Abstract
Surgical pathology specimens are an important resource for medical research, particularly for cancer research. Although research studies would benefit from information derived from the surgical pathology reports, access to this information is limited by use of unstructured free-text in the reports. We have previously described a pipeline-based system for automated annotation of surgical pathology reports with UMLS concepts, which has been used to code over 450,000 surgical pathology reports at our institution. In addition to coding UMLS terms, it annotates values of several key variables, such as TNM stage and cancer grade. The object of this study was to evaluate the potential and limitations of automated extraction of these variables, by measuring the performance of the system against a true gold standard – manually encoded data entered by expert tissue annotators. We categorized and analyzed errors to determine the potential and limitations of information extraction from pathology reports for the purpose of automated biospecimen annotation.
INTRODUCTION
Thousands of paraffin embedded surgical pathology specimens as well as frozen and prepared tissue are archived every year in paraffin archives and tissue banks throughout the country. These specimens (especially tumor specimens) are useful resources for the research community. The utility of these resources depends greatly on the degree of tissue annotation that accompanies the material. In the case of paraffin embedded and archived clinical material, there is currently no way to automatically annotate the existing specimens. In the case of banked tissue it is possible to have highly trained experts annotating the findings, but at a great labor cost. In both cases, useful information can be derived from the Surgical Pathology Report (SPR). While laboratory and microbiology reports are now commonly available in the form of structured data, surgical pathology reports are generally only available as free-text.
SPRs contain an abundance of important information including: cancer type, location, pathological stage, metastasis status, values of prognostic attributes, tumor size and weight, etc. Pratt started pioneer work on auto coding pathology reports12 since 1970th. Other previous work in other domains have established methods for encoding free-text clinical reports4,5,6. The long-term goal of this project is to utilize natural language processing methods to extract information from free-text SPRs in order to annotate biospecimens.
As part of the Shared Pathology Informatics Network (SPIN) we developed a pipeline-based system7 for automatic annotation of surgical pathology reports using GATE – an open source architecture for language engineering. Other existing large-scale initiatives such as the Cancer Bioinformatics Grid (caBIG)8 and National Biospecimen Network (NBN)9 require access to data and tissue resources including those derived from the SPR. Specifically, there is a need to improve and increase the annotation of biospecimens in order to further the research goals of these initiatives.
Can existing clinical reports be used to automatically annotate biospecimens? This paper describes an evaluation study on the existing SPIN system to determine the feasibility of extending it to automate annotation by comparison against existing manual methods.
METHODS
System
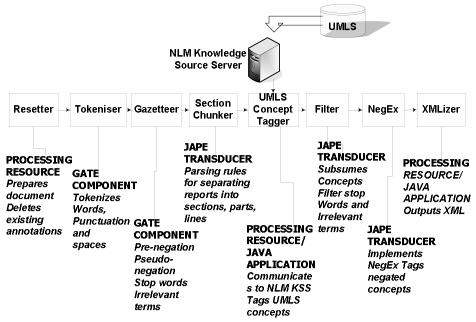
The system uses GATE - an open-source framework for language engineering1,2,3. The architecture enables a pipeline-based approach, in which sequential processing is performed to accomplish the following tasks: (1) tokenization of words and punctuation; (2) annotation of the sections of the surgical pathology report (e.g. final diagnosis, gross description, comment); (3) annotation of concepts using a subset of UMLS semantic types; (4) differential annotation of negated concepts with the NegEx10 negation algorithm; (5) identification of attribute values using JAPE rules. Information from the annotated reports is converted to XML using the CHIRPS Schema – a representation of the semantics of the clinical document, for a set of key concepts including part, organ, procedure, diagnoses and findings 11 (Figure 1).
Figure 1.
GATE pipeline
Case Selection
A total of 465 free-text surgical pathology reports matching cases in the Pennsylvania Cancer Alliance Bionformatics Consortium (PCABC) database (http://pcabc.upmc.edu) were obtained from the Medical Archival Record System (MARS) – a clinical data repository at the University of Pittsburgh. We applied (De-ID) - a de-identification application - to remove HIPAA mandated identifiers. Use of de-identified patient records was approved by the Institutional Review Board under protocols #0304081 and # 03057.
Variable Selection
Our goal was to determine the feasibility of utilizing the existing GATE pipeline for extraction of key variables from free-text pathology reports. In order to assess this, we performed a manual comparison of automated extraction versus gold-standard for a limited set of variables: (a) Gleason score (a measure of tumor grade), (b) tumor stage and (3) status of lymph node metastasis. These variables were selected because they: (1) represent three of the most generally important variables that can be obtained from the surgical pathology report in cancer cases; (2) represent scenarios where extraction was expected to be simple (Gleason Score and TNM) as well as complex (lymph node status); and (3) have significant practical importance, for example in automating determination of eligibility for clinical trials.
Gold Standard
We evaluated the automated extraction of the three variables listed above against a previously existing database of manually encoded data for patients with Prostate Cancer. PCABC contains structured data entered after histologic review of the specimen by a set of pathologists with special expertise in Prostate Pathology. The data in the PCABC database was entered by different pathologists than the pathologists dictating the pathology report.
Data Processing
After completion of processing, XML SPY was used to convert the CHIRPS XML into an Access database. Values for Gleason score and TNM score were extracted from the database using UMLS concept identifiers for Gleason and TNM. To determine whether any lymph node metastasis were present in a report, we identified UMLS concepts that were indicators of metastases, including METASTATIC ADENOCARCINOMA, METASTATIC CARCINOMA, MALIGNANCY and TUMOR. When any of these concepts were asserted in the text, metastasis was said to be present. When none these concepts were asserted in the text, metastasis was said to be present. When any concept was both asserted and negated in different parts of the report, the assertion was selected over the negation. Thus, we reproduced the heuristic we anticipated using in a future extraction system.
Error Coding
The modular nature of the GATE framework was exploited to determine the root cause of each error. We developed a coding scheme that categorized each error into the following subtypes:
Chunker Error: Wrong Section: The actual information we were looking for was not in the coded section. For example, the Gleason Grade which was expected in the Final Diagnosis section but was reported in the Addendum.
Chunker Error: Section Truncation: The correct information was in the anticipated document section but the Chunker either missed this section or truncated relevant text.
UMLS Concept Tagger Error: The component which tags UMLS concepts failed to tag the variable name. Therefore the values for these variables could not be identified. For example, if the report indicates “histologic grade 3+3 =6” the value of 6 would not be annotated as a Gleason Score, because the system does not identify the more generic term “histologic grade” as related to Gleason Score.
Over Scrubbing Error: A keyword required for concept annotation was improperly scrubbed out by the de-identifier. For example, “Gleason” might be removed as a patient identifier.
Semantic Disagreement: The program annotated values for the finding that could be extracted, but the values did not agree with the gold standard database.
No Mention of Desired Information: The pathology report did not contain any information about the variable.
Incomplete Information: The report provided incomplete information. For example, the report only contained a primary score (i.e. Gleason Score 3) as opposed to the complete three-value score, which indicates the primary pattern, secondary pattern and sum (Gleason Score 3+3 = 6).
Ambiguous Information due to Topology: The terms MALIGNANCY or TUMOR are coded as UMLS concepts. But this information was not associated with tissue origin in the final output. For example, if the assertion represented tumor in the main specimen and the negation represented absence of tumor in the lymph nodes, our data processing rule (select assertion over negation) would erroneously result in a false positive for LN metastasis.
We determined counts and frequencies for each error subtype, and computed performance metrics such as recall and precision when applicable.
RESULTS
Gleason Score
Results of the error analysis for the variable Gleason score are shown in Table 1. Over half of the errors were not related to the information extraction system, but rather reflected disagreement between the reporting pathologist and the expert annotator. Less frequently, there was incomplete information in the original report. For example, the original report might contain only a primary score (i.e. Gleason Score 3) as opposed to the complete three-value score, which indicates the primary pattern, secondary pattern and sum (Gleason Score 3+3 = 6). The remaining errors for Gleason score were system related. Over-scrubbing was the most common cause of system error (19.4%), followed by errors in UMLS tagging (15.5%), and chunking of the document errors (3.7%).
Table 1.
Errors related to Gleason Score
| Non- system related | 61.30% |
| Semantic Disagreement | 145 (40.8%) |
| No Mention of Information | 43 (12.1%) |
| Incomplete information | 30 (8.4 %) |
| System related | 38.70% |
| Over-scrubbing | 69 (19.4%) |
| Chunker Error: Wrong Section | 13 (3.7%) |
| Concept Tagging Error | 55 (15.5%) |
| Total error rate | 355(76.3%) |
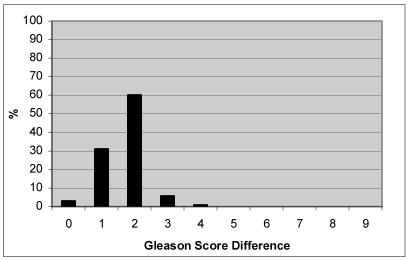
To determine the degree of divergence between report and gold-standard database values, we calculated the total Gleason Score difference for all value pairs (Figure 2). In the majority of cases (93.8%), there was a 2-point difference or lower between values out of a possible 9 points, meaning the score between the report and the gold standard annotation had a difference of 2. For example, the report had a sum of Gleason as 7 and the gold standard had the sum as 9.
Figure 2.
Degree of difference for Gleason Score
One advantage of automated extraction is that it could potentially leverage enormous existing data-sources such as clinical information systems, thereby including very large numbers of data-points. In this scenario, semantic disagreements between expert annotators and clinical data sources might be less important because differences could cancel out over huge N.
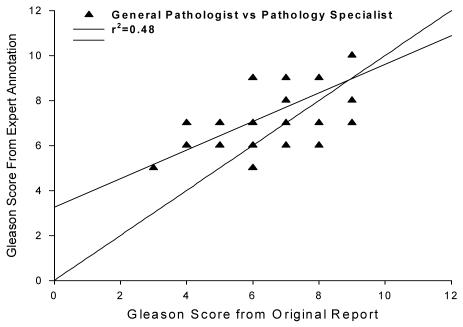
Figure 3 shows the correlation of Gleason scores between clinical report and expert annotator. There are two important observations. First, the correlation is not very high (r2=0.48), indicating that expert and reporting pathologist observations of Gleason Score did not closely agree. Second, experts tend to up-grade low Gleason scores and down-grade the high Gleason scores. The mean and SD is 6.0 ± 1.1 for the general pathologist group and 7.1 ± 1 for the expert group which were significantly different (p=.001).
Figure 3.
Gleason Score correlation between general pathologist and expert annotator
TNM Stage
Results of the error analysis for TNM Stage are shown in Table 2. System related errors occurred more than non-system related errors. Again semantic disagreement was the most frequent non-system related error. For system related errors, chunker errors were most frequent because of the position of the TNM stage information in the report. TNM stage information is most likely to be found at the end of the Final Diagnosis section, and is thus more susceptible to truncation errors.
Table 2.
Errors related to TNM Stage
| Non- system related | 41.30% |
| Semantic Disagreement | 115 (29%) |
| No Mention of Desired Information | 49 (12.3%) |
| System related | 58.80% |
| Chunker Error: Missed Out | 107 (27%) |
| Chunker Error: Wrong Section | 21 (5.3%) |
| Concept Tagging Error | 105 (26.5%) |
| Total Error Rate | 397 (85.3%) |
Lymph Node Metastases
All errors for this variable were system related. The total error was very low compared with the other two variables (Table 3). Six reports did not give the desired information due to a failure to chunk the diagnosis section from the reports. Twenty-three cases were due to ambiguous topology. The recall was 94% and the precision was 100%. For those reports who gave LN metastasis information, Table 4 shows the agreement on positive or negative status for LN metastases between the extracted value from the reports and the annotation from the pathology specialist.
Table 3.
Errors related to LN Metastases
| System related | |
| Chunker Error | 6 (20.6 %) |
| Ambiguous Information due to topology | 23 (79.4 %) |
| Total Error Rate | 29(6.2%) |
Table 4.
LN Metastasis status
| Extracted Value | |||
|---|---|---|---|
| Negative | Positive | ||
| Gold Standard | Negative | 393 | 23 |
| Positive | 0 | 14 | |
DISCUSSION
In this study, we investigated two broad categories of errors that could hinder attempts to use Information Extraction as a method for automated tissue annotation: (1) errors related to the text processing and extraction of values from the report, and (2) errors related to semantic disagreement between the report and the gold standard. By including both of these measures in our evaluation, we sought to (1) understand the inherent limitations of information extraction for providing values for the relevant variables. (2) characterize the reasons for disagreement between extracted values and report values, (3) identify the relative prevalence of these failures, and (4) compare the rates of processing and semantic errors across extracted variables. This information would be of significant value in establishing priorities for development of an Information Extraction System for the Cancer Bioinformatics Grid (caBIG).
We detected errors related to both system and observer variation. The relative contribution of these errors differed among the three studies variables. For Gleason Score and TNM stage, more than half of the errors were not system related. Both of these variables require complex multi-faceted judgments, so it is not surprising that there were many disagreements between general pathologists and specialist pathologist. This problem intrinsically exists within pathology reports and cannot be corrected with system improvements. Furthermore, we detected evidence of a systematic bias between the extracted (generalist) grading and the gold-standard (specialist) grading. Averaging across a large dataset would not yield a comparable result. In contrast to Gleason Score and TNM stage, results for LN Metastasis status showed that all errors were system related.
The results suggest that automated extraction for the purpose of tissue annotation may be more valid for some variables, and less valid for others. In spite of disagreements between information in the reports and in the gold standard, automatic information extraction could still be beneficial for quick access to information, depending on the error threshold a user can withstand. For example, a user may only need to retrieve a superset of cases that include the target cases, and would be willing to check the cases manually to match the exact value of the query. Furthermore, nearly 94% the Gleason score disagreement were only 1 or 2 degrees different, so the information extracted could still be extremely useful.
Numerous system errors were observed. Chunker errors were the most prevalent system failure for TNM stage extraction. Pathology reports have been considered to be relatively well formatted and structured compared with other medical reports, and this structural character has been exploited by many system developers. The Gate pipeline looks for keywords and spacing to delimit report sections, but in real pathology report practice, standardization is not strictly followed by all pathologists, and there are also institutional variations in reporting styles. Therefore, some pathology reports are less consistently formatted. Fortunately, chunker problems may be relatively simple to correct. For example, if we included other sections like Addendum and Comment sections when tagging UMLS concepts, we may substantially improve the TNM score recall. An alternative solution is to eliminate the chunker entirely.
For Gleason score extraction, the most common failure was due to errors in UMLS concept tagging. If the Gleason score is not tagged as a UMLS concept, we will not be able to extract the Gleason score. Many of the missed UMLS concepts were due to over-scrubbing by the de-identifier, which extracted the proper name “Gleason”. This error has since been corrected.
A more complex kind of error is apparent for extraction of LN Metastasis status. In order to determine the status of LN metastasis, we needed to determine the presence or absence of UMLS concepts that represent LN metastasis, such as “Tumor”or “Metastatic Adenocarcinoma” within the part of the report describing the lymph nodes. NegEx performed very well on detecting negations. In fact, almost all the errors were the result of ambiguous topology – for a given concept (e.g. “tumor”) we could not identify if it is associated with the main specimen itself or with the lymph nodes. This is a current limitation of the CHIRPS XML Schema that is used to represent the data extracted from the surgical pathology reports. One way of correcting this type of error is to have diagnosis concepts associated with topology in the final representation.
CONCLUSIONS
This study provides an analysis of some of the difficulties that could be encountered in extraction of information from free-text surgical pathology reports to automatically annotate biospecimens. Each of the variables had a different profile of errors, suggesting that some variables may be ‘better targets’ for information extraction and auto-annotation than others. In particular, more complex judgments requiring fine distinctions along a spectrum (such as Gleason Score) or multiple discrete decisions (such as TNM stage) may be associated with more inherent intra-observer disagreement which confounds attempts to auto-annotate from clinical free-text. For other variables, even simple NLP methods appear capable of reliably extracting information that is highly correlated with expert annotator judgments in the same case.
The current SPIN annotation system could be extended for automating annotation in several ways. First, it could be used as a method for accessing cases. Given that the user can identify a range of values (Gleason score = 4) the system could return a set that is highly enriched for that finding, but must indicate the need for further manual review. Second, the system could provide highly reliable and valid auto-annotation for a subset of variables. Third, the system could be used to provide a “rough draft’ for expert manual annotators – in order to limit the burdens of manual annotation.
ACKNOWLEDGEMENTS
Kaihong Liu is supported by National Library of Medicine Medical Informatics Training Grant number 5-T15-LM07059. Support for this project was provided by grants from the National Cancer Institute for the Shared Pathology Informatics Network (UO1 CA91343 and U01 CA091429). We would like to thank Manoj Ramachandran for his technical support.
REFERENCES
- 1.Cunningham H, Wilks Y, and Gaizauskas R. GATE -- a General Architecture for Text Engineering. In: Proceedings of the 16th Conference on Computational Linguistics (COLING-96), 1996.
- 2.Cunningham H, Humphreys, K, Gaizauskas R, and Wilks Y. Software Infrastructure for Natural Language Processing. In: Proceedings of the Fifth Conference on Applied Natural Language Processing (ANLP-97), 1997.
- 3.http://gate.ac.uk/
- 4.Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A general natural-languagetext processor for clinical radiology. Journal of the American Medical Informatics Association. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee M. Christensen, Peter J. Haug, and Marcelo Fiszman. MPLUS: A Probabilistic Medical Language Understanding System. Proceedings of the Workshop on Natural Language Processing in the Biomedical Domain, Philadelphia, July 2002, pp. 29–36.
- 6.Hahn U, Romackera M, Schulzb S. MEDSYNDIKATE –a natural language system for the extraction of medical information from findings reports. International Journal of Medical Informatics. 2002;67:63–74. doi: 10.1016/s1386-5056(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell K.J, Becich,M.J, Berman J.J, Chapman W.W, Gilbertson J, Gupta D, Harrison J, Legowski E, Crowley RS. Implementation and Evaluation of a Negation Tagger in a Pipeline-based System for Information Extraction from Pathology Reports. p. 663–667 Medinfo 2004. [PubMed]
- 8.http://caBIG.nci.nih.gov/
- 9.http://www.ndoc.org/about_ndc/reports/pdfs/FINAL_NBN_Blueprint.pdf
- 10.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of Biomedical Informatics. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 11.Namini AH, Berkowicz DA, Kohane IS, Chueh H. A submission model for use in the indexing, searching, and retrieval of distributed pathology case and tissue specimens. Medinfo 2004. 2004:1264. [PubMed] [Google Scholar]
- 12.Dunham SG, Pacak GM, Pratt WA. Automatic Indexing of Pathology Data. Journal of the American Society for Information Science. 1978;29(2):81–90. doi: 10.1002/asi.4630290207. [DOI] [PubMed] [Google Scholar]