP kinase Kpp2 (Müller et al., 1999). Despite these similarities, Kpp6 and Kpp2 control discrete developmental processes. While Kpp2 is primarily involved in mating, Kpp6 plays an important role during penetration of the plant cuticle.
Transcriptional regulation of kpp6
kpp6 was identified because its transcription is elevated in cells that express an active bW/bE heterodimer while kpp2 is transcribed constitutively (not shown). A close inspection of the kpp6 promoter has not revealed the presence of sites related to the two known binding sites for the bW/bE complex (Romeis et al., 2000; Brachmann et al., 2001). Although the possibility that the bW/bE heterodimer binds sites with significant sequence divergence from the two sites identified so far cannot be excluded, we consider it more likely that kpp6 is not a direct target for the b proteins but is regulated by a transcription factor in the b regulatory cascade.
Pheromone stimulation triggers the induction of shorter kpp6 transcripts whose 5′ ends were mapped between –205 and –184. This pheromone-dependent expression requires the transcription factor Prf1 and two PREs (putative binding sites for Prf1) in the kpp6 promoter. In contrast, maximum levels of the longer transcripts with 5′ ends between –432 and –415 are seen only when the bW/bE complex has formed. Surprisingly, weak induction of the longer transcript is also observed in a prf1Δ strain stimulated with pheromone and in the absence of an active bW/bE heterodimer. This could suggest the existence of a second pheromone response factor. In the related basidiomycete Coprinus cinereus, a regulatory gene encoding the HMG domain protein Pcc1 has been identified that is induced by both mating type loci (Murata et al., 1998).
The pheromone-dependent regulation of kpp6 ensures that Kpp6 protein is already produced in mating structures on the leaf surface. Subsequent b-dependent expression guarantees maintenance of high levels of transcription during filamentous growth when pheromone signaling is downregulated (Urban et al., 1996). At present, it is unclear whether the persistence of kpp6 transcription throughout the sexual life cycle is solely dependent on b.
The YERK1 class of MAP kinases in phytopathogenic fungi
In yeasts and filamentous fungi, members of the YERK1 class of MAP kinases are involved in different developmental processes (Kültz, 1998). MAP kinases identified in phytopathogenic fungi control early stages of infection (Xu, 2000). For example, in M.grisea, pmk1 mutants are defective in differentiating appressoria (Xu and Hamer, 1996), and a similar phenotype is observed in cmk1 mutants of Colletotrichum lagenarium (Takano et al., 2000) as well as in chk1 mutants of Cochliobolus heterostrophus (Lev et al., 1999). Ustilago maydis differs from M.grisea in having two MAP kinases of the YERK1 subfamily: Kpp2 and Kpp6. kpp6Δ kpp2Δ double mutants are still able to fuse with wild-type cells (albeit inefficiently), indicating that at least one additional pathway is involved in pheromone-dependent fusion in U.maydis. Interestingly, these double mutants have lost their ability to induce tumor formation or anthocyanin production, while single mutants show only attenuated pathogenic development. This suggests that the two MAP kinases can partially substitute for each other during pathogenesis. While Kpp2 functions in pheromone signaling (Müller et al., 1999) this process is unaffected in kpp6Δ mutants, showing that kpp2 and kpp6 are involved in distinct processes in wild-type cells.
Having two MAP kinases of the YERK1 group is reminiscent of the situation in S.cerevisiae (Madhani and Fink, 1998), where Kss1p and Fus3p display a high degree of similarity but control different processes. While Fus3p is necessary for cell cycle arrest after pheromone stimulation, Kss1p regulates invasive growth and filamentation (Madhani et al., 1997; Breitkreutz et al., 2001). When fus3 is deleted, Kss1p can complement the missing function during mating, but this is not possible in a strain expressing the inactive variant Fus3pT180A,Y182F (Breitkreutz et al., 2001). A U.maydis strain carrying the analogous mutation in kpp6 arrested at the stage of appressoria, while the corresponding mutant in kpp2 failed to produce appressoria (P.Müller and R.Kahmann, in preparation). Analyzing the available fungal genome sequences revealed that the ascomycetes Aspergillus fumigatus, Candida albicans, M.grisea, Neurospora crassa, Pneumocystis carinii and Schizosaccharomyces pombe possess only one YERK1 MAPK, while the genomes of the basidiomycetes Cryptococcus neoformans, Phanerochaete chrysosporium and U.maydis contain two different YERK1 MAPK genes. This suggests that ascomycetes, excluding S.cerevisiae, which has a partially duplicated genome (Wong et al., 2002), have one while basidiomycetes have two different MAP kinases of the YERK1 family. The work presented here is the first indication that the two YERK1 MAP kinases present in U.maydis have distinct functions.
Activation of Kpp6 is required for pathogenesis
Maize plants infected with mixtures of compatible kpp6Δ strains or with a solopathogenic strain carrying the kpp6Δ allele show a significant reduction of tumor formation. Pathogenic development was almost completely abolished by replacing the resident allele with the inactive allele kpp6T355A,Y357F. The strong phenotype of kpp6T355A,Y357F mutant strains and the use of a novel staining procedure for U.maydis allowed us to pinpoint the defect in pathogenicity to a block in appressorium function. This is the first identified mutant in U.maydis that is still able to form appressoria, but is unable to penetrate the plant surface.
In M.grisea, mutants in mps1 are also blocked in appressorial penetration (Xu et al., 1998). Mps1 belongs to the YERK2 family (Supplementary figure 1) and is highly homologous with the S.cerevisiae MAPK Slt2p, which regulates cell wall integrity and cytoskeleton reorganization (Lee et al., 1993). An altered cell wall composition may explain the failure of mps1 mutants to penetrate (Xu, 2000). Superficially, we observe a similar penetration defect in infections with the kpp6T355A,Y357F mutants. However, mps1 mutants still induce a plant defense response and can infect plant cells through wounds (Xu et al., 1998), while kpp6T355A,Y357F mutant strains are unable to elicit anthocyanin production and cannot cause disease after infiltration into leaf tissue (J.Schirawski, unpublished data). Therefore, it seems unlikely that Kpp6 is a functional homolog of Mps1.
On the plant surface, dikaryotic filaments of U.maydis differentiate into appressoria, from which penetrating hyphae emerge (Snetselaar and Mims, 1994). These processes are not exclusively regulated by the bW and bE genes, since a strain expressing the bW/bE complex constitutively still requires kpp6 for efficient penetration. MST12, a transcription factor homologous to Ste12p, has recently been identified in M.grisea (Park et al., 2002). Interestingly, mst12 deletion mutants in M.grisea show the same phenotype as kpp6Δ mutants in U.maydis, i.e. appressorium formation is unaffected but they fail to penetrate and are defective in infectious growth (Park et al., 2002). However, no homolog of this gene could be identified in the available sequence of 93% of the U.maydis genome.
The MAPK Kpp6 is most likely activated by phosphorylation through a MAPK kinase (MEK). The only characterized MEK in U.maydis is Fuz7, a homolog of Ste7p of S.cerevisiae. fuz7Δ mutants are non-pathogenic, most likely because of a defect in appressorium formation (Banuett and Herskowitz, 1994b; P.Müller and R.K.Kahmann, in preparation). Thus, the fuz7Δ mutants display the same phenotype as the kpp2Δ kpp6Δ double mutants, which are unable to form appressoria (J.Schirawski and P.Müller, unpublished data). This suggests that Fuz7 acts upstream of Kpp2 and Kpp6 during pathogenic development. Furthermore, the U.maydis genome appears not to harbor other MEKs homologous to kinases of the STE7 family, while it contains at least two additional MEK genes that code for homologs of Mkk1p and Pbs2p of S.cerevisiae, respectively (not shown).
The nature of the signal that is transmitted through the MAPK module containing Kpp6 is currently unknown. So far, the only molecules known to lead to the activation of fungal MAP kinases are pheromones. Since kpp6 is required only after fungal contact with the plant, it is possible that plant signals trigger its activation. It has been proposed for M.grisea that cutin monomers and surface hydrophobicity, which are both necessary for appressorium formation, may be sensed (Lee and Dean, 1994; Gilbert et al., 1996). However, so far it has not been possible to link this with downstream signaling modules. It will be a challenge to determine how the MAPK cascade containing Kpp6 is activated, whether plant compounds are involved and how the downstream targets of this pathway facilitate plant penetration following appressorium formation.
Materials and methods
Plasmids and plasmid constructions
Plasmid constructions are described in detail in Supplementary data. To obtain genomic fragments of kpp6, the cloned amplicon frb92 was used to screen a genomic λEMBL3 library (Schulz et al., 1990). A 6782 bp BamHI fragment containing 2667 bp of the 5′ UTR, the 1599 bp ORF of kpp6, 627 bp of intergenic region and 1929 bp of the 5′ part of a gene without homologs in the databases (Figure 1A) was cloned into pUC19 (Yanisch-Perron et al., 1985) to generate pkpp6B. Amplification and sequencing of overlapping portions from the transcribed region of kpp6 from a λgt10 cDNA library (Schauwecker et al., 1995) indicated that no introns were present.
Plasmid pkpp6Δ-Hyg(-) contains kpp6 disrupted by a hygromycin resistance cassette. pkpp6-myc contains a triple c-myc tag inserted before the stop codon of kpp6. pkpp6-Cbx contains the wild-type kpp6 gene fused to a carboxin resistance cassette for replacement in the kpp6 locus. pkpp6T355A,Y357F-Cbx and pPkpp6(PREmut):kpp6-Cbx are both derivatives of pkpp6-Cbx generated by site-directed mutagenesis. p423ADH-Kpp6, p423ADH-Kpp6C, p423ADH-Fus3 and p423ADH-Kss1 are derived from p423-ADH (Mumberg et al., 1995) and express the complete Kpp6 protein, the C-terminal 367 amino acids of Kpp6 and the complete Fus3 and Kss1 proteins, respectively. In pJS2ble, the kpp6 promoter is fused to eGFP followed by a phleomycin resistance cassette and sequences downstream of kpp6.
Strains and growth conditions
The E.coli K-12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes. Saccharomyces cerevisiae and U.maydis strains employed in this study are listed in Table III. Ustilago maydis strains were constructed by transformation of progenitor strains with linearized plasmids (Table III). Integration into either the ip locus (Loubradou et al., 2001) or the endogenous kpp6 gene was verified by diagnostic PCR and subsequent Southern blot analysis.
Table III. Strains used in this study.
| Strain | Relevant genotype | Reference | Plasmid transformed | Progenitor strain | Integration into |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae | |||||
| YM107 | MATa kss1::hisG fus3::TRP1ura3-52 | Madhani and Fink (1997) | |||
| his3::hisG leu2::hisG trp1::hisG | |||||
| 1686 | Matα thr1 arg1 | R.Wickner, personal communication | |||
| Ustilago maydis | |||||
| FB1 | a1 b1 | Banuett and Herskowitz (1989) | |||
| FB2 | a2 b2 | Banuett and Herskowitz (1989) | |||
| FB6a | a2 b1 | Banuett and Herskowitz (1989) | |||
| FBD11 | a1a2 b1b2 | Banuett and Herskowitz (1989) | |||
| FBD12-3 | a1a2 b1b1 | Banuett and Herskowitz (1989) | |||
| SG200 | a1mfa2 bW2bE1 | Bölker et al. (1995) | |||
| HA103 | a1 bW2bE1con | Hartmann et al. (1996) | |||
| SG200Δkpp2-1 | a1mfa2 bW2bE1 kpp2-1Δ | Müller et al. (1999) | |||
| FB1Δkpp2-1 | a1 b1 kpp2-1Δ | Müller et al. (1999) | |||
| FB2Δkpp2-1 | a2 b2 kpp2-1Δ | Müller et al. (1999) | |||
| PM291 | a2pra2con b2 | P.Müller and R.Kahmann, in preparation | |||
| PM124 | a2pra2con b2 prf1Δ | P.Müller and R.Kahmann, in preparation | |||
| AB33 | a2 Pnar:bW2,bE1 | Brachmann et al. (2001) | |||
| AB34 | a2 Pnar:bW2,bE2 | Brachmann et al. (2001) | |||
| AB81 | a1 b1 kpp6Δ | This study | pkpp6Δ-Hyg(-) | FB1 | kpp6 locus |
| AB82 | a2 b2 kpp6Δ | This study | pkpp6Δ-Hyg(-) | FB2 | kpp6 locus |
| AB83 | a1mfa2 bW2bE1 kpp6Δ | This study | pkpp6Δ-Hyg(-) | SG200 | kpp6 locus |
| AB84 | a1 bcon kpp6Δ | This study | pkpp6Δ-Hyg(-) | HA103 | kpp6 locus |
| AB91 | a1 b1 Δkpp2-1 kpp6Δ | This study | pkpp6Δ-Hyg(-) | FB1Δkpp2-1 | kpp6 locus |
| AB92 | a2 b2Δkpp2-1 kpp6Δ | This study | pkpp6Δ-Hyg(-) | FB2Δkpp2-1 | kpp6 locus |
| AB93 | a1mfa2 bW2bE1Δkpp2-1 kpp6Δ | This study | pkpp6Δ-Hyg(-) | SG200Δkpp2-1 | kpp6 locus |
| JS11 | a1 b1 kpp6Δ::eGFP-BleR | This study | pJS2ble | AB81 | kpp6 locus |
| JS12 | a2 b2 kpp6Δ::eGFP-BleR | This study | pJS2ble | AB82 | kpp6 locus |
| AB251 | a1a2 b1b2 ipr[kpp6-myc(1599)]ips | This study | pkpp6-myc(1599) | FBD11 | ip locus |
| AB252 | a1a2 b1b1 ipr[kpp6-myc(1599)]ips | This study | pkpp6-myc(1599) | FBD12-3 | ip locus |
| AB301 | a1 b1 kpp6-CbxR | This study | pkpp6-Cbx | AB81 | kpp6 locus |
| AB302 | a2 b2 kpp6-CbxR | This study | pkpp6-Cbx | AB82 | kpp6 locus |
| AB303 | a1mfa2 bW2bE1 kpp6-CbxR | This study | pkpp6-Cbx | AB83 | kpp6 locus |
| AB311 | a1 b1 kpp6T355A,Y357F-CbxR | This study | pkpp6T355A,Y357F-Cbx | AB81 | kpp6 locus |
| AB312 | a2 b2 kpp6T355A,Y357F-CbxR | This study | pkpp6T355A,Y357F-Cbx | AB82 | kpp6 locus |
| AB313 | a1mfa2 bW2bE1 kpp6T355A,Y357F-CbxR | This study | pkpp6T355A,Y357F-Cbx | AB83 | kpp6 locus |
| AB332 | a2 b2 Pkpp6(PREmut):kpp6-CbxR | This study | pPkpp6(PREmut):kpp6-Cbx | AB82 | kpp6 locus |
Quantitative mating assays in S.cerevisiae were performed as described previously (Sprague, 1991).
Ustilago maydis strains were grown at 28°C in liquid CM (Holliday, 1974), YEPSL (0.4% yeast extract, 0.4% peptone, 2% sucrose), potato dextrose (2.4% potato dextrose broth; Difco), NM or AM minimal medium (Banks et al., 1993) on a rotary shaker at 220 r.p.m. or on solid potato dextrose agar. Charcoal-containing CM and PD plates were used for RNA preparations and mating assays, respectively (Holliday, 1974). Hygromycin B was from Roche, phleomycin from Cayla and carboxin from Riedel de Haën. All other chemicals were of analytical grade and were obtained from Sigma or Merck.
DNA, RNA and protein procedures
Standard molecular techniques were followed (Sambrook et al., 1989). Transformation of S.cerevisiae (Guthrie and Fink, 1991) and of U.maydis (Schulz et al., 1990) was performed as described previously. Ustilago maydis DNA was isolated as described previously (Hoffman and Winston, 1987). RNA was isolated from strains growing on charcoal-containing CM plates (Schauwecker et al., 1995) or from liquid culture (Krüger et al., 1998). Total RNA (10 µg/lane) was separated on MOPS-buffered 1% agarose gels and transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech). Double-stranded probes were used for northern blot analysis: bW2 (Urban et al., 1996); bE1 (Kämper et al., 1995); mfa1 and mfa2 (Bölker et al., 1992); dik6 (Bohlmann, 1996); rRNA (Bottin et al., 1996); kpp6, the 752 bp frb92 amplicon originally isolated in the RNA fingerprint. Radioactive labeling was performed with the NEBlot kit (New England Biolabs). A PhosphorImager (Storm 840; Molecular Dynamics) and the program IMAGEQUANT (Molecular Dynamics) were used to visualize radioactive signals. The 3′ and 5′ ends of kpp6 transcripts were mapped by isolating cDNA clones from a U.maydis λgt10 cDNA library (Schauwecker et al., 1995) by PCR. 5′-RACE analysis was performed using the GeneRacer Kit (Invitrogen), the kpp6 gene-specific primer AGCTTCTTCTCCGTAACCGTTAGC and RNA isolated from FBD11 and FB12-3, which were grown for 48 h on charcoal-containing CM plates.
Protein extracts were prepared as described previously (Straube et al., 2001). Proteins (100 µg/lane) were separated on 10% polyacrylamide gels and transferred to Hybond-P nitrocellulose membranes (Amersham Pharmacia Biotech) for 1 h at 1 mA/cm2 in a semi-dry blot chamber (UniEquip). Antibody detection was performed using polyclonal rabbit anti-Myc A14 (Santa Cruz) and the ECL+ detection kit (Amersham Parmacia Biotech).
In vitro kinase assay
GST and GST–Kpp6C fusion protein were purified from E.coli and subjected to kinase assay with myelin basic protein (MBP) as substrate. Details are described in Supplementary data.
Mating, confrontation assay, pheromone stimulation and plant infection
To test for mating, compatible strains were co-spotted on charcoal-containing PD plates, which were sealed with parafilm and incubated at 21°C for 48 h. Confrontation assays were performed as described previously (Müller et al., 1999). For pheromone stimulation, strains were grown in liquid CM medium to an OD600 of 0.6. Synthetic a1 pheromone (Koppitz et al., 1996) dissolved in dimethyl sulfoxide was added to a final concentration of 2.5 µg/ml and cells were incubated for 5 h at 28°C in a 15 ml plastic tube on a tissue culture roller prior to harvest.
Plant infections of the corn variety Early Golden Bantam (Olds Seeds, Madison, WI) were performed essentially as described previously (Gillissen et al., 1992) (see Supplementary data for details).
Light microscopy and image processing
For microscope observation we used a Zeiss Axiophot with differential interference contrast (DIC) optics. The pictures were taken using a CCD camera (C4742-95, Hamamatsu). GFP fluorescence was detected by a specific filter set (BP 470/20, FT 493, BP 505–530) (Zeiss). Image processing and measurements were performed using AXIOVISION 3.1 (Zeiss) and CANVAS 7.0 (Deneba). Details of staining procedures are given in the Supplementary data.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr Manuel Tönnis and Professor Dr Horst Kessler for providing synthetic pheromone, Dr Hiten Madhani for strain YM107, Dr Reed Wickner for supporting additional experiments and Dr Michael Feldbrügge for helpful discussions. We are grateful to Bayer CropScience for providing access to the U.maydis genome sequence. This work was supported through SFB369.
References
- Andrews D.L., Egan,J.D., Mayorga,M.E. and Gold,S.E. (2000) The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant Microbe Interact., 13, 781–786. [DOI] [PubMed] [Google Scholar]
- Banks G.R., Shelton,P.A., Kanuga,N., Holden,D.W. and Spanos,A. (1993) The Ustilago maydis nar1 gene encoding nitrate reductase activity: sequence and transcriptional regulation. Gene, 131, 68–78. [DOI] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1989) Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl Acad. Sci. USA, 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1994a) Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp. Mycol., 18, 247–266. [Google Scholar]
- Banuett F. and Herskowitz,I. (1994b) Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev., 8, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1996) Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development, 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Bohlmann R. (1996) Isolierung und Charakterisierung von filamentspezifisch exprimierten Genen aus Ustilago maydis. PhD thesis, Fakultät für Biologie, Ludwig-Maximilian-Universität, München, Germany.
- Bohlmann R., Schauwecker,F., Basse,C. and Kahmann,R. (1994) Genetic regulation of mating and dimorphism in Ustilago maydis. In Daniels,M.J. (ed.), Advances in Molecular Genetics of Plant–Microbe Interactions, Vol. 3. Kluwer, Dordrecht, The Netherlands, pp. 239–245.
- Bölker M., Urban,M. and Kahmann,R. (1992) The a mating type locus of U.maydis specifies cell signaling components. Cell, 68, 441–450. [DOI] [PubMed] [Google Scholar]
- Bölker M., Genin,S., Lehmler,C. and Kahmann,R. (1995) Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot., 73, 320–325. [Google Scholar]
- Bottin A., Kämper,J. and Kahmann,R. (1996) Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet., 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl,G., Kämper,J. and Kahmann,R. (2001) The bW/bE heterodimer regulates a distinct set of genes in Ustilago maydis. Mol. Microbiol., 42, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A., Boucher,L. and Tyers,M. (2001) MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol., 11, 1266–1271. [DOI] [PubMed] [Google Scholar]
- Brundrett M., Bougher,N., Dell,B., Grove,T. and Malajczuk,N. (1996) Working with Mycorrhizas in Forestry and Agriculture. Australian Centre for International Agricultural Research, Canberra, Australia, Monograph 32.
- D’Souza C.A. and Heitman,J. (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev., 25, 349–364. [DOI] [PubMed] [Google Scholar]
- Gilbert M.S., Johnson,A. and Dean,R. (1996) Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol. Mol. Plant Pathol., 48, 335–346. [Google Scholar]
- Gillissen B., Bergemann,J., Sandmann,C., Schröer,B., Bölker,M. and Kahmann,R. (1992) A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell, 68, 647–657. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA.
- Hartmann H.A., Kahmann,R. and Bölker,M. (1996) The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J., 15, 1632–1641. [PMC free article] [PubMed] [Google Scholar]
- Hartmann H.A., Krüger,J., Lottspeich,F. and Kahmann,R. (1999) Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell, 11, 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C.S. and Winston,F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E.coli. Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Holliday R. (1974) Ustilago maydis. In King,R.C. (ed.), Handbook of Genetics, Vol. 1. Plenum Press, New York, NY, pp. 575–595.
- Huber S.M., Lottspeich,F. and Kämper,J. (2002) A gene that encodes a product with similarity to dioxygenases is highly expressed in teliospores of Ustilago maydis. Mol. Genet. Genomics, 267, 757–771. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Steinberg,G., Basse,C., Feldbrügge,M. and Kämper,J. (2000) Ustilago maydis, the causative agent of corn smut disease. In Kronstad,J.W. (ed.), Fungal Pathology. Kluwer, Dordrecht, The Netherlands, pp. 347–371.
- Kämper J., Reichmann,M., Romeis,T., Bölker,M. and Kahmann,R. (1995) Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell, 81, 73–83. [DOI] [PubMed] [Google Scholar]
- Koppitz M., Spellig,T., Kahmann,R. and Kessler,H. (1996) Lipoconjugates: structure-activity studies for pheromone analogues of Ustilago maydis with varied lipophilicity. Int. J. Pept. Protein Res., 48, 377–390. [DOI] [PubMed] [Google Scholar]
- Kronstad J., De Maria,A., Funnell,D., Laidlaw,R.D., Lee,N., Moniz de Sá,M. and Ramesh,M. (1998) Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch. Microbiol., 170, 395–404. [DOI] [PubMed] [Google Scholar]
- Krüger J., Loubradou,G., Regenfelder,E., Hartmann,A. and Kahmann,R. (1998) Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet., 260, 193–198. [DOI] [PubMed] [Google Scholar]
- Kültz D. (1998) Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol., 46, 571–588. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Irie,K., Gotoh,Y., Watanabe,Y., Araki,H., Nishida,E., Matsumoto,K. and Levin,D.E. (1993) A yeast mitogen-activated protein kinase homolog (Mpk1) mediates signalling by protein kinase C. Mol. Cell. Biol., 13, 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-H. and Dean,R. (1994) Hydrophobicity of contact surface induces appressorium formation in Magnaporthe grisea. FEMS Microbiol. Lett., 115, 71–76. [Google Scholar]
- Lev S., Sharon,A., Hadar,R., Ma,H. and Horwitz,B.A. (1999) A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl Acad. Sci. USA, 96, 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubradou G., Brachmann,A., Feldbrügge,M. and Kahmann,R. (2001) A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol., 40, 719–730. [DOI] [PubMed] [Google Scholar]
- Madhani H.D. and Fink,G.R. (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science, 275, 1314–1317. [DOI] [PubMed] [Google Scholar]
- Madhani H.D. and Fink,G.R. (1998) The riddle of MAP kinase signaling specificity. Trends Genet., 14, 151–155. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Styles,C.A. and Fink,G.R. (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell, 91, 673–684. [DOI] [PubMed] [Google Scholar]
- Mayorga M.E. and Gold,S.E. (1999) A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol., 34, 485–497. [DOI] [PubMed] [Google Scholar]
- Müller P., Aichinger,C., Feldbrügge,M. and Kahmann,R. (1999) The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol., 34, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous protein in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Murata Y., Fujii,M., Zolan,M.E. and Kamada,T. (1998) Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics, 149, 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Xue,C., Zheng,L., Lam,S. and Xu,J.R. (2002) MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact., 15, 183–192. [DOI] [PubMed] [Google Scholar]
- Romeis T., Brachmann,A., Kahmann,R. and Kämper,J. (2000) Identification of a target gene for the bE/bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol., 37, 54–66. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Frisch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez-Martinez C. and Perez-Martin,J. (2001) Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis—similar inputs, different outputs. Curr. Opin. Microbiol., 4, 214–221. [DOI] [PubMed] [Google Scholar]
- Schauwecker F., Wanner,G. and Kahmann,R. (1995) Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe Seyler, 376, 617–625. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett,F., Dahl,M., Schlesinger,R., Schäfer,W., Martin,T., Herskowitz,I. and Kahmann,R. (1990) The b alleles of U.maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell, 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Snetselaar K.M. and Mims,C.W. (1992) Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia, 84, 193–203. [Google Scholar]
- Snetselaar K.M. and Mims,C.W. (1993) Infection of maize stigmas by Ustilago maydis: light and electron microscopy. Phytopathology, 83, 843–850. [Google Scholar]
- Snetselaar K.M. and Mims,C.W. (1994) Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res., 98, 347–355. [Google Scholar]
- Sprague J.R. Jr (1991) Assay of yeast mating reaction. Methods Enzymol., 194, 77–93. [DOI] [PubMed] [Google Scholar]
- Straube A., Enard,W., Berner,A., Wedlich-Söldner,R., Kahmann,R. and Steinberg,G. (2001) A split motor domain in cytoplasmic dynein. EMBO J., 20, 5901–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y., Kikuchi,T., Kubo,Y., Hamer,J.E., Mise,K. and Furusawa,I. (2000) The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant Microbe Interact., 13, 374–383. [DOI] [PubMed] [Google Scholar]
- Urban M., Kahmann,R. and Bölker,M. (1996) Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet., 251, 31–37. [DOI] [PubMed] [Google Scholar]
- Wong S., Butler,G. and Wolfe,K.H. (2002) Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl Acad. Sci. USA, 99, 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten H.A., Bohlmann,R., Eckerskorn,C., Lottspeich,F., Bölker,M. and Kahmann,R. (1996) A novel class of small amphipathic peptides affect aerial hyphal growth and surface hydrophobicity in Ustilago maydis. EMBO J., 15, 4274–4281. [PMC free article] [PubMed] [Google Scholar]
- Xu J.R. (2000) MAP kinases in fungal pathogens. Fungal Genet. Biol., 31, 137–152. [DOI] [PubMed] [Google Scholar]
- Xu J.R. and Hamer,J.E. (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev., 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Xu J.R., Staiger,C.J. and Hamer,J.E. (1998) Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defence responses. Proc. Natl Acad. Sci. USA, 95, 12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]