Abstract
The POU transcription factors Oct1 and Oct2 bind to DNA in various monomer and dimer configurations. Depending on the DNA sequence to which they bind, the dimers are arranged in configurations that are either accessible (PORE sequence) or inaccessible (MORE sequence) to the B-cell-specific cofactor OBF1 (OcaB, Bob1). As shown previously, the MORE and related sequences (such as the heptamer/octamer motif) are found in immunoglobulin heavy chain promoters. Here we show that the expression of Osteopontin, which contains a PORE sequence in its enhancer region, depends on the presence of OBF1 in B cells. OBF1 alleviates DNA sequence requirements of the Oct1 dimer on PORE-related sequences in vitro. Furthermore, OBF1 stabilizes POU dimer–DNA interactions and overrides Oct1 interface mutations, which abolish PORE-mediated dimerization without OBF1. Our data indicate that the PORE-type Oct1 or Oct2 dimer, rather than the monomer, is the primary target of the cofactor OBF1. Based on our biochemical data, we propose a mode of OBF1–Oct1 dimer interaction, suggesting a novel arrangement of the subdomain connectivities.
Keywords: gene regulation/OBF1/POU/transcription factor
Introduction
Specificity in the transcriptional regulation of gene expression is necessary to enable the correct temporospatial expression pattern during development. The combination of multiple factors represents an efficient way to integrate different signal pathways and to coordinate cell type and cell cycle specificity. To this end, transcription factors bind to DNA directly and assemble with each other and coactivators and/or corepressors. This leads to the formation of specific transcriptional complexes with distinct characteristics based on the DNA-binding sequence of the regulatory regions and the particular factors involved.
The POU family of transcription factors is involved in the transcriptional regulation of a wide array of ubiquitous and tissue-specific genes. Oct1, often regarded as the prototype POU factor, is broadly expressed. Oct2 and Oct4 are exemplary POU genes with a narrow expression profile. Oct2 is mainly found in cells of the lymphoid system and Oct4 is limited to the mammalian germline, including stem cells of the early embryo and germ cells (Ryan and Rosenfeld, 1997).
Members of the POU transcription factor family share a conserved bipartite DNA-binding domain called the POU domain, containing the modular POU-specific domain (POUS) and the POU-homeodomain (POUH). The subdomains are connected by a flexible linker, which is variable in sequence and length (15–56 amino acid residues). POU factors bind to DNA elements, such as the octamer motif (ATGCAAAT), as monomers (Staudt et al., 1986; Schöler et al., 1989). More recently, POU factors have been shown to homo- and heterodimerize on specific DNA motifs (Jacobson et al., 1997; Botquin et al., 1998; Rhee et al., 1998; Scully et al., 2000; Tomilin et al., 2000; Reményi et al., 2001). The Oct-factor subgroup binds two of these octamer-related sequences: the PORE (Palindromic Oct-factor Recognition Element: ATTTGAAATGCAAAT) and the MORE (More PORE: ATGCATATGCAT). The PORE was first identified as an Oct4 binding sequence in the first intron of the osteopontin (OPN) gene in embryonic carcinoma (EC) cells. Homo- and heterodimers of various POU factors, including Oct1, Oct2, Oct4 and Oct6, can assemble on the PORE in vitro. Dimerization on the PORE mediates strong transcriptional activation, supporting the notion of dimer formation in vivo (Botquin et al., 1998). MOREs and related sequences (e.g. the heptamer/octamer motif) are found in immunoglobulin heavy chain promoters (VH). Oct family members can also bind cooperatively as homo- and heterodimers on the MORE in vitro (Tomilin et al., 2000).
Furthermore, POU proteins form heterodimers with transcription factors of other families. Several POU factors have been shown to interact with members of the HMG family, such as Sox2, in vitro (Yuan et al., 1995; Botquin et al., 1998; Nishimoto et al., 1999) or to assemble with cofactors (Sauter and Matthias, 1998; Scully et al., 2000). The most extensively studied cofactor is OBF1, which is expressed in lymphoid cells and interacts specifically with Oct1 and Oct2 monomers and dimers (Luo et al., 1992; Gstaiger et al., 1995; Luo and Roeder, 1995; Strubin et al., 1995; Tomilin et al., 2000). The interaction with the dimer is dependent on the conformation of the POU/DNA complex, as OBF1 can only bind to PORE-mediated Oct1 dimers but not to those on MOREs. The recently solved crystal structures of the POU domains of the Oct1 dimer bound to the MORE and PORE revealed the structural basis for this selectivity of OBF1. The two POU dimers adopt two different configurations, using different surface patches to dimerize on each element. As a result, the same amino acids that are available to interact with OBF1 in the PORE configuration form part of the POUS–POUH dimer interface on the MORE, precluding an interaction with OBF1 (Tomilin et al., 2000; Reményi et al., 2001).
Osteopontin (OPN) is expressed in various tissues (Denhardt et al., 1995) and the immune system (Weber and Cantor, 1996; O’Regan and Berman, 2000). Secreted OPN stimulates B cells to produce immunoglobulins and, in conjunction with an unidentified 14 kDa peptide, to proliferate (Weber and Cantor, 1996). Moreover, B lymphocytes have been shown to play a role in new bone formation in which OPN is also involved (Marusic et al., 2000). One common theme that has emerged from several of these studies is that OPN is involved in cell migration and adhesion.
In this study, we first demonstrate a regulatory link between POU proteins and OBF1 and OPN expression in B cells. We then describe the effect of OBF1 on the PORE-mediated dimerization of the POU domain of Oct1. We show that OPN expression in lymphoid cells depends on OBF1 and reveal that the PORE element is active in B cells. Consequently, lymphoid cells provide an environment conducive to the differential expression of genes via different conformations of the same transcription factor and cofactor recruitment that are dictated by the DNA sequence. Using Oct1 interface mutants that specifically inhibit dimerization on the PORE, we show that OBF1 can compensate for the loss of the PORE-specific dimer interface. Besides overriding the effect of the mutations, OBF1 alleviates DNA sequence requirements by clamping the dimer to the DNA, significantly stabilizing the protein–DNA complex. Our results suggest that an arrangement of the POU subdomains is adopted by the Oct1–OBF1 complex on the PORE that is different from that proposed for Oct1 alone.
Results
Osteopontin is a target gene of OBF1
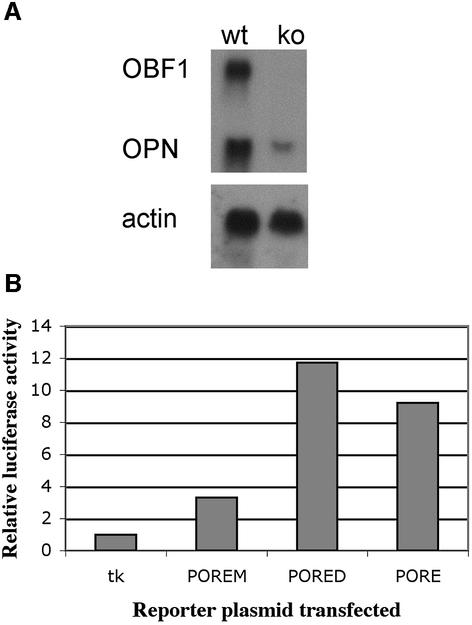
So far, MOREs have been shown to mediate transcriptional activation in B cells while POREs are active in pluripotent embryonal cells (Botquin et al., 1998; Tomilin et al., 2000). To determine whether both regulatory elements can be functional in the same cell type in vivo, we examined whether POREs are active in B cells. To this end, we studied the PORE located in the first intron of OPN, which is known to regulate OPN expression in embryonic stem cells and EC cells (Botquin et al., 1998). First, we analyzed the level of OPN transcripts by northern blot analysis to compare the mRNA of normal and OBF1-deficient splenocytes stimulated in vitro (Figure 1A). OPN transcription was far stronger in wild-type splenocytes than in OBF1-deficient cells. We concluded that OPN is a target gene of this transcriptional coactivator in vivo. However, indirect stimulation of OPN via activation of another gene cannot be excluded.
Fig. 1. Osteopontin is regulated by OBF1 in lymphoid cells. (A) Analysis of OPN expression in wild-type (wt) and OBF1–/–(ko) splenocytes. Cells were stimulated in vitro and total RNA was analyzed by northern blotting. Osteopontin (OPN) transcripts were found at high levels in wild-type splenocytes. OBF1–/– splenocytes show reduced OPN transcript levels. Actin, control for comparable mRNA levels in both samples; OBF1, control to show the presence of transcripts in wild-type cells and absence in knockout cells. (B) Comparison of enhancer activities of PORE-derived elements in transient transfection experiments. BJA-B cells were transfected with different luciferase reporter plasmids (x-axis). The y-axis shows activation of transcription, expressed as relative luciferase activities. The tk minimal promoter served as a control. BJA-B cells naturally express OBF1. PORED and POREM are derivatives of the PORE, to which only the POU dimer and monomer, respectively, can bind.
To determine whether OBF1 has the potential to stimulate the OPN gene directly, we analyzed several PORE variants in B cells. For this purpose, three different PORE-variant reporter plasmids were transfected into BJA-B cells known to express high levels of OBF1 protein. Hexamers of the PORE sequence were cloned in front of the thymidine kinase minimal promoter (tk) driving the luciferase gene. The mutation in the PORED restricted binding to POU dimers in vitro while only monomers bind the POREM (referred to as O, O–1, O–3 in Botquin et al., 1998). In F9 EC cells, both the PORE and PORED were more active than POREM, which was only slightly more active than the tk promoter alone (Botquin et al., 1998). In B cells, reporter activity compared with that of the tk reporter alone was ∼9-fold higher for the PORE, ∼12-fold higher for PORED and ∼3-fold higher for POREM (Figure 1B). These levels of increased reporter activity are comparable to those obtained in cotransfection experiments with OBF1 using 293 cells. OBF1 stimulates PORE activity, with the PORED mediating higher transcriptional activity than the PORE and POREM (Tomilin et al., 2000). This result suggests that the OPN PORE can be activated by different sets of POU factors and their coactivators, namely by Oct1 (Oct2) with OBF1 (B cells) and by Oct4 possibly with a yet unknown coactivator (early pluripotent embryonic cells). Our results also show that, in this experimental set-up, a POU dimer is required for high OPN expression levels, whereas a POU monomer appears to be insufficient to support full transcriptional activation.
Here, we extend the finding that the PORE in the first intron of the OPN gene is important for the transcriptional activation of OPN not only in pluripotent embryonal cells but also in lymphoid cells. Moreover, in addition to our previous findings (Botquin et al., 1998; Tomilin et al., 2000), these data further imply that the activation is modulated through synergism between the Oct1 (Oct2) dimer formed on this PORE and the lymphoid-specific cofactor OBF1. They also confirm our earlier hypothesis that OPN is a target gene of an Oct1 (and/or Oct2) dimer in conjunction with OBF1 in lymphoid cells.
Identification of potential PORE-interface mutants
The experiments mentioned above suggest that an Oct dimer together with OBF1 plays a critical role in regulating OPN expression in lymphoid cells. To analyze the molecular interaction between these proteins, we introduced a set of specific mutations into the POU domain of Oct1 (POU1) based on the information provided by the crystal structure of the PORE-mediated POU1 dimer (Reményi et al., 2001). The non-overlapping nature of the MORE and the PORE dimerization interfaces allowed us to design mutants that were able to affect one type of POU dimer formation selectively while leaving the other intact (Reményi et al., 2001; data not shown).
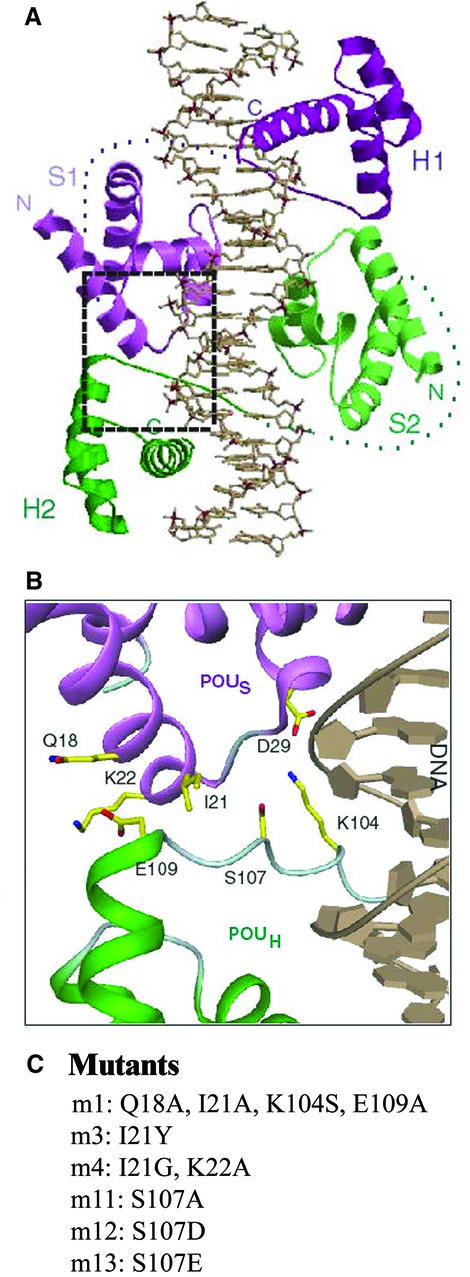
In the crystal structure of the POU–PORE complex, the POUS–POUH dimer interface is formed by three types of interactions: (i) POUS–POUH salt bridges (D29-K104 and K22-E109); (ii) specific hydrogen bonds of R20 (POUS) and S107 (POUH) to a common phosphate group in the minor groove of the DNA; (iii) van der Waals interactions within the POUS–POUH interface (by Q18, I21, K22, K104, S107 and E109) (Figure 2A and B). In order to affect dimer formation on the PORE element, amino acid residues that play a prominent role in the PORE-type interface (POUS: Q18, I21 and K22; POUH: K104, S107 and E109) were mutated into either small amino acids (A, G or S), to remove side chain-specific interactions, or amino acids containing bulky or charged side chains (Y, D or E) to cause steric clashes or electrostatic repulsion within the interface (Figure 2C). Two of these mutations are particularly interesting since they imitate phospho serines (m12, S107D; m13, S107E).
Fig. 2. POUS–POUH interface in the PORE-type dimer. (A) Representation of the POU1 dimer on the PORE. The coordinates were taken from the published crystal structure (Reményi et al., 2001). One POU1 molecule is colored purple and the other green. (B) Close-up of the POUS–POUH interface focusing on the intermolecular protein–protein interactions. All amino acid residues mentioned are predicted to play a role in forming the dimer interface. POUS is colored purple and POUH is green. The critical residues are indicated in yellow and the DNA is colored gray. Two-digit amino acids are part of POUS; three-digit amino acids belong to POUH. (C) Mutants used in this study.
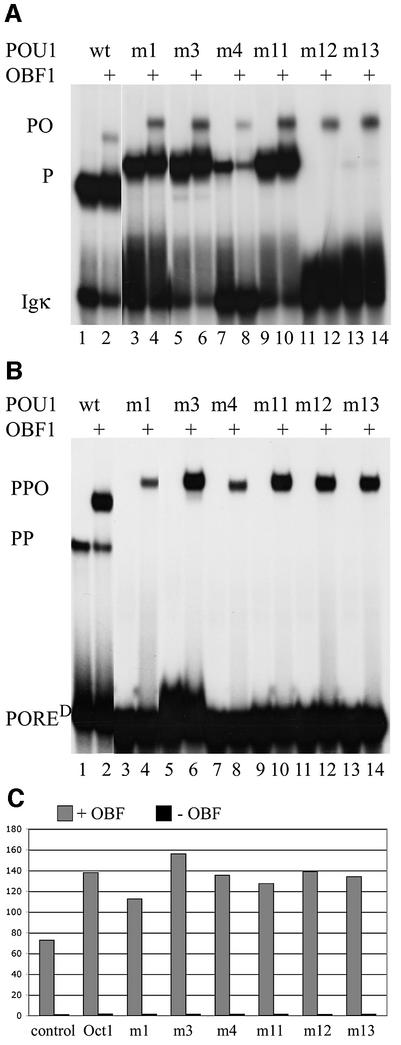
The POU1 dimer-interface mutants were expressed in Escherichia coli and their protein–DNA and protein– protein interactions were analyzed by electrophoretic mobility shift assays (EMSAs). First, monomer formation on the canonical octamer motif was tested using an oligonucleotide derived from the Igκ promoter (Bergman et al., 1984) (Figure 3A, odd lanes). Two mutants [m3 (I21Y) and m11 (S107A)] had only a slight effect on DNA binding. In m3, monomer binding was only affected slightly, as I21 is not involved in DNA binding and this residue is far from the double helix (Figure 2) (Klemm et al., 1994). In m11, the serine at position 107 (S107) was changed to alanine, a mutation that has no effect on DNA binding as this residue is not involved in POU monomer contacts to Igκ (Klemm et al., 1994). In m1, which was generated to preclude dimerization between POUS and POUH, residues Q18, I21, K104 and E109 were mutated into alanines and one serine (Figure 2C), which affected monomer binding very slightly. The effect of the mutations in m4 was slightly stronger. It is unlikely that the I21G mutation resulted in the weaker interaction with the DNA, since the m3 mutant (I21Y) does not affect DNA– monomer binding. However, the K22A mutation may prevent an interaction with E109. In marked contrast, the phosphoserine-mimicking mutants m12 and m13, in which S107 is mutated into an aspartic or a glutamic acid (Maciejewski et al., 1995), abolished monomer binding to Igκ (Figure 3A). Since S107 is located close to the DNA helix when it is bound to Igκ, replacing it with the bulky and negatively charged residues probably interferes with DNA binding due to electrostatic repulsion and steric clash. In summary, m1, m3, m4 and m11 mutants bind to Igκ as monomers, whereas m12 and m13 do not.
Fig. 3. Monomer and dimer binding activities of POU1 and its derivatives under the influence of OBF1. (A) EMSA showing the different effects of POU1 mutants on monomer binding to Igκ, an octamer- containing promoter. Odd lanes, binding pattern of POU1 and its mutants on the octamer site; even lanes, binding pattern of OBF1 plus POU1 and its mutants on the octamer site. P, POU1–DNA complex; PO, POU1–OBF1–DNA complex. (B) EMSA showing the effects of mutants and OBF1 on dimer binding on PORED. Set-up as in (A). PP, POU1–POU1–DNA complex; PPO, POU1–POU1–OBF1–DNA complex. The OBF1-containing complex on the PORED consists of two POU1 molecules and one OBF1 molecule (Tomilin et al., 2000; Reményi et al., 2001). The unmutated POU1 protein exerts a slightly higher mobility than the mutant proteins, which have an additional 3 kDa histidine tag. The tag slows down the migration in the gel, but does not affect interaction with the DNA. (C) Comparison of the trans-activities of the Oct1 variants in transient transfection experiments; 293 cells were transfected with PORED luciferase reporter plasmid, wild-type or mutant Oct1 expression vectors (none in the control) with (+) or without (–) OBF1 expression vector (x-axis). The y-axis shows activation of transcription, expressed as relative luciferase activities.
Several studies had previously described a clamping effect of OBF1 on the binding of POU1 monomer to DNA (Babb et al., 1997; Sauter and Matthias, 1998). To assess whether OBF1 can overcome the compromised DNA binding activities of some of these POU1 mutant proteins, we compared their DNA binding in the presence and absence of the OBF1 peptide used for solving the Oct1–OBF–DNA crystal structure (Figure 3A) (Chasman et al., 1999). None of the mutations had a negative effect on the POU1–OBF1 interaction. OBF1-assisted binding was equally weak for all POU1 proteins that showed normal DNA binding and did not correlate with their intensity of monomer binding to Igκ DNA (compare even and odd lanes). Furthermore, m12 and m13, which cannot bind to Igκ alone, also formed an OBF1-containing complex. These latter results indicate that OBF1 enables Oct1 binding to Igκ DNA when monomer binding is severely compromised.
OBF1 rescues mutations that impair POU1 dimerization
We then examined the effect of various POU1 mutations on dimerization using a PORE variant, PORED, that only binds POU dimers but not monomers (Botquin et al., 1998; Tomilin et al., 2000). While the unmutated POU1 domain formed a dimer on PORED, confirming previous data, none of the six mutants retained this capability (Figure 3B), as predicted by the structural analysis of the POUS–POUH dimer interface in the POU1–PORE complex (Reményi et al., 2001).
The N-terminal OBF1 peptide (Chasman et al., 1999) was mixed with each of the seven Oct1 derivatives to examine its effect on dimerization. A strong heterotrimer was formed when OBF1 was added to unmutated POU1 and PORED (Figure 3B, lane 2). This is in contrast to a weak OBF1-induced ternary complex formation with POU1 on Igκ (Figure 3A, lane 2). This finding supports the notion that the POU1 dimer is a better substrate for interaction with OBF1 than the monomer (Tomilin et al., 2000). Strikingly, all POU1 mutants, which failed to dimerize in the absence of OBF1, formed ternary complexes on the PORED in the presence of OBF1 (Figure 3B, even lanes). The complexes formed with m3, m11, m12 and m13 POU1 mutants were as strong as those with unmutated POU1; the m1 and m4 mutants showed ∼6- and 4-fold reduced capability for heterotrimer formation, respectively. Similar results were obtained when the original PORE motif was used as a probe (data not shown). These results demonstrate that OBF1 rescues the detrimental effect of all POU1 mutations tested on binding to PORED. Similar to the POU1–OBF1 (PO) complex formed by the Oct1 derivatives, the POU1–POU1–OBF1 (PPO) complex showed relatively little variation in binding intensity. This is in striking contrast to the high variability of POU1 monomer binding intensities.
To address the question whether the Oct1 mutants could also be rescued in vivo, we performed cotransfection experiments of a PORED luciferase reporter plasmid (Botquin et al., 1998) together with OBF1 and Oct1 expression vectors into 293 cells (Figure 3C). Oct1 alone activated luciferase activity very little compared with the control (bars 2 and 4; Tomilin et al., 2000) and activation by the mutant Oct1 proteins was even smaller (even bars). Upon addition of the OBF1 expression vector, reporter activity was highly stimulated. The unmutated Oct1 caused the activity to double compared with OBF1 transfected alone (bars 1 and 3). In agreement with the results obtained in the EMSA (Figure 3B), the activity of the mutated and unmutated Oct1 cotransfected with OBF1 showed little variation in transactivation intensity. All Oct1 variants increase OBF1-induced activity ∼2-fold. As in the gel shift assay, m1 has the smallest potential. The high background of OBF1 alone might be due to its synergy with endogenous Oct1 in 293 cells.
OBF1 alleviates DNA sequence requirements
The above experiments showed that mutations abolishing dimerization of POU1 on PORED could be rescued by OBF1. Another interface involved in the complex formation is that between DNA and POU1. We investigated whether the OBF1 peptide may not only overcome mutations in the POUS–POUH interface but may also exert positive effects if the protein–DNA interface is unfavorably mutated. To this end, we generated various oligonucleotides containing mutations in the octamer site within the PORE based on sequence requirements of the octamer motif for POU1 binding (Verrijzer et al., 1992). We then examined the formation of POU1 monomers, dimers and OBF1-induced complexes on these PORE derivatives.
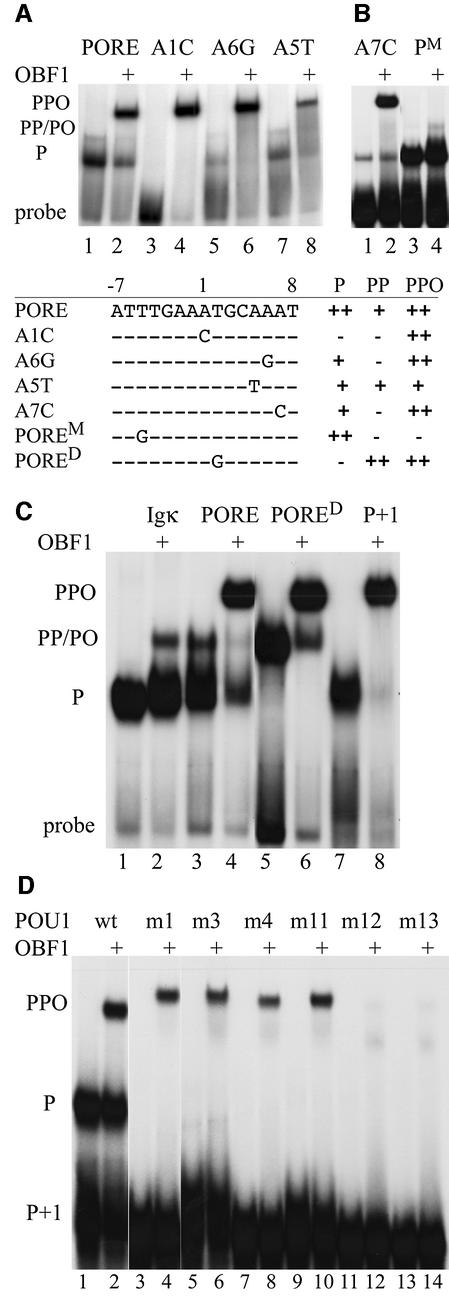
PORED is one of these unfavorable mutations. Here, the T at the second position of the octamer motif is mutated into a G (T2G). The POU1 monomer cannot bind, whereas the POU1 dimer can; a heterotrimer with OBF1 binds even more efficiently (Figure 3B). Further DNA mutants are shown in Figure 4A and B. The A1C PORE mutant generally impairs POU1 binding in the absence of OBF1. Upon addition of OBF1, however, a heterotrimer is formed which binds as strongly as to the wild-type PORE. Similar data are obtained for the PORE mutants A6G and A7C (referred to as O–4 in Botquin et al., 1998) (Figure 4A and B), although some weak monomer binding is obtained even in the absence of OBF1. In addition, on the POREM, where the octamer is intact but the T 5 bp upstream of the octamer within the PORE is mutated into a G, a weak heterodimer, but no heterotrimer, was formed (Figure 4B). Furthermore, on PORE A5T, a heterotrimer bound significantly more weakly than on the wild-type PORE (Figure 4A, lanes 7 and 8). This supports previous data (Cepek et al., 1996; Gstaiger et al., 1996; Chasman et al., 1999) showing that the A5T mutation generally reduces OBF1 binding. None of these octamer-related motifs has been identified as target sequences (Verrijzer et al., 1992), suggesting that OBF1 dramatically alleviates the DNA sequence requirements of POU1 on PORE-derived elements.
Fig. 4. Sequence requirement alleviation mediated by OBF1. (A and B) EMSA showing OBF1 rescuing POU1 binding on mutant POREs. Odd lanes, binding pattern of POU1 to the PORE and its derivatives; even lanes, binding pattern of OBF1 plus POU1 to the PORE and its derivatives. P, POU1–DNA complex; PP, POU1–POU1–DNA complex; PPO, POU1–POU1–OBF1–DNA complex. The table shows the specific point-mutated PORE derivatives and their abilities to bind the POU1 monomer (P), the POU1 homodimer (PP) and the POU1–POU1–OBF1 complex (PPO). (C) EMSA comparing POU1 with (even lanes) and without OBF1 (odd lanes) on Igκ, PORE and derivatives thereof used to demonstrate differences and similarities in their mobilities through the gel. P+1, PORE with one nucleotide inserted between the two half-sites; P, POU1–DNA complex; PP, POU1–POU1–DNA complex; PO, POU1–OBF1–DNA complex; PPO: POU1–POU1–OBF1–DNA complex. (D) EMSA with oligonucleotide P+1. Odd lanes, binding patterns of POU1 and its mutants; even lanes, binding patterns of OBF1 plus POU1 and its mutants. P, POU1–DNA complex; PPO, POU1–POU1–OBF1–DNA complex. The unmutated POU1 protein exerts a slightly higher mobility than the mutant proteins, which have an additional 3 kDa histidine tag.
OBF1–POU1 dimer complex tolerates changes in PORE binding site separation
The PORE P+1 sequence contains an insertion of one nucleotide between the two halves of the element. The Oct4 monomer binds to P+1 as well as to the wild-type PORE, whereas Oct4 dimers cannot form (Botquin et al., 1998). Here we extend this finding to Oct1, showing that its POU domain forms a monomer only, and no dimer, on P+1 (Figure 4C, lane 7). For both POU1 and Oct4, these data are explained by the loss of the POUS–POUH dimer interface across the two binding sites observed in the POU1–PORE crystal structure and modeled in the Oct4–PORE complex (Reményi et al., 2001). When OBF1 is added, it forms a higher order complex with POU1 (lane 8). There are two reasons why we think that this complex contains two molecules of POU1 and one molecule of OBF1. First, it migrates to the position of the higher order complex with PORE and PORED (Figure 4C, lanes 4 and 6), which was previously described as a POU1–POU1–OBF1 heterotrimer on DNA (Tomilin et al., 2000; Reményi et al., 2001). Secondly, the complex that is composed of only one POU1 molecule and one OBF1 migrates faster (lane 2) to the position of the POU1 dimer (Figure 4C, lanes 3 and 5). Therefore, OBF1 promotes binding of two POU1 molecules on P+1. This result was surprising, as we had expected only a POU1–OBF1 complex to form on the octamer motif, similar to the one on Igκ DNA (Figure 4C, compare lanes 2 and 7). The question remains: how can the second POU1 molecule bind strongly enough to the non-octamer half of the PORE sequence (ATTTG) without the bridging interface that is required to link the two POU1 molecules in the absence of OBF1 (Botquin et al., 1998; Reményi et al., 2001)?
To reveal a molecular rational for this apparent paradox, we carried out several experiments. First, binding of POU1 to POREM was tested in the presence and absence of OBF1. The position of the POU1–OBF1 complex on POREM was identical to that formed on Igκ, but the intensity was even weaker than that for Igκ (cf. Figure 4B and C). This indicates that the same octamer within the PORE can be a good or a poor substrate for the POU1–OBF1 complex, depending on whether the second POU1 is able to bind or not. The m4 mutant protein bound weakly to Igκ (Figure 3A, lanes 7 and 8), whereas no binding was observed on POREM (data not shown). Therefore, we considered it unlikely that m4 would bind to P+1 comparably with the other POU1 variants if a mere structural distortion were responsible for the POU1 dimer–OBF1 complex formation on P+1. In contrast, if the two POU1 molecules are arranged parallel to the DNA axis in the presence of OBF1, and this new arrangement represents a better substrate for OBF1, m4 may bind well on P+1 in the presence of OBF1. Therefore, the seven POU1-variant proteins were tested for binding on P+1 in the presence and absence of OBF1 (Figure 4D). None of the mutants could form a monomer on P+1 (Figure 4D, odd lanes), in contrast to monomer binding on the Igκ site (Figure 3A). This was not due to the nucleotide insertion, as they were also unable to form on the PORE, except for m3 and m11, which bound very weakly (data not shown). Only when OBF1 was added did complexes form with the unmutated POU1, m1, m3, m4 and m11 which, according to their position on the gel, presumably contain two POU1 molecules and one OBF1 (Figure 4D, even lanes; cf. Figure 4C). Since the complexes of all four mutant proteins are similar in binding intensity to P+1, binding of m4 due to structural distortion seems to be an unlikely explanation. We consider it more likely that the POU subdomains are positioned so that the linkers between them are arranged parallel to the DNA axis (see Discussion).
Only m12 and m13 binding to P+1 could not be rescued by OBF1 (Figure 4D, lanes 11–14). These mutant proteins have an aspartic and a glutamic acid, respectively, instead of a serine at position 107. While OBF1 helped tolerate these substitutions on the PORE, it appears to have reached its limit on clamping proteins with these bulky and negatively charged side chains to P+1.
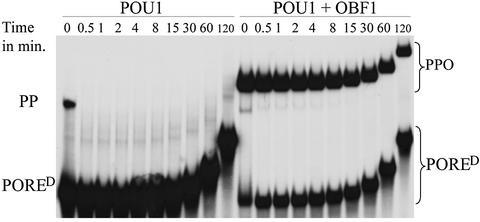
OBF1 stabilizes the POU1 dimer DNA complex by reducing its dissociation rate
The data presented so far indicate that OBF1 has a stabilizing effect on the POU1 dimer on the PORE. We performed off-rate experiments to assess this effect further. To this end, a binding mix was prepared to which a 500-fold excess of unlabeled oligonucleotide was added. The reaction was loaded onto a gel at regular time intervals. An increasingly weaker intensity of the bands with time reflects proteins dissociating from the DNA and being adsorbed by the excess of unlabeled oligonucleotide, thus becoming undetectable.
The POU1 dimer dissociates from the PORED within 30 s of binding to it in the absence of OBF1 (Figure 5, left panel). This indicates a high off-rate and kinetic instability of the protein–DNA complex. The heterotrimer containing the OBF1 peptide forms a much higher intensity band than the POU1 dimer alone (compare 0 min lanes in right and left panels). Furthermore, the coactivator dramatically reduces the off-rate of the POU1 dimer (right panel). Even 2 h after addition of the unlabeled oligonucleotide, ∼50% of the complex is still bound to the labeled probe, indicating that it has not dissociated from the DNA during that time. This reflects a >200-fold reduction in the off-rate with OBF1. This is in striking contrast to the lack of stabilization of the Oct1 monomer by OBF1 in a similar experiment (Strubin et al., 1995), again indicating that the PORE-type Oct1 dimer is a preferred substrate of the coactivator compared with the POU1 monomer.
Fig. 5. Off-rate EMSA showing the stability of the POU1 dimer and the POU1–POU1–OBF1 complex on DNA. A binding reaction was prepared into which a 500-fold excess of unlabeled oligonucleotide was added after the proteins had been allowed to bind the labeled probe. Aliquots of the reaction mixture were loaded onto a gel at regular time intervals between 30 s and 120 min after addition of the unlabeled oligonucleotide. Dissociating proteins become undetectable on the gel as they re-associate to the unlabeled oligonucleotide.
Discussion
Osteopontin in B cells: gene regulation and function
POU factors exert a high level of flexibility in regulating gene expression, attributed in part to their ability to bind DNA as monomers, homodimers and heterodimers. In addition, these dimers can adopt different configurations depending on the DNA sequence to which they bind (Tomilin et al., 2000). In doing so, they expose different surface patches, which in consequence can recruit different cofactors. In the case of Oct4, the two dimer configurations that form on the PORE and MORE elements also react differently to phosphorylation of the POU factor (Reményi et al., 2001).
Within a given cell, genes cannot be separated spatially from one another and thus are exposed to the same transcription factors and cofactors. In such an environment, the differential regulation of many genes by the same transcription factor must occur. This may be at least partially attributed to the selective recruitment of cofactors. We previously reported that the heavy chain of the immunoglobulins is regulated by the MORE-type dimer in lymphoid cells (Tomilin et al., 2000), and in this study we established that osteopontin is a target gene of the PORE-like dimer and OBF1 in the same cells. Thus, we found an environment conducive to various degrees of differential gene regulation.
Secreted OPN stimulates B cells to produce immunoglobulins and, together with an unidentified 14 kDa peptide, to proliferate (reviewed by Weber and Cantor, 1996). However, the precise function of osteopontin in B cells remains elusive. In the absence of OBF1, osteopontin is expressed at very low levels (Figure 1A) and might consequently not be able to stimulate B cells to proliferate, leading to fewer mature B cells in OBF1-deficient mice. This reduction of B-cell number in OBF1-deficient mice has been reported (Kim et al., 1996; Nielsen et al., 1996; Schubart et al., 1996). Thus, osteopontin, together with the 14 kDa peptide, might have an autocrine effect on B cells.
OBF1 alleviates DNA sequence requirements for POU dimerization and stabilizes the dimer on the DNA
The binding specificity of POU1 has been analyzed by screening a library of randomly synthesized oligonucleotides with POU1 for high-affinity targets (Verrijzer et al., 1992). Consequently, a sequence similar to the octamer motif was defined as the Oct-binding consensus. Based on this consensus, we generated various oligonucleotides with unfavorable mutations within the PORE and tested them for monomer, dimer and OBF1-induced complex formation. We found that even though monomer and dimer binding were often impaired, OBF1 would discount these unfavorable sequences and clamp the POU1 dimer onto the DNA (Figure 4A). The lack of an OBF1-induced complex on the POREM and the presence of a weak one on A5T proved that the cofactor does not bind indiscriminately. Nonetheless, OBF1 alleviates sequence requirements substantially. This implies that a weak promoter might only be activated by the Oct1 dimer in the presence of OBF1, whereas a strong PORE-type binding site may not require a cofactor to activate the target gene.
The results obtained with P+1, an element in which the two Oct-factor binding sites have been moved apart by the insertion of one nucleotide, indicate that OBF1-induced dimerization does tolerate changes in the separation of the two half binding sites on PORE (Figure 4C). Botquin et al. (1998) showed that dimerization does not occur on P+1, and proposed that this was a consequence of the interface having been lost between the two protein molecules. OBF1 overcomes these binding difficulties and mediates heterotrimer binding with the POU1 dimer to the oligonucleotide. There are two possible explanations to account for this ability of OBF1: (i) OBF1 may clamp the two molecules onto the DNA at a less favorable binding site so that the POU factors are bound as on the PORE, relative to each other, ignoring the phasing mutation; (ii) the POU1 molecules bind to the same nucleotides on P+1 as on the PORE, but protein–protein interaction is not required because OBF1 tethers the two molecules to the DNA. This has to be such a strong interaction that the POU1 molecules do not have to support each other to remain bound to the DNA.
We favor the second explanation partly because of the off-rate results (Figure 5), which revealed that the OBF1 peptide has a strong stabilizing effect on the POU1 dimer. This solid stabilizing potential of OBF1 has not been detected on the POU1 monomer. A slight OBF1 stabilizing effect on the POU1 monomer has been shown by DNase I footprint assays (Babb et al., 1997). However, titration studies were unable to demonstrate an effect of OBF1 on POU1 stability (Luo and Roeder, 1995). Furthermore, the dissociation rate of the Oct1 monomer from the octamer binding site was not influenced by the presence of OBF1 (Strubin et al., 1995), even though the cofactor exhibits a clamping effect by securing the two separate POUS and POUH domains onto the DNA, as shown by EMSA.
In addition to the PORE-like sequence, the flanking nucleotides also influence the binding ability of the POU1 dimer and OBF1. POU1 does not bind PORED as a monomer, but if the two nucleotides 3′ of the element are mutated from GG to TA it does (data not shown). The fact that none of the 56 target sites selected for POU1 binding (Verrijzer et al., 1992) contained two Gs following the octamer motif may reflect how POU proteins need to interact as monomers and dimers with the PORE. This supports the notion that the two Gs following the PORE are disadvantageous for monomer formation. Moreover, POU1 and its mutants m1, m3, m4 and m11 (Figure 2) can form monomers on BCL1 (Tomilin et al., 2000; data not shown), which contains a consensus octamer motif followed by the nucleotides CA. Indeed, the TA and CA combinations 3′ of the octamer motif, which allow monomer binding, were obtained as binding sequences in the experiment reported by Verrijzer et al. (1992).
The genes that should only be expressed in small quantities or when high levels of POU1 are present in the cell might be regulated by elements to which OBF1 cannot clamp the POU1 dimer. In such a case, the dimer can dissociate from the regulatory element rapidly and thus discontinue transcription of the gene. Genes regulated by the OBF1–Oct1 heterotrimer may need to be expressed at high levels once they are activated. This can be provided for, since the complex may remain bound to DNA for a longer time and may thus allow multiple rounds of transcription to occur. For example, the osteopontin gene may require high levels of mRNA to be generated continuously, as the protein concentration is decreasing when it is secreted from the confined cell to the extracellular milieu.
Mimicking phosphorylation at the PORE dimer interface prohibits POU1–DNA interaction
One of the most frequently used mechanisms to regulate the activity of transcription factors in response to different extra- and intracellular signals is phosphorylation and dephosphorylation (Whitmarsh and Davis, 2000). Activities of several members of the POU factor family are controlled by this mechanism. Oct1 is hyperphos phorylated as cells enter mitosis. This correlates with strongly reduced Oct1 binding to the octamer site and a concomitant inhibition of transcription. Phosphorylation of Oct1 is rapidly reversed as cells exit mitosis and enter the G1 phase of the cell cycle (Roberts et al., 1991). It was shown that mitosis-specific phosphorylation of S107 in the homeodomain of Oct1 was sufficient to inhibit the DNA-binding ability of this factor (Segil et al., 1991). This serine is situated in a KRTSIE motif, which is a potential site for phosphorylation by a cAMP- or cGMP-dependent protein kinase.
The mutations in m12 (S107D) and m13 (S107E) imitate phosphoserines (Maciejewski et al., 1995). In agreement with previous observations, these mutants are unable to bind POU binding sites as both monomers and dimers (Figure 3). The bulky and negatively charged aspartic and glutamic acids get close to the negatively charged DNA and inhibit contact between the amino acid and the phosphate backbone. This implies that the S107-phosphorylated Oct1 cannot bind the elements that it bound in the unphosphorylated state.
OBF1 overcomes the steric clash of the phosphoserine mimic with the DNA. When the cofactor is added to the binding reaction, it mildly rescues the phosphorylation-mimicked POU1 mutant monomer on Igκ. Binding is more enhanced on the PORE and PORED. This difference correlates with the fact that OBF1 stabilizes the dimer more than the monomer (Figure 5) (Luo and Roeder, 1995; Strubin et al., 1995). However, on P+1 the phosphorylation-mimicked POU1 dimer cannot be rescued by OBF1. Thus, phosphorylation of S107 could inhibit transcription of a gene regulated by a P+1-type element but not one regulated by a PORE-type sequence. By this distinction, genes could be differentially regulated by the same transcription factor dimer depending on the presence or absence of a coactivator, the phosphorylation state of the transcription factor and the DNA sequence of the regulatory element.
There are multiple events that trigger kinases and phosphatases, changing the expression patterns of various genes, such as cAMP elevation, membrane depolarization/calcium influx and growth factor reception. More specifically, cAMP levels play an important role in the costimulation of lymphoid cells, which is critical for an appropriate immune response. In T cells, elevation of cAMP levels is inhibitory. CD28 costimulation induces expression of a cAMP phosphodiesterase, leading to reduced cAMP levels and thus stimulation of T cells (reviewed by Frauwirth and Thompson, 2002). Similarly, cAMP could result in phosphorylated Oct1 in B cells. Owing to its phosphorylation, the transcription factor might not be able to activate all genes required to stimulate the cell (e.g. OPN). Upon costimulation mediated by a receptor resulting in a reduction of cAMP, Oct1 could be dephosphorylated and regulate a wider variety of genes. Incidentally, serine 107, which is phosphorylated in vivo, is situated within a KRTSIE motif, which is a potential site for phosphorylation by a cAMP-dependent protein kinase.
Novel POU1 dimer configuration upon interaction with OBF1
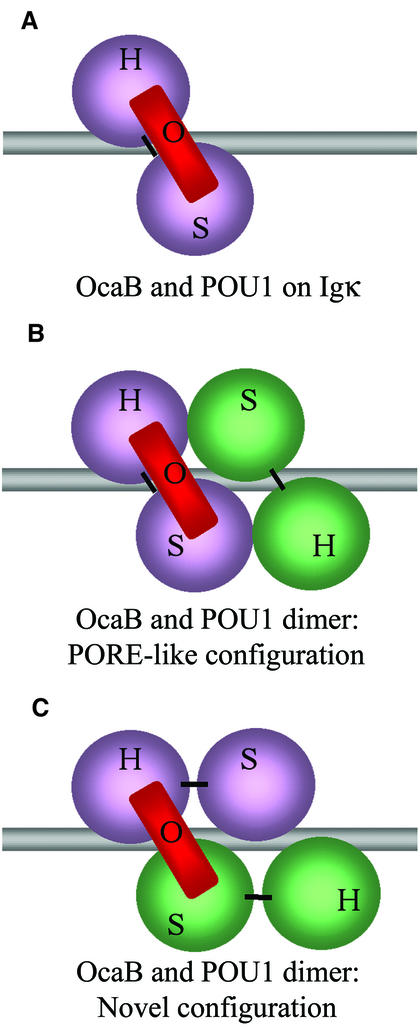
Our data show that OBF1 compensates for PORE interface mutations. We also found that the cofactor stabilizes the POU1 dimer, locking it onto the DNA. Based on our observations, we speculate that OBF1 may induce the Oct1 dimer to adopt yet another arrangement than that proposed for the PORE- and MORE-type binding (Botquin et al., 1998; Tomilin et al., 2000; Reményi et al., 2001). If the dimer binds as proposed for the PORE (Figure 6B), why does OBF1 have such a strong effect on two POU1 molecules, clamping only one of them to the DNA? How does this help the second molecule bind to the first and the DNA, especially if these molecules are mutated in the PORE dimer interface and bind to P+1?
Fig. 6. Model of POU1–OBF1 complexes bound to specific DNA elements in different configurations. (A) POU1 and OBF1 binding the octamer motif as described previously (Chasman et al., 1999). (B) Proposed POU1 dimer configuration (Botquin et al., 1998) with OBF1 binding to the POU1 molecule at the octamer half-site (Reményi et al., 2001). (C) POU1 dimer as in (B), but with a different linker connectivity. Here, OBF1 binds to the POUS domain of one POU1 molecule and to the POUH of another on the octamer half-site, with the individual subdomains arranged as in the crystal structure described previously (Reményi et al., 2001).
We propose two possibilities for the novel configuration. One may be a scenario in which the interaction with OBF1 slightly modifies the overall configuration of the POU1 dimer from that of the PORE-type dimer. In this case, the binding of OBF1 would result in a minor shuffle of the POU1 molecules on the DNA, so that they interact differently from the PORE-type dimer in order to allow for all unfavorable binding conditions discussed above. This would appear as in Figure 6B, but would show differences from the PORE-like dimer in a more detailed illustration. An alternative scenario would be that the POU1 subdomains POUS and POUH are bound to the DNA sequence motif as observed for the PORE-type dimer (Figure 6B) (Botquin et al., 1998; Reményi et al., 2001).
In the PORE-type dimer, the two subdomains of one molecule are bound to one half-site, whereas we propose that, with OBF1 binding, the two subdomains belonging to one molecule bind parallel to the DNA strand (Figure 6C). Thus, OBF1 would bind to the POUH and POUS from two different POU1 molecules, leading to both POU1 molecules being clamped to the DNA. This model could provide a rationale as to why POU1 is able to bind P+1 in the presence of OBF1 but not in its absence, and how the cofactor overcomes mutations in the protein–protein and protein–DNA interfaces (Figures 3B and 4).
Furthermore, the fact that OBF1 reduces the POU1 dimer off-rate from the PORED so severely (Figure 5) supports a novel arrangement whose interaction between the different molecules is stronger than that of the PORE-type dimer. Moreover, this experiment does not show any PO intermediate complex forming as time progresses. A PO intermediate would have been supportive of a PORE-type configuration, with both linkers being perpendicular to the DNA axis (Figure 6B). If OBF1 had clamped the POU1 dimer in the PORE configuration, the POU1 molecule associated with OBF1 might have remained bound to the DNA even after the first POU1 had dissociated. In addition, the novel configuration supports the idea of the clamping effect, which is much more pronounced for the POU1 dimer (Figure 5) than for the monomer (Luo and Roeder, 1995; Strubin et al., 1995).
The flexible nature of the linker connecting the POU subdomains argues against resolving this issue by a structural approach, since the linker region has remained invisible in all POU–DNA crystal structures solved so far. It might still be possible to elucidate which of the two arrangements is true by determining the crystal structure of the POU1 dimer with OBF1 on the PORE. If the POU1 molecules bind as in the PORE-type dimer, the novel dimer configuration can be assumed in order to accommodate the stabilizing impact of OBF1 on the dimer. If the subdomains cannot be superimposed on the coordinates of the PORE-type dimer, the induced-fit model would be more likely.
These results change our view on differential regulation and its fine tuning via the various POU dimers in the context of coactivators. Most importantly, our results highlight the limitations of a mutational in vitro analysis when it comes to applying the results to biological questions in cells or even living organisms. Prior to this study, we assumed that introducing specific dimer mutations into an endogenous POU gene might have exerted a strong phenotype due to the total loss of dimerization in vivo. Now, we might anticipate a more subtle or even absent phenotype. However, only the introduction of PORE and MORE interface mutations into endogenous POU genes will provide an idea of whether associated factors that facilitate dimerization in vitro may also help to maintain the full regulatory program in vivo.
Materials and methods
Site-directed mutagenesis, protein purification and protein expression
POU1 and mutants thereof were prepared as described previously (Reményi et al., 2001). OBF1 was chemically synthesized by Sigma and was identical to the peptide used for crystallization in earlier work (Chasman et al., 1999).
EMSA
Approximately 30 ng of POU1 (wild-type or mutant) protein were incubated with a radiolabeled oligonucleotide in the presence or absence of 10 ng of chemically synthesized OBF1 polypeptide (Chasman et al., 1999) in the following protein–DNA binding buffer: 25 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM EDTA, 0.04% Triton-X, 10% glycerol.
Igκ: 5′-ctgactcctgccttcagggtATGCAAATtattaagtctcgag 3′; other: 5′-ctgaaagttaaaatcac-X-ggaaaagcaag 3′, where X stands for the sequences provided in Figure 4A. For P+1, X is ATTTGATAATGCAAAT.
For the off-rate experiment (Figure 5), a 500-fold excess of unlabeled oligonucleotide was added to the reaction. Aliquots of the reaction mixture were loaded onto the gel at the time intervals indicated.
Cotransfection assays
Transient transfections into 293 cells (200 ng of reporter plasmid, 100 ng of human β-actin lacZ plasmid as an internal control, 700 ng of OBF1 expression vector and 1 ng of Oct1 expression vector when appropriate) and assays were performed as described previously (Tomilin et al., 2000).
BJA-B cells were grown exponentially in suspension with gentle stirring in RPMI 1640 medium supplemented with 10% fetal calf serum and standard amounts of l-glutamine, penicillin/streptomycin and non-essential amino acids. Then, 8 × 105 cells were electroporated in 0.9 ml of phosphate-buffered saline (400 V, 500 µF; Bio-Rad 0.4 cm cuvettes; GenePulser) with 10 µg of reporter plasmid and 1 µg of human β-actin lacZ plasmid. Cells were harvested after 24–36 h and lysed and assayed as described for the 293 cells.
RNA isolation and northern blotting
RNA of splenocytes was prepared as described previously (Schubart et al., 2001). A 1 kb HindIII fragment from the 2AR plasmid (kindly provided by D.Denhardt, Rutgers University, Piscataway, NJ) (HindIII–HindIII OPN probe fragment position +157 to +1144 of the OPN cDNA) containing mouse osteopontin cDNA was used as a probe.
Acknowledgments
Acknowledgements
We would like to thank Winship Herr for the pCG-Oct1 plasmid and David Denhardt for the 2AR plasmid. K.L. is a student of the Open University, UK. This work was supported by HFSP grant RGP 62/2002 to H.R.S., P.M. and M.W. Additional support was provided by the EMBL, the Novartis Research Foundation, the Marion Dilley and David George Jones Funds and the Commonwealth and General Assembly of Pennsylvania.
References
- Babb R., Cleary,M.A. and Herr,W. (1997) OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol. Cell. Biol., 17, 7295–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Y., Rice,D., Grosschedl,R. and Baltimore,D. (1984) Two regulatory elements for immunoglobulin κ light chain gene expression. Proc. Natl Acad. Sci. USA, 81, 7041–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botquin V., Hess,H., Fuhrmann,G., Anastassiadis,C., Gross,M.K., Vriend,G. and Schöler,H.R. (1998) New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev., 12, 2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepek K.L., Chasman,D.I. and Sharp,P.A. (1996) Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev., 10, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Chasman D., Cepek,K., Sharp,P.A. and Pabo,C.O. (1999) Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein–DNA interface. Genes Dev., 13, 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D.T., Butler,W.T., Chambers,A.F. and Senger D.R. (eds) (1995) Osteopontin: Role in Cell Signaling and Adhesion. New York Academy of Sciences, New York, NY.
- Frauwirth K.A. and Thompson,C.B. (2002) Activation and inhibition of lymphocytes by costimulation. J. Clin. Invest., 109, 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M., Knoepfel,L., Georgiev,O., Schaffner,W. and Hovens,C.M. (1995) A B-cell coactivator of octamer-binding transcription factors. Nature, 373, 360–362. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Georgiev,O., van Leeuwen,H., van der Vliet,P. and Schaffner,W. (1996) The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J., 15, 2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Jacobson E.M., Li,P., Leon-del-Rio,A., Rosenfeld,M.G. and Aggarwal,A.K. (1997) Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev., 11, 198–212. [DOI] [PubMed] [Google Scholar]
- Kim U., Qin,X.F., Gong,S., Stevens,S., Luo,Y., Nussenzweig,M. and Roeder,R.G. (1996) The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature, 383, 542–547. [DOI] [PubMed] [Google Scholar]
- Klemm J.D., Rould,M.A., Aurora,R., Herr,W. and Pabo,C.O. (1994) Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell, 77, 21–32. [DOI] [PubMed] [Google Scholar]
- Luo Y. and Roeder,R.G. (1995) Cloning, functional characterization and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol., 15, 4115–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Fujii,H., Gerster,T. and Roeder,R.G. (1992) A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell, 71, 231–241. [DOI] [PubMed] [Google Scholar]
- Maciejewski P.M., Peterson,F.C. Anderson,P.J. and Brooks,C.L. (1995) Mutation of serine 90 to glutamic acid mimics phosphorylation of bovine prolactin. J. Biol. Chem., 270, 27661–27665. [DOI] [PubMed] [Google Scholar]
- Marusic A., Grcevic,D., Katavic,V., Kovacic,N., Lukic,I.K., Kalajzic,I. and Lorenzo,J.A. (2000) Role of B lymphocytes in new bone formation. Lab. Invest., 80, 1761–1774. [DOI] [PubMed] [Google Scholar]
- Nielsen P.J., Georgiev,O., Lorenz,B. and Schaffner,W. (1996) B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol., 26, 3214–3218. [DOI] [PubMed] [Google Scholar]
- Nishimoto M., Fukushima,A., Okuda,A. and Muramatsu,M. (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol., 19, 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan A. and Berman,J.S. (2000) Osteopontin: a key cytokine in cell-mediated and granulomatous inflammation. Int. J. Exp. Pathol., 81, 373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reményi A., Tomilin,A., Pohl,E., Lins,K., Philippsen,A., Reinbold,R., Schöler,H.R. and Wilmanns,M. (2001) Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell, 8, 569–580. [DOI] [PubMed] [Google Scholar]
- Rhee J.M., Gruber,C.A., Brodie,T.B., Trieu,M. and Turner,E.E. (1998) Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem., 273, 34196–34205. [DOI] [PubMed] [Google Scholar]
- Roberts S.B., Segil,N. and Heintz,N. (1991) Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science, 253, 1022–1026. [DOI] [PubMed] [Google Scholar]
- Ryan A.K. and Rosenfeld,M.G. (1997) POU domain family values: flexibility, partnerships and developmental codes. Genes Dev., 11, 1207–1225. [DOI] [PubMed] [Google Scholar]
- Sauter P. and Matthias,P. (1998) Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol., 18, 7397–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H.R., Balling,R., Hatzopoulos,A.K., Suzuki,N. and Gruss,P. (1989) Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J., 8, 2551–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart D.B., Rolink,A., Kosco-Vilbois,M.H., Botteri,F. and Matthias,P. (1996) B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature, 383, 538–542. [DOI] [PubMed] [Google Scholar]
- Schubart K., Massa,S., Schubart,D., Corcoran,L.M., Rolink,A.G. and Matthias,P. (2001) B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat. Immunol., 2, 69–74. [DOI] [PubMed] [Google Scholar]
- Scully K.M. et al. (2000) Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science, 290, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Segil N., Roberts,S.B. and Heintz,N. (1991) Cell-cycle-regulated phosphorylation of the transcription factor Oct-1. Cold Spring Harb. Symp. Quant. Biol., 56, 285–292. [DOI] [PubMed] [Google Scholar]
- Staudt L.M., Singh,H., Sen,R., Wirth,T., Sharp,P.A. and Baltimore,D. (1986) A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature, 323, 640–643. [DOI] [PubMed] [Google Scholar]
- Strubin M., Newell,J.W. and Matthias,P. (1995) OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell, 80, 497–506. [DOI] [PubMed] [Google Scholar]
- Tomilin A., Reményi,A., Lins,K., Bak,H., Leidel,S., Vriend,G., Wilmanns,M. and Schöler,H.R. (2000) Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell, 103, 853–864. [DOI] [PubMed] [Google Scholar]
- Verrijzer C.P., Alkema,M.J., van Weperen,W.W., Van Leeuwen,H.C., Strating,M.J. and van der Vliet,P.C. (1992) The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J., 11, 4993–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.F. and Cantor,H. (1996) The immunology of Eta-1/osteopontin. Cytokine Growth Factor Rev., 7, 241–248. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davis,R.J. (2000) Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci., 57, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Corbi,N., Basilico,C. and Dailey,L. (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev., 9, 2635–2645. [DOI] [PubMed] [Google Scholar]