Abstract
Intracellular lipid traffic is mediated both by membrane vesicles and by a number of non-vesicular pathways facilitated by cytoplasmic lipid binding proteins. For these proteins to act effectively they must be targeted accurately to specific membranes. Here we identify a novel short conserved determinant called the FFAT motif that is shared by several seemingly unrelated lipid binding proteins and is also found in Opi1p, a transcriptional regulator of phospholipid synthesis in yeast. FFAT motifs act as membrane- targeting determinants by their direct interaction with homologues of VAMP-associated protein (VAP), a conserved endoplasmic reticulum (ER) protein. In budding yeast, all four proteins with FFAT motifs interact with Scs2p, a homologue of VAP, to target the ER to some extent. The precise intracellular distribution of each of these proteins depends on the integration of the FFAT–Scs2p interaction with other targeting determinants, and the interaction is functionally significant. We conclude that binding to a VAP homologue is a common mechanism by which proteins with FFAT motifs, most of which are involved in lipid metabolism, target ER membranes.
Keywords: FFAT motif/lipid traffic/membrane contact sites/oxysterol binding protein (OSBP)/peripheral membrane proteins
Introduction
A major role of intracellular lipid binding proteins is to move hydrophobic lipids across the aqueous environment of the cytoplasm independent of vesicular membrane traffic (Wirtz, 1991, 1997). The need to interact with more than one membrane compartment may explain why most lipid binding proteins are cytoplasmic, with peripherally associated membrane pools (Ridgway et al., 1992; Aikawa et al., 1999), often at multiple intracellular sites (Aikawa et al., 1999; Gallegos et al., 2001; Wyles et al., 2002). The endoplasmic reticulum (ER) is a pivotal organelle in lipid traffic since it is the site of synthesis of most lipids (Baumann and Walz, 2001). In addition, the ER is one organelle for which non-vesicular lipid traffic has been demonstrated biochemically (Vance, 1990; Voelker, 2000). However, only a small number of lipid binding proteins have been shown to target the ER, including a family of phosphatidylinositol transfer proteins (PITPs) related to retinal degeneration type B protein in Drosophila (rdgB) (Vihtelic et al., 1993; Aikawa et al., 1999; Litvak et al., 2002), and also oxysterol binding protein (OSBP) (Wyles et al., 2002).
OSBP was identified by its ability to bind with high affinity to 25-hydroxycholesterol (25-HC) (Levanon et al., 1990). This oxysterol, a hydrophilic derivative of cholesterol produced by a specific sterol hydroxylase in the ER (Lund et al., 1998), is a highly potent inhibitor of sterol regulatory element binding protein (SREBP), a transcription factor anchored to the ER (Taylor et al., 1984; Wang et al., 1994). However, 25-HC does not interact with either SREBP or its binding partner (Hua et al., 1996; Brown et al., 2002). While the effects of 25-HC on SREBP might be mediated by OSBP, or any of the other 11 members of the family of OSBP-related proteins (ORPs) (Jaworski et al., 2001; Lehto et al., 2001), no molecular mechanism linking OSBP or the ORPs to SREBP has yet been elucidated.
OSBP predominantly targets the trans-Golgi network (TGN), mediated by its pleckstrin homology (PH) domain (Ridgway et al., 1992; Levine and Munro, 2002). However, it has recently been shown that OSBP can also target the ER by binding the integral membrane protein VAP (Skehel et al., 2000; Wyles et al., 2002). In addition, Osh1p and Osh2p, two yeast homologues of OSBP, are among eight proteins found to coprecipitate with Scs2p, a homologue of VAP in Saccharomyces cerevisiae also on the ER (Kagiwada et al., 1998; Gavin et al., 2002). VAP was originally cloned by its ability to bind vesicle associated membrane protein (VAMP; VAP stands for VAMP-associated protein), and VAP has been reported to interact with other proteins involved in vesicular fusion (Skehel et al., 1995; Weir et al., 2001). Despite many suggested roles for VAP and Scs2p, mainly in relation to vesicle trafficking, their functions remain unknown (Nikawa et al., 1995; Kagiwada et al., 1998; Soussan et al., 1999; Pennetta et al., 2002).
Here we describe a short conserved peptide motif consisting of two phenylalanines (FF) in an acidic tract (the FFAT motif) that targets proteins to the ER by binding directly to VAP and its homologues. The motif is found in a diverse range of lipid binding proteins in humans: OSBP and seven other ORPs, all three human homologues of rdgB and Goodpasture’s antigen binding protein (GPBP). We studied the in vivo role of FFAT motifs in protein targeting for the four yeast proteins with the motif: Opi1p, a transcriptional regulator specific to phospholipid synthesis, and three homologues of OSBP. In this physiological context, all four proteins showed a degree of Scs2p-dependent targeting which was integrated with other targeting signals to produce an overall distribution specific for each protein. Lipid binding proteins with FFAT motifs may use their common interaction with VAP to gain access to the ER membrane, either for lipid traffic or for lipid sensing.
Results
Yeast OSBP homologues contain ER targeting determinants
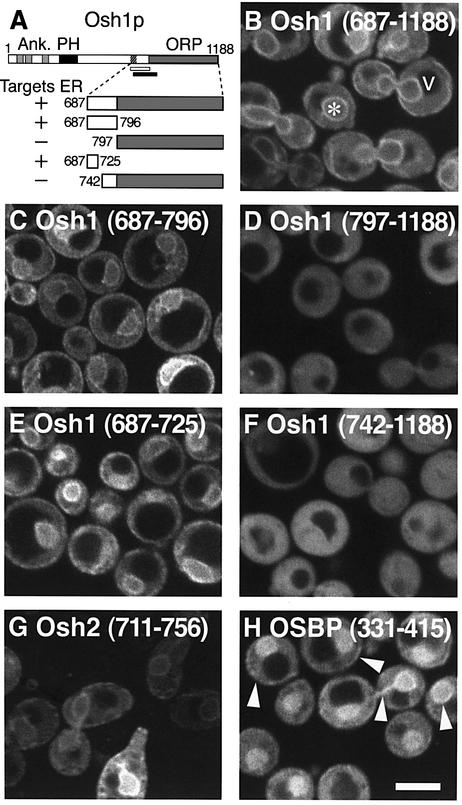
We have previously identified a region at the C-terminus of Osh1p, consisting of its ORP domain together with 110 amino acids upstream, that is able to complement the phenotype of the whole protein in a Δosh1 strain (Levine and Munro, 2001). Since intracellular targeting determines OSBP homologue function (Levine and Munro, 2001), we examined the targeting of this region of Osh1p using a GFP fusion in live yeast (constructs summarized in Figure 1A). This fusion protein localized both to the nuclear envelope and in peripheral patches, characteristic of the yeast ER (Figure 1B) (Pichler et al., 2001; Voeltz et al., 2002). This pattern is significant because a region of OSBP was recently shown to bind the human ER protein VAP (Wyles et al., 2002).
Fig. 1. Dissection of an ER targeting determinant in OSBP homologues. (A) Diagram of the domain structure of Osh1p and summary of the five segments expressed in (B)–(F) and their ER targeting. Previously identified domains in Osh1p are three ankyrin repeats (Ank.), a PH domain and an ORP domain. (B–F) Confocal micrographs of live wild-type yeast (RS453B) expressing the indicated residues of Osh1p fused to GFP from plasmids pTL376–380, respectively. As shown in (B), the ER in yeast consists of the nuclear envelope (asterisk) and discontinuous patches in the cell periphery. In addition, these constructs show some diffuse cytoplasmic fluorescence which is excluded from the vacuole (v). ER targeting is seen in (B), (C) and (E), and correlates with the inclusion of residues 687–725 of Osh1p [hatched area in (A)]. (G and H) Homologous regions from Osh2p and OSBP, respectively, were expressed as indicated from plasmids pTL381 and pTL382. Both constructs targeted the ER. At higher levels of expression (H), membrane targeting (arrowheads) was accompanied by increasing diffuse fluorescence, indicating saturation of a membrane binding site. The region of OSBP in (H) is homologous to the portion of Osh1p identified by the open bar in (A). For comparison, the solid bar in (A) marks the boundaries of the VAP binding site in OSBP defined by Ridgway and co-workers (Wyles et al., 2002). Scale bar: 5 µm.
Analysis of the requirements for ER localization showed that the upstream region of the Osh1 construct (residues 687–796) was necessary and sufficient for ER targeting (Figure 1C), while the ORP domain alone (residues 797–1188) was diffusely cytoplasmic (Figure 1D). Further dissection indicated that just 39 residues (687–725) targeted a reporter construct to the ER (Figure 1E), while their omission led to a diffuse pattern (Figure 1F). To investigate whether this targeting was conserved in other OSBP homologues, we expressed fusion proteins containing homologous regions of either Osh2p or OSBP, and both these constructs targeted the ER (Figure 1G and H). These results imply that there is a highly conserved ER targeting determinant in this segment of OSBP homologues.
Osh1 and OSBP share a motif found in other lipid binding proteins
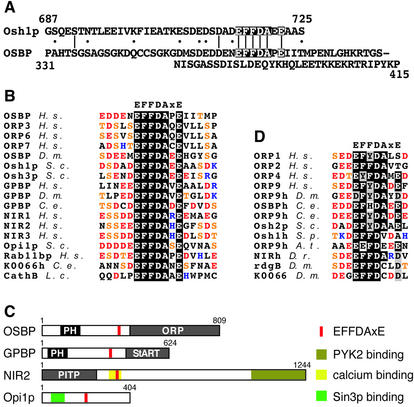
The finding of ER targeting by both OSBP and Osh1p led us to look for a shared targeting determinant. Alignment of the relevant segments of primary sequence revealed a single area of similarity, EFFDAxE, immediately upstream of which is an acidic tract (Figure 2A). A search of protein sequence databases identified the sequence EFFDAxE in a total of 17 distinct eukaryotic proteins (Figure 2B), of which 14 are directly implicated in lipid binding or lipid sensing. Apart from OSBP and Osh1p, there are five other OSBP homologues from different species including three ORPs and Osh3p. EFFDAxE is also found in GPBP and its homologues, and in both human homologues of rdgB (NIR2 and NIR3) as well as in NIR1, a homologue of rdgB that lacks the PITP domain (Lev et al., 1999). Interestingly, the same motif occurs in one other S.cerevisiae protein, the phospholipid regulator Opi1p (Figure 2B).
Fig. 2. Alignment of a conserved motif identified in OSBP homologues. (A) Alignment of 39 residues from Osh1p and 85 residues from OSBP that target the ER (single-letter code). Identities and homologies are indicated by bars and dots, respectively. Six identical residues in a run of seven (EFFDAxE) are shaded black. (B) Seventeen eukaryotic proteins contain EFFDAxE as identified by a PROSITE search (http://ca.expasy.org/tools/scanprosite/). Non-conserved residues are coloured to highlight glutamic acid/aspartic acid (red), serine/threonine (orange) and basic residues (blue). The proteins are homologues of OSBP (× 7), GPBP homologues (× 3), three human homologues of rdgB (NIR1–3), Opi1p, rab11 binding protein, a worm homologue of a protein of unknown function (KIAA0066) and a cathepsin B protease from Leishmania. While the latter is luminal, all the other proteins are cytoplasmic and these EFFDAxE sequences all have acidic flanking regions. (C) Position of EFFDAxE in four representative proteins: OSBP, GPBP, NIR2 and Opi1p. Domains are either indicated in the key or as in Figure 1A. Lipid binding domains of three different types are present: OSBP, ORP domain; GPBP, StART domain; NIR2, PITP domain (Ridgway et al., 1992; Vihtelic et al., 1993; Ponting and Aravind, 1999). N-terminal PH domains are seen in 11 out of 12 ORPs (Jaworski et al., 2001; Lehto et al., 2001), and also in GPBP. While NIR3 is highly similar to NIR2, the PITP domain is missing in NIR1. The EFFDAxE of NIR proteins occurs in a region previously shown to bind calcium (see Discussion). NIR proteins also contain a domain that interacts with the tyrosine kinase PYK2 (Lev et al., 1999). A region near the N-terminus of Opi1p binds to Sin3p, which is part of a histone acetylase complex (Wagner et al., 2001). (D) Sequences closely related to EFFDAxE found in 13 homologues of proteins in (B). Ten OSBP homologues, two rdgB homologues (including rdgB itself) and a homologue of KIAA0066 all contain sequences closely related to EFFDAxE, with conservative and non-conservative changes indicated by grey and white shading, respectively. The flanks are all highly acidic [coloured as in (B)]. Abbreviations for species of origin are as follows: H.s., Homo sapiens; D.m., Drosophila melanogaster; C.e., Caenorhabditis elegans; S.c., S.cerevisiae; S.p., S.pombe; A.t., Arabidopsis thalania; D.r., Danio rerio; L.c., Leishmania chagasi. Where there are multiple entries of near-identical homologues in different mammals, only the human protein has been included.
A conserved feature in all 16 cytoplasmic occurrences of EFFDAxE is the predominance of acids and serines/threonines in adjacent residues, with a depletion of basic residues (Figure 2B). These effects extend 10 residues upstream and four residues downstream. The domain structures of proteins with EFFDAxE shows that, while OSBP and GPBP are similar in form, OSBP, NIR2 and Opi1p are structurally unrelated (Figure 2C). None of the EFFDAxE sequences occur in motifs previously recognized in the PROSITE database. The finding of a novel short motif in a wide variety of unrelated lipid binding proteins, as well as in the lipid regulator Opi1p, is highly suggestive of involvement of the motif in a conserved aspect of lipid metabolism.
EFFDAxE targets the ER by interacting with VAP homologues
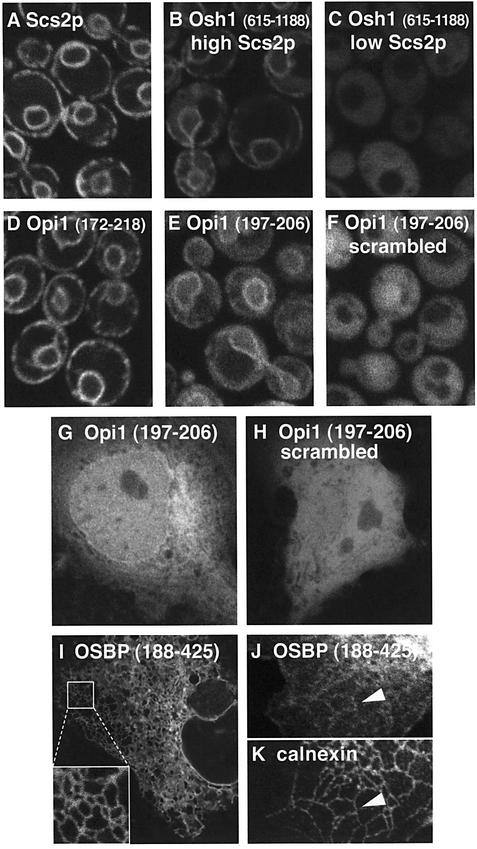
A recent study of 230 multiprotein complexes in S.cerevisiae found that Scs2p, a yeast homologue of VAP, coprecipitates with eight other proteins: Fks1p, Num1p, Opi1p, Osh1p, Osh2p, Rpn10p, Stt4p and YGR086Cp (Gavin et al., 2002). Therefore, in addition to the interaction of OSBP with VAP, two out of the three yeast proteins containing EFFDAxE are potential interactors with Scs2p, a yeast homologue of VAP. This led us to investigate whether the motif binds to Scs2p, initially in live cells. First, we confirmed that Scs2p localizes to the ER (Kagiwada et al., 1998), using a GFP-tagged Scs2p construct, which targeted the nuclear envelope, and large peripheral patches (Figure 3A), characteristic of the yeast ER.
Fig. 3. The effects of Scs2p on targeting by Osh1 and Opi1 constructs. (A) GFP-tagged Scs2p expressed from pTL201 in strain TLY252. Unlike constructs containing FFAT motifs, GFP–Scs2p targets the ER without cytoplasmic background fluorescence. (B and C) The C-terminus of Osh1, including the ER targeting determinant and the ORP domain, was expressed as a GFP fusion (plasmid pTL375) in strain TLY251, in which Scs2p levels are regulatable by carbon source (see Materials and methods). (B) High levels of Scs2p mediated tight localization of the Osh1 construct to ER membranes (cf. Figure 1). (C) In contrast, repression of Scs2p production made the same construct diffuse. (D and E) Forty-seven and 10 amino acid segments from Opi1p tagged with GFP were expressed from plasmids pTL383 and pTL384, respectively, in strain TLY251 expressing high levels of Scs2p. Both constructs targeted ER membranes, with the longer construct showing stronger membrane targeting. (F) Plasmid pTL385, which expresses the same 10 residues from Opi1p as pTL384 [see (E)], but in a scrambled order, was expressed in TLY251. Loss of native order abolished ER targeting. (G and H) Plasmids pCL384 and pCL385, expressing the same fusion proteins as (E) and (F) under the CMV promoter, were transfected into COS-7 cells and visualized after 48 h. In both cases there was diffuse cytoplasmic and intranuclear fluorescence. In addition the short Opi1-derived sequence, but not its scrambled counterpart, targeted the nuclear envelope. (I–K) Plasmid pCL386 expressing GFP-tagged OSBP (188–425) was expressed in COS-7 cells. A prominent reticular pattern was seen in the cytoplasm of live cells (I), which was partly preserved after fixation (J), colocalizing with the luminal ER protein calnexin [(K), arrowheads].
We next tested whether ER localization of Osh1p constructs is mediated by Scs2p by replacing the SCS2 promoter with the inducible GAL1 promoter. With high levels of Scs2p, the ORP domain and flanking sequence of Osh1p localized to the ER with less diffuse cytoplasmic staining than in a wild-type strain (Figure 3B, cf. Figure 1B). In the same strain grown to inhibit Scs2p production, the construct was entirely delocalized (Figure 3C), as was the 85 residue segment of OSBP (data not shown). This indicates that Scs2p can mediate ER targeting of OSBP homologues in vivo.
To test whether the same motif is functional in proteins beyond the OSBP family, we then examined localization by the sequence identified in Opi1p. A GFP fusion to 47 amino acids including the motif was tightly localized to the ER in cells overexpressing Scs2p (Figure 3D). To test the minimal requirements for targeting in vivo, we then expressed a GFP fusion protein containing 10 amino acids (DDEEFFDASE) from Opi1p. This sequence targeted a reporter fusion protein to the ER in cells with high Scs2p (Figure 3E), but did not localize either in cells with repressed Scs2p production (data not shown) or when the order of the Opi1-derived residues was scrambled (Figure 3F), excluding the possibility that a strong negative charge is responsible for targeting of this region. Therefore the motif common to OSBP homologues and Opi1p is an Scs2p-dependent ER-targeting domain.
Since most of the proteins identified with this motif are mammalian, we determined whether targeting occurs in mammalian fibroblasts. The minimal 10 amino acid motif from Opi1p showed weak targeting predominantly to the nuclear envelope (Figure 3G). As a control, scrambling the order of these residues abrogated targeting (Figure 3H). Although the short Opi1p construct did show weak reticular localization in the cytoplasm, this was faint and entirely lost during fixation. In order to visualize such targeting more clearly, we expressed segments of OSBP containing EFFDAxE in the same cells. Short segments of OSBP (e.g. 41 amino acids) gave similar results to the 10 residues of Opi1p (data not shown). In contrast, much longer constructs showed a very prominent reticular pattern in vivo (Figure 3I) which colocalized with an ER marker after fixation (Figure 3J and K). The stronger membrane targeting of this construct was completely ablated by mutation of the FFAT motif (EFFDA→ EAFDA) and may result from the inclusion of a putative dimerization domain of OSBP (residues 260–280) (Ridgway et al., 1992), as constructs missing this segment only showed weak nuclear envelope targeting (data not shown).
Since the most striking features of the motif we have identified are its occurrence in a diverse range of proteins involved in lipid metabolism and the presence of two phenylalanines (FF) in an acidic tract, we propose the name FFAT motif for the sequence EFFDAxE, and closely related sequences, occurring in a locally acidic environment.
The FFAT motif directly binds VAP homologues in vitro
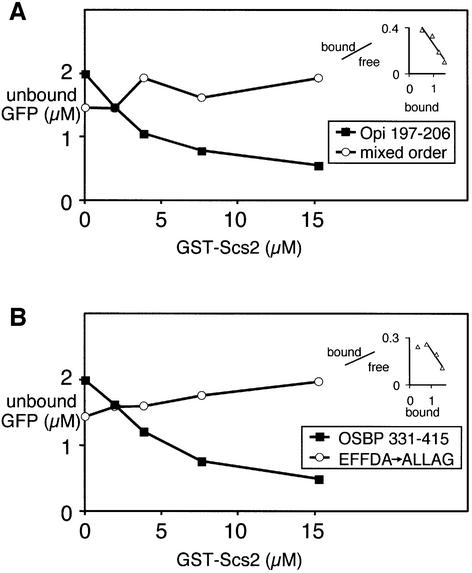
To determine whether the interaction of proteins with FFAT motifs and VAP homologues is direct, we carried out in vitro binding studies. A fusion protein containing GFP and the 10 residue minimal FFAT motif from Opi1p was purified from bacteria, as was a GST–Scs2 fusion protein with the transmembrane domain missing. The FFAT motif of Opi1p bound saturably to GST–Scs2 with a dissociation constant of 5 ± 1 µM (n = 3) (Figure 4A), and also to GST–VAP with similar affinity (data not shown). As a control, protein in which the Opi1 residues were scrambled showed no interaction. Therefore the FFAT–VAP interaction is direct and has an affinity comparable to other interactions that recruit peripheral membrane proteins (Lemmon et al., 1995).
Fig. 4. Direct binding of the FFAT motif of Opi1p to Scs2p. (A) Increasing amounts of GST–Scs2, from which the transmembrane domain is deleted, immobilized on glutathione–Sepharose were incubated with constant amounts of GFP fusion protein that contained the Opi1p FFAT motif or its scrambled counterpart. Bound GFP fusion protein was removed by centrifugation, and unbound GFP was assayed by fluorimetry of the supernatant. The native Opi1 FFAT motif showed saturable binding with an estimated Kd of 4.5 ± 1.4 µM (n = 3), while the scrambled sequence showed no interaction. (B) In vitro binding assay, as in (A), of GST–Scs2 with GFP fusion proteins containing an 85 amino acid segment of OSBP (331–415) and a mutant version of this construct in which the FFAT motif is mutated EFFDA→ALLAG. The OSBP segment bound the yeast VAP homologue with an estimated Kd of 8.9 ± 1.1 µM (n = 4), similar to the interaction of Scs2 and Opi1p FFAT motif, but the mutated sequence did not bind. Representative experiments are shown, with dissociation constants estimated by Scatchard plots (inset) from three and four independent experiments, respectively.
To generalize the finding that FFAT motifs bind VAP homologues, we examined the interaction of the OSBP FFAT with Scs2p. A GFP fusion protein containing 85 residues from OSBP bound with a dissociation constant of 9 ± 1 µM (n = 4), while substitution of five residues of the FFAT motif (EFFDA→ALLAG) completely inhibited binding (Figure 4B). These results indicate that FFAT motifs from divergent proteins bind similarly to VAP homologues, implying that the FFAT–VAP interaction is highly conserved.
FFAT motifs have a single invariant position
Sequences other than those fitting the strict definition of the FFAT motif might also bind VAP and its homologues. To determine the parameters for interaction with VAP more accurately, we studied the differences in FFAT motifs that are tolerated in vivo. First, we examined homologues of proteins with EFFDAxE to see whether they had related sequences. We identified 13 proteins with motifs similar to EFFDAxE (Figure 2D), one of which we had already found to target the ER (Figure 1G, Osh2p). Within each protein family, the position of the motif in the domain structure was very well conserved.
To define the requirements for binding by FFAT motifs further, we examined the effect of substituting individual residues (Table I). Alanine replacement of residues EFFD showed that position 1 tolerated change, but substitutions at positions 2, 3 or 4 disrupted localization. Replacement of the alanine at position 5 with proline had a partial effect. In addition, while substitution of either all the acidic residues in the N-terminal flanking region, or both residues at positions 6 and 7 did not affect targeting, removing both these acidic flanks prevented localization (Table I).
Table I. The effect of point mutations in FFAT motif on ER targeting.
| N-terminus | Core | C-terminus | Both flanks | |||||
|---|---|---|---|---|---|---|---|---|
| No. in FFAT | –3 to –1 | 1 | 2 | 3 | 4 | 5 | 6 + 7 | –3 to –1 and 6 + 7 |
| Plasmid pTL number | 397 | 391 | 392 | 393 | 394 | 395 | 396 | 398 |
| Wild-type residues | D–D–E | E | F | F | D | A | E–E | D–D–E … and … S–E |
| Mutated to | A–A–A | A | A | A | A | P | K–Q | A–A–A … and … R–N |
| Localized at ER | + | + | – | – | – | ± | + | – |
ER localization was scored in cells expressing plasmids pTL391–397 in cells with high levels of Scs2p (strain TLY251 grown in galactose).
Our experimental results agree with the indications gained from natural variants of the FFAT motif (Figure 2D). Conservative changes appear to be tolerated at positions 1–3, although just one of the Fs may be exchanged for a Y, and even non-conservative changes may be acceptable at positions 5 and 7. However, the aspartate (D) at position 4 appears invariant, as is the presence of flanking acidic residues, implying that these residues may be essential for FFAT motifs.
A role for Scs2p in the targeting of Osh proteins
A theme that emerges from previous studies of the intracellular localizations of OSBP and its homologues is that targeting is tightly regulated in the context of the whole protein. For example, OSBP and Osh proteins do not have obvious pools on the ER and are diffuse in the cytoplasm under some conditions, i.e. apparently untargeted even though they contain multiple targeting domains (Ridgway et al., 1992; Levine and Munro, 2001; Wyles et al., 2002). Therefore, we extended our previous studies of the localizations of Osh proteins in yeast to determine the role that Scs2p plays in the targeting of full-length proteins.
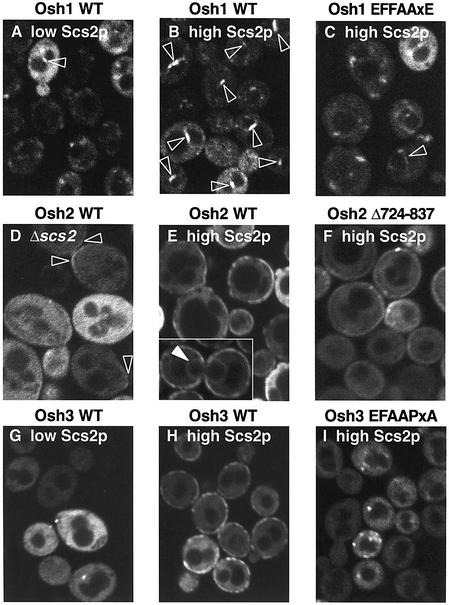
Osh1p has been shown to have a dual localization: Golgi membranes and the nucleus–vacuole junction (NVJ) (Levine and Munro, 2001). This small subcompartment of the ER is formed by the binding of one vacuolar protein to one nuclear envelope protein (Pan et al., 2000), and Osh1p is the only cytoplasmic protein so far identified at this site, with the ankyrin repeats at the N-terminus of Osh1p being necessary and sufficient for targeting to the NVJ (Levine and Munro, 2001). Localization of GFP–Osh1p was affected by reduction of Scs2p levels: Golgi targeting was maintained, but NVJs were targeted only in occasional cells (Figure 5A). Similar weak NVJ targeting was also seen in Δscs2 cells, indicating that it does not depend on residual Scs2p (data not shown), but rather implicating the ankyrin repeats in this targeting. In contrast, cells with high levels of Scs2p showed far more prominent targeting of GFP–Osh1p to the NVJ (Figure 5B). However, this was lost by introducing a point mutation in the FFAT motif (D719→A) (Figure 5C). Hence, targeting of Osh1p to the NVJ by its ankyrin repeats is enhanced by the interaction of its FFAT motif with Scs2p.
Fig. 5. Role of Scs2p in the localization of Osh proteins. GFP fusions to the N-terminus of full-length Osh proteins, either wild type (WT) or with the specified mutations, were expressed from CEN plasmids which vary in copy number between cells. (A–C) Osh1p (pTL310 and pTL316) expressed in TLY251. Localization to the nucleus–vacuole junction (NVJ), a single discoid structure between nucleus and vacuole, is indicated by arrowheads. (A) In cells with low Scs2p, Osh1p predominantly targeted punctae, characteristic of the yeast Golgi. (B) In cells with high Scs2p, NVJ targeting of wild-type Osh1p was prominent. (C) NVJ targeting was almost completely lost as a result of a point mutation in the FFAT motif D719→A. (D–F) Osh2p (pTL312 and pTL317) expressed in Δscs2 and inducible Scs2p yeast strains Y16119 and TLY251. The latter was grown to induce high levels of Scs2p. (D) Without Scs2p, wild-type Osh2p was largely diffuse but also showed occasional weak linear peripheral localization, particularly at incipient bud sites (arrowheads). (E) High levels of Scs2p restored the localization of wild-type Osh2p to its normal non-uniform pattern at the cell periphery, and in a small minority of cells led to weak localization to the nuclear envelope (filled arrowhead in inset). (F) Deletion of 114 residues including the FFAT motif led to weaker and more uniform peripheral targeting, similar to (D). (G–I) Osh3p (pTL313 and pTL318) expressed in TLY251 as indicated. (G) With low Scs2p, targeting of wild-type Osh3p was diffuse with occasional cytoplasmic punctae in highly expressing cells only. (H) High Scs2p levels led to strong non-uniform peripheral targeting of wild-type Osh3p. (I) In contrast, Osh3 with a mutated FFAT showed no peripheral localization in cells expressing high levels of Scs2p.
We have found previously that Osh2p is non-uniformly distributed around the cell periphery, enriched in small buds and near the neck region of mother cells (Levine and Munro, 2001). Here, we expressed GFP–Osh2p in a Δscs2 strain and found that its peripheral localization was now very faint indeed (Figure 5D). The same was seen in strain TLY251 grown to repress Scs2p expression (data not shown). By comparison, in cells with high levels of Scs2p, GFP–Osh2p showed a non-uniform peripheral distribution (Figure 5E), similar in nature to wild-type cells (Levine and Munro, 2001). In some cells, expression of high levels of Scs2 induced additional weak targeting of GFP–Osh2p to the nuclear envelope (Figure 5E inset). To investigate the role of the FFAT in targeting by Osh2p, we expressed a mutant version of Osh2p in which the entire region including the FFAT motif was deleted. Even in cells with high Scs2p levels, peripheral targeting was weaker and its distribution was now far more uniform around the entire cell periphery (Figure 5F). These results indicate that normal targeting of full-length Osh2p depends on Scs2p.
Osh3p has been found to be diffuse in the cytoplasm in wild-type cells (Levine and Munro, 2001), and this was not affected by reduction of Scs2p (Figure 5G). Overexpression of Scs2p induced strong patchy peripheral targeting of GFP–Osh3p, not seen in wild-type cells (Figure 5H). However, mutation of the FFAT motif prevented this targeting completely (Figure 5I). These results indicate that Osh3p targets the periphery in an Scs2p-dependent fashion similarly to Osh2p, although this targeting is weaker since it is less obvious under normal growth conditions.
By comparison, high levels of Scs2p had no effect either on ER morphology, as assessed by a general ER marker (Are2p), or on the prominence of the NVJ, as assessed by a specific NVJ marker (Tsc13p) (data not shown), indicating that the effect of Scs2p on Osh proteins is specific (Zweytick et al., 2000; Kohlwein et al., 2001). Therefore our results indicate that Scs2p plays a role in the targeting of all three Osh proteins with FFAT motifs and hence that this interaction is likely to be functionally important in vivo.
Targeting of the transcriptional regulator Opi1p to the ER
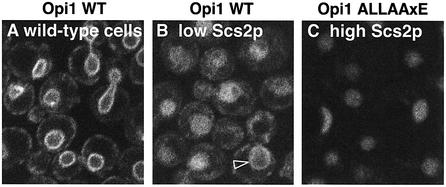
Although the function of OPI1 and its genetic interactions have been studied in detail (Greenberg and Lopes, 1996; Graves and Henry, 2000), and some of its interactions have been studied biochemically (Wagner et al., 2001), the protein has not been localized previously. Having established that the FFAT motif of Opi1p targets the ER, we investigated the localization of full-length Opi1p as a GFP fusion protein. The GFP–Opi1p construct rescued the phenotype of Δopi1 cells in the same manner as untagged Opi1p (data not shown) (see Materials and methods). Under normal growth conditions, GFP–Opi1p targeted the ER more on the nuclear envelope than in the periphery (Figure 6A). To investigate the role of Scs2p in targeting by GFP–Opi1p, this was expressed in cells with low Scs2p. Under these conditions, targeting was largely diffuse in both the cytoplasm and the nucleoplasm (Figure 6B), although some cells still showed slight localization to the nuclear envelope (Figure 6B, arrowhead). The weak membrane association of Opi1p that persists with low Scs2p was also seen in Δscs2 cells (data not shown). Mutation of the FFAT motif of Opi1 completely delocalized the protein from membranes, with the vast majority of the mutant protein being intranuclear (Figure 6C). These results confirm that Scs2p plays a significant part in the localization of Opi1p, acting via its FFAT motif.
Fig. 6. Role of Scs2p in Opi1p localization. Confocal images of cells expressing GFP-tagged Opi1p, either wild type or with a mutated FFAT motif (pTL211 and pTL219). (A) In wild-type cells (RS453B) under normal growth conditions, GFP–Opi1p was present at the nuclear envelope and to a lesser extent in peripheral patches. At high levels only, some cells contained bright punctae on the nuclear envelope (data not shown). (B) In strain TLY251, grown in dextrose to repress production of Scs2p, Opi1p was only weakly targeted to membranes (arrowhead) and showed a prominent diffuse intranuclear pool. (C) In the same strain grown in galactose, so that wild-type Opi1p localized to membranes as in (A), Opi1 with multiple mutations in its FFAT motif was almost entirely diffusely intranuclear.
Functional role for the FFAT motif in Osh1p
It has been shown previously by ourselves and others that deletion of OSH1 reduces the uptake of tryptophan at low temperatures (Jiang et al., 1994; Levine and Munro, 2001). Although the mechanism of this phenotype is unknown, it is also seen with a variety of defects in ergosterol metabolism and may be related to a defect in sterol sensing or traffic. We compared the ability of Osh1p, either wild type or with a point mutant (EFFDAxE→EFFAAxE), to rescue growth on low tryptophan. While wild-type Osh1p restored growth, the mutated protein clearly failed to do so (Figure 7). Since there are no growth phenotypes for Δosh2 or Δosh3 strains, we were unable to test the effect of similar mutations in the FFAT motifs of Osh2p and Osh3p. These results with Osh1p indicate that the interaction of a FFAT motif with a VAP homologue is functionally significant.
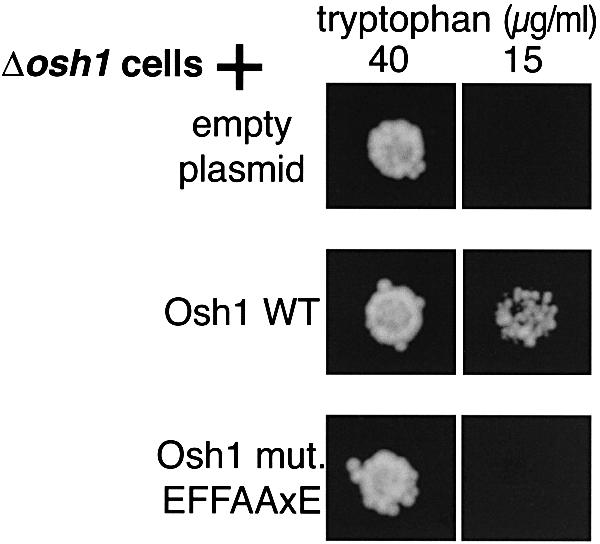
Fig. 7. Role of FFAT motif in OSH1 phenotype. Δosh1 cells (TLY221) were transformed with the indicated plasmids (pRS416, pTL314 and pTL315) and ∼100 cells were spotted onto plates containing either 40 µg/ml (normal levels) or 15 µg/ml (reduced levels) tryptophan and incubated at 25°C for 2 days. While wild-type Osh1p rescued growth on low tryptophan, Osh1 with a point mutation in the most conserved residue of its FFAT motif (D719→A) was unable to rescue growth. Similar results were obtained for 10 colonies of each genotype.
Discussion
Correct targeting of lipid binding proteins to donor and acceptor membranes allows specific lipid transfer while maintaining the unique lipid profiles of individual organelles (Schneiter et al., 1999). Here we describe the FFAT motif, a short conserved linear sequence that allows a large number of cytoplasmic lipid binding proteins (12 in human cells) to target the ER via VAP and its homologues. This work both identifies the precise means by which OSBP binds VAP (Wyles et al., 2002), and confirms earlier results from a proteomic study that found complexes between Scs2p and Opi1p, Osh1p and Osh2p (Gavin et al., 2002). None of the other five Scs2p interactors identified in that study have FFAT motifs.
The FFAT–VAP interaction may be involved in the traffic of a variety of different lipids from or to the ER. The lipid specificities of the seven homologues of OSBP that have FFAT motifs have not yet been established, but may include other oxysterols or even phospholipids (Moreira et al., 2001; Xu et al., 2001; Laitinen et al., 2002; Wang et al., 2002). Two other families of lipid binding proteins also have FFAT motifs. GPBP has a similar domain architecture to OSBP, although by homology its lipid binding domain is predicted to bind phospholipids (Ponting and Aravind, 1999). RdgB and its human homologues NIR2 and NIR3 have PITP domains and are known to target the ER, but the means of targeting is unknown (Vihtelic et al., 1993; Litvak et al., 2002). The only proteins in which FFAT motifs overlap with a previously described domain are rdgB/NIRs, since a calcium binding activity has been mapped to a 55 amino acid region including the FFAT motif, and has been proposed to be a function of the highly acidic 15 residues around the FFAT motif (Lev et al., 1999). In our in vitro binding assay, 100 µM calcium caused a 2-fold increase in the affinity of the Opi1p FFAT motif for Scs2p, indicating either that calcium binding has only a minor effect on the FFAT–VAP interaction or that it is not a general property of FFAT motifs.
Opi1p, a transcriptional repressor of phospholipid metabolism, is the sole member of a fourth class of proteins involved in lipid metabolism that has a FFAT motif. A genetic interaction between SCS2 and OPI1 has long been established (Nikawa et al., 1995; Greenberg and Lopes, 1996; Kagiwada et al., 1998), and a direct interaction between Scs2p and Opi1p was suggested by a recent proteomic study (Gavin et al., 2002). Here, we find that the Scs2p–Opi1p interaction is important for the intracellular targeting of Opi1p in vivo, in that Opi1p accumulates inside the nucleus in the absence of an interaction with Scs2p. This suggests that Scs2p may function to reduce the intranuclear activity of Opi1p, which might explain why deletion of SCS2 causes a phenotype associated with overactivity of Opi1p (Kagiwada et al., 1998). Our current work is to investigate the functional relationship between Scs2p and Opi1p. The finding that Opi1p targets Scs2p on the ER has interesting parallels, since other transcription factors regulating lipid metabolism are found in the ER, including SREBP in humans (Wang et al., 1994; Hoppe et al., 2000).
ER targeting by OSBP and its homologues is normally tightly regulated and occurs at such a low level as to be undetectable (Ridgway et al., 1992; Levine and Munro, 2001). However, using yeast as a model system, we have found a role for the FFAT motifs of OSBP homologues in vivo. Targeting of all three Osh proteins with FFAT motifs is affected by Scs2p. The effects of Scs2 are partial, suggesting that Scs2p is no more than partly responsible for normal targeting. The redistribution caused by high levels of Scs2p is also selective, in that all three Osh proteins are redistributed only to a subset of the sites where Scs2p is found (Figures 3A and 5). This implies that the interaction with Scs2p is integrated with other targeting signals. For Osh1p, the other targeting determinant is the ankyrin repeat region targeting the NVJ. For Osh2p and Osh3p, a second site is distributed uniformly around the entire cell periphery, possibly associated with the plasma membrane (data not shown). Thus, the interaction with Scs2p appears to restrict Osh proteins to regions of the ER adjacent to the other sites they target (Figure 5). In addition to the effect on targeting, we found that the FFAT motif of Osh1p is required for its ability to rescue the Δosh1 phenotype (Figure 7). This is an important confirmation of the functional significance of the interaction of FFAT motifs with a VAP homologue.
FFAT motifs allow proteins to target ER membranes. But why should the vast majority of FFAT motifs occur in lipid binding proteins? For Osh proteins, we found that multiple targeting determinants combine to target the protein to sites where the ER apposes other organelles. Such contact sites between the ER and other organelles have been described for the plasma membrane (Metuzals et al., 1997; Staehelin, 1997; Takeshima et al., 2000; Pichler et al., 2001), the vacuole in yeast (Pan et al., 2000), peroxisomes (Mullen et al., 1999), the TGN (Ladinsky et al., 1999) and in particular for mitochondria, where lipids traffic in both directions (Vance, 1990; Voelker, 2000). These membrane contact sites are ideal subdomains for energy-efficient non-vesicular transfer of small molecules such as lipids and calcium ions (Voelker, 2000; Voeltz et al., 2002). Since the ER is the site of synthesis of most lipids, including those which bind OSBP, NIRs and GPBP (Lund et al., 1998; Baumann and Walz, 2001; Rolls et al., 2002), an important function of FFAT motifs may be to facilitate non-vesicular traffic of newly synthesized lipids from the ER.
In addition to using VAP as an ER anchor for lipid transfer, there may be more specific roles for the interactions between lipid binding proteins and VAP on the ER. In particular, they may affect the regulation of cholesterol synthesis by SREBP, since VAP has been shown to interact with the binding partner of SREBP on the ER (Yang et al., 2002). Thus, the FFAT–VAP interaction provides a previously unappreciated route for oxysterols and other lipids to signal to SREBP.
In summary, the FFAT motif mediates ER targeting of a large number of proteins involved in the regulation of lipid metabolism, and our results suggest a significant role for VAP homologues in regulating lipid metabolism.
Materials and methods
Plasmid construction
Plasmids used in this study are described in Table II. Plasmids including the natural ends of Osh proteins also include the gene’s own terminator, as indicated by a + entry in Table II. Plasmids containing 10 amino acids from Opi1p (197–206, underlined in Table II) have these residues inserted in either native or scrambled order between GFP and a cassette consisting of the tobacco etch virus cleavage site and a tandem repeat of the Z domain of protein A (tZZ) (Levine and Munro, 2001). Residue numbers for Osh1p relate to the sequence of the gene obtained from the yeast strain SEY6210 (DDBJ/EMBL/GenBank accession No. AY241177).
Table II. Plasmids used in this study.
| pRS406 | Integrates at URA3 | Sikorski and Hieter (1989) |
| pRS416 | CEN URA3 (low copy) | Sikorski and Hieter (1989) |
| pTL201 | pRS406 PHO5 promoter (168 bp), GFP, Scs2p(1–244) | |
| pTL211 | pRS406 PHO5 promoter, GFP, Opi1p (1–404) | |
| pTL212 | pRS416 OSH1 promoter (944 bp), GFP, Opi1p | |
| pTL213 | pRS416 OSH1 promoter, Opi1p | |
| pTL219 | pRS406 PHO5 promoter, GFP, Opi1p (EFFD→ALLA) | |
| pTL310 | pRS416 PHO5 promoter, GFP, Osh1p (1–1188) + terminator | |
| pTL312 | pRS416 PHO5 promoter, GFP, Osh2p (1–1283) + terminator | Levine and Munro (2001) |
| pTL313 | pRS416 PHO5 promoter, GFP, Osh3p (1–997) + terminator | Levine and Munro (2001) |
| pTL314 | pRS416 OSH1 promoter (370 bp), Osh1p + terminator | |
| pTL315 | pRS416 OSH1 promoter, Osh1p (D719→A) + terminator | |
| pTL316 | pRS416 PHO5 promoter, GFP, Osh1p (D719→A) + terminator | |
| pTL317 | pRS416 PHO5 promoter, GFP, Osh2p (Δ724–838) + terminator | |
| pTL318 | pRS416 PHO5 promoter, GFP, Osh3p (EFFDAeEe→EFAAPAAA) + terminator | |
| pTL375 | PRS406 PHO5 promoter, GFP, Osh1p (615–1188) + terminator | |
| pTL376 | PRS406 PHO5 promoter, GFP, Osh1p (687–1188) + terminator | Levine and Munro (2001) |
| pTL377 | pRS406 PHO5 promoter, GFP, Osh1p (687–796) | |
| pTL378 | pRS406 PHO5 promoter, GFP, Osh1p (797–1188) + terminator | |
| pTL379 | pRS406 PHO5 promoter, GFP, Osh1p (687–725) | |
| pTL382 | pRS406 PHO5 promoter, GFP, OSBP (331–415) | |
| pTL383 | pRS406 PHO5 promoter, GFP, Opi1p (172–218) | |
| pTL384 | pRS406 PHO5 promoter, GFP, MTSENLDDEEFFDASE, tZZ | |
| pTL385 | pRS406 PHO5 promoter, GFP, MTSFEELDDEDNFASE, tZZ | |
| pTL391–395 | pTL379 with following changes: pTL391, E716→A; pTL392, F717→A; pTL393, F718→A; pTL394, D719→A; pTL395, A720→P | |
| pTL396 | pTL379 721EEAAS-stop→KQLLARSSSFCSL-stop | |
| pTL397 | pRS406 PHO5 promoter, GFP, MTSANLAAAEFFDASE, tZZ | |
| pTL398 | pRS406 PHO5 promoter, GFP, MTSANLAAAEFFDARN, tZZ | |
| pTL413 | pRS414 (TRP1 CEN) PHO5 promoter, Tsc13p (1–559), GFP | |
| Are2-EGFP | pRS416 MET25 promoter, Are2p (1–642), EGFP | Zweytick et al. (2000) |
| pCL384 | CMV immediate early gene promoter, then as pTL384 | |
| pCL385 | CMV immediate early gene promoter, then as pTL385 | |
| pCL386 | CMV immediate early gene promoter, GFP, OSBP (188–425) | |
| pCL387 | CMV immediate early gene promoter, GFP, OSBP (188–425) EFFDA→ALLAG | |
| pCL388 | CMV immediate early gene promoter, GFP, OSBP (245–425) | |
| pCL389 | CMV immediate early gene promoter, GFP, OSBP (289–425) | |
| pCL390 | pGEX-4T-3 (Amersham Pharmacia) GST, Scs2p (1–225) | |
| pCL391 | pTrcHisA(Invitrogen) His6, GFP, MTSENLDDEEFFDASE, tZZ | |
| pCL392 | pTrcHisA His6, GFP, MTSFEELDDEDNFASE, tZZ | |
| pCL393 | pTrcHisA His6, GFP, OSBP (331–415) | |
| pCL394 | pTrcHisA His6, GFP, OSBP (331–415) EFFDA→EAFDA | |
| pFRILZ4 | pRS414 (TRP CEN) INO1 promoter (372 bp), lacZ |
Yeast strains
Yeast strains used were as follows: RS453B (wild type) and TLY221 (Δosh1) (Levine and Munro, 2001); TLY251 derived from RS453B by PCR integration upstream of the first codon of SCS2: the heterologous marker gene HIS5 from Schizosaccharomyces pombe, the GAL1 promoter and a myc epitope (correct integration was checked by PCR and by western blotting detecting a myc-tagged galactose-inducible protein migrating at 28 kDa); TLY252 derived by integration upstream of SCS2 in RS453B of HIS5, the PHO5 promoter and GFP. Y16119 and Y10943 are haploid strains YER120w::kanMX4 (Δscs2) and YHL020c::kanMX4 (Δopi1), respectively, obtained from Euroscarf (Frankfurt, Germany).
Cell imaging
Yeast cells growing in log phase at 30°C were visualized as described previously (Levine and Munro, 2002). GFP fusions other than those with full-length Osh proteins (see Figure 5) were expressed from integrating plasmids based on pRS406 to produce uniform expression within individual colonies. In cells derived from strain TLY251 (see Figures 3 and 5), Scs2p levels were regulated by carbon source: low levels were achieved by growth in dextrose and high levels were typically induced by overnight growth in galactose, although similar results were obtained 4 h after addition of galactose to cells grown in raffinose (data not shown). Comparison of Scs2p levels by western blotting under these two conditions indicated a >20-fold induction (data not shown). COS-7 cells were grown to confluence on glass coverslips, transfected with 5 µg of plasmid DNA per 3 cm well using Lipofectamine 2000 (Invitrogen) and visualized 48 h later. For immunofluorescent colocalization, COS-7 cells were fixed and permeabilized to preserve ER architecture with 2% paraformaldehyde and 0.2% glutaraldehyde (Lewis and Pelham, 1992), and calnexin was detected with rabbit primary antibodies (Stressgen Technologies, Canada) and rhodamine-conjugated secondary antiserum.
In vitro binding assay
A fusion protein of GST and Scs2 (residues 1–225, pCL390) was purified from bacterial lysates to >95% purity on glutathione–Sepharose using standard protocols. His6-tagged GFP-tagged proteins (pCL391–394) were purified to >90% purity as described previously (Levine and Munro, 2002). Binding was carried out in a 100 µl reaction volume for 15 min at room temperature in phosphate-buffered saline pH 7.4 with 0.5 mg/ml soybean trypsin inhibitor. GST–Scs2p beads and bound GFP-tagged fusion proteins were removed by centrifugation, and unbound GFP was detected by fluorimetry of the supernatant using a LS50 spectrophotometer (excitation 485 nm, emission 515 nm, slit widths 5 nm).
Assay for functionality of GFP-tagged Opi1p
Rescue of Δopi1 phenotype (unrepressible overexpression of INO1) was monitored according to previously published protocols (Sreenivas et al., 2001). A Δopi1 strain (Y10943) was transformed with two CEN plasmids: pFRILZ4, expressing lacZ under the INO1 promoter, and either pTL212 (GFP tagged Opi1p) or pTL213 (untagged Opi1p). Cells were grown to mid-log phase with or without 200 µM inositol and 1 mM choline. Five OD600 units of cells were collected by centrifugation, resuspended in 500 µl of 100 mM phosphate buffer pH 7.2 containing 10 mM potassium chloride, 1 mM magnesium sulphate, 50 mM β-mercaptoethanol, 0.005% w/w SDS and 3% w/w chloroform, mixed with glass beads (425–600 µm diameter) and shaken vigorously for 30 s in a Ribolyser (Hybaid). Lysate was mixed with O-nitrophenyl-β-d-galactopyranoside solution (final concentration 0.67 mg/ml) and incubated for 6 h at 30°C before stopping the reaction with sodium carbonate (final concentration 300 mM). β-galactosidase activity was measured by the absorbance at 420 nm and normalized to the number of cells added, as assessed by the absorbance at 600 nm.
Assay for rescue of Δosh1 phenotype
Cells were tested as described previously (Levine and Munro, 2001).
Acknowledgments
Acknowledgements
We would like to thank Fulvio Reggiori, Neale Ridgway and Guenther Daum for kind gifts of plasmids, Steve Moss and members of his laboratory for advice, and Karl Matter and Sean Munro for comments on the manuscript. This work was funded by grants from the BBSRC (grant number 31/C15982 to C.J.R.L.), the Wellcome Trust (grant number 060537 to A.R.) and Fight For Sight.
References
- Aikawa Y., Kuraoka,A., Kondo,H., Kawabuchi,M. and Watanabe,T. (1999) Involvement of PITPnm, a mammalian homologue of Drosophila rdgB, in phosphoinositide synthesis on Golgi membranes. J. Biol. Chem., 274, 20569–20577. [DOI] [PubMed] [Google Scholar]
- Baumann O. and Walz,B. (2001) Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol., 205, 149–214. [DOI] [PubMed] [Google Scholar]
- Brown A.J., Sun,L., Feramisco,J.D., Brown,M.S. and Goldstein,J.L. (2002) Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell, 10, 237–245. [DOI] [PubMed] [Google Scholar]
- Gallegos A.M. et al. (2001) Gene structure, intracellular localization and functional roles of sterol carrier protein-2. Prog. Lipid Res., 40, 498–563. [DOI] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Graves J.A. and Henry,S.A. (2000) Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics, 154, 1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.L. and Lopes,J.M. (1996) Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev., 60, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski,K., Rape,M., Schlenker,S., Ulrich,H.D. and Jentsch,S. (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell, 102, 577–586. [DOI] [PubMed] [Google Scholar]
- Hua X., Nohturfft,A., Goldstein,J.L. and Brown,M.S. (1996) Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell, 87, 415–426. [DOI] [PubMed] [Google Scholar]
- Jaworski C.J., Moreira,E., Li,A., Lee,R. and Rodriguez,I.R. (2001) A family of 12 human genes containing oxysterol-binding domains. Genomics, 78, 185–196. [DOI] [PubMed] [Google Scholar]
- Jiang B., Brown,J.L., Sheraton,J., Fortin,N. and Bussey,H. (1994) A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding protein. Yeast, 10, 341–353. [DOI] [PubMed] [Google Scholar]
- Kagiwada S., Hosaka,K., Murata,M., Nikawa,J. and Takatsuki,A. (1998) The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol., 180, 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S.D., Eder,S., Oh,C.S., Martin,C.E., Gable,K., Bacikova,D. and Dunn,T. (2001) Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky M.S., Mastronarde,D.N., McIntosh,J.R., Howell,K.E. and Staehelin,L.A. (1999) Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol., 144, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen S., Lehto,M., Lehtonen,S., Hyvarinen,K., Heino,S., Lehtonen,E., Ehnholm,C., Ikonen,E. and Olkkonen,V.M. (2002) ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J. Lipid Res., 43, 245–255. [PubMed] [Google Scholar]
- Lehto M., Laitinen,S., Chinetti,G., Johansson,M., Ehnholm,C., Staels,B., Ikonen,E. and Olkkonen,V.M. (2001) The OSBP-related protein family in humans. J. Lipid Res., 42, 1203–1213. [PubMed] [Google Scholar]
- Lemmon M.A., Ferguson,K.M., O’Brien,R., Sigler,P.B. and Schlessinger,J. (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl Acad. Sci. USA, 92, 10472–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Hernandez,J., Martinez,R., Chen,A., Plowman,G. and Schlessinger,J. (1999) Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol., 19, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Hsieh,C.L., Francke,U., Dawson,P.A., Ridgway,N.D., Brown,M.S. and Goldstein,J.L. (1990) cDNA cloning of human oxysterol-binding protein and localization of the gene to human chromosome 11 and mouse chromosome 19. Genomics, 7, 65–74. [DOI] [PubMed] [Google Scholar]
- Levine T.P. and Munro,S. (2001) Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus–vacuole junction. Mol. Biol. Cell, 12, 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T.P. and Munro,S. (2002) Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and independent, components. Curr. Biol., 12, 695–704. [DOI] [PubMed] [Google Scholar]
- Lewis M.J. and Pelham,H.R. (1992) Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell, 68, 353–364. [DOI] [PubMed] [Google Scholar]
- Litvak V., Shaul,Y., Shulewitz,M., Amarilio,R., Carmon,S. and Lev,S. (2002) Targeting of Nir2 to lipid droplets is regulated by a specific threonine residue within its PI-transfer domain. Curr. Biol., 12, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Lund E.G., Kerr,T.A., Sakai,J., Li,W.P. and Russell,D.W. (1998) cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem., 273, 34316–34327. [DOI] [PubMed] [Google Scholar]
- Metuzals J., Chang,D., Hammar,K. and Reese,T.S. (1997) Organization of the cortical endoplasmic reticulum in the squid giant axon. J. Neurocytol., 26, 529–539. [DOI] [PubMed] [Google Scholar]
- Moreira E.F., Jaworski,C., Li,A. and Rodriguez,I.R. (2001) Molecular and biochemical characterization of a novel oxysterol-binding protein (OSBP2) highly expressed in retina. J. Biol. Chem., 276, 18570–18578. [DOI] [PubMed] [Google Scholar]
- Mullen R.T., Lisenbee,C.S., Miernyk,J.A. and Trelease,R.N. (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell, 11, 2167–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., MurakamiA., Esumi,E. and Hosaka,K. (1995) Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J. Biochem. (Tokyo), 118, 39–45. [DOI] [PubMed] [Google Scholar]
- Pan X., Roberts,P., Chen,Y., Kvam,E., Shulga,N., Huang,K., Lemmon,S. and Goldfarb,D.S. (2000) Nucleus–vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell, 11, 2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetta G., Hiesinger,P., Fabian-Fine,R., Meinertzhagen,I. and Bellen,H. (2002) Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron, 35, 291–306. [DOI] [PubMed] [Google Scholar]
- Pichler H., Gaigg,B., Hrastnik,C., Achleitner,G., Kohlwein,S.D., Zellnig,G., Perktold,A. and Daum,G. (2001) A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem., 268, 2351–2361. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. and Aravind,L. (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci., 24, 130–132. [DOI] [PubMed] [Google Scholar]
- Ridgway N.D., Dawson,P.A., Ho,Y.K., Brown,M.S. and Goldstein,J.L. (1992) Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol., 116, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Hall,D.H., Victor,M., Stelzer,E.H. and Rapoport,T.A. (2002) Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell, 13, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R. et al. (1999) Electrospray ionization tandem mass spectrometry (ESI–MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol., 146, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel P.A., Martin,K.C., Kandel,E.R. and Bartsch,D. (1995) A VAMP-binding protein from Aplysia required for neurotransmitter release. Science, 269, 1580–1583. [DOI] [PubMed] [Google Scholar]
- Skehel P.A., Fabian-Fine,R. and Kandel,E.R. (2000) Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl Acad. Sci. USA, 97, 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussan L., Burakov,D., Daniels,M.P., Toister-Achituv,M., Porat,A., Yarden,Y. and Elazar,Z. (1999) ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J. Cell Biol., 146, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A., Villa-Garcia,M.J., Henry,S.A. and Carman,G.M. (2001) Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase C. J. Biol. Chem., 276, 29915–29923. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J., 11, 1151–1165. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Komazaki,S., Nishi,M., Iino,M. and Kangawa,K. (2000) Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell, 6, 11–22. [DOI] [PubMed] [Google Scholar]
- Taylor F.R., Saucier,S.E., Shown,E.P., Parish,E.J. and Kandutsch,A.A. (1984) Correlation between oxysterol binding to a cytosolic binding-protein and potency in the repression of hydroxymethylglutaryl coenzyme-a reductase. J. Biol. Chem., 259, 12382–12387. [PubMed] [Google Scholar]
- Vance J.E. (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem., 265, 7248–7256. [PubMed] [Google Scholar]
- Vihtelic T.S., Goebl,M., Milligan,S., O’Tousa,J.E. and Hyde,D.R. (1993) Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J. Cell Biol., 122, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker D.R. (2000) Interorganelle transport of aminoglycero phospholipids. Biochim. Biophys. Acta, 1486, 97–107. [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Rolls,M.M. and Rapoport,T.A. (2002) Structural organization of the endoplasmic reticulum. EMBO Rep., 3, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Dietz,M., Wittmann,J., Albrecht,A. and Schuller,H.J. (2001) The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol., 41, 155–166. [DOI] [PubMed] [Google Scholar]
- Wang C., JeBailey,L. and Ridgway,N.D. (2002) Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem. J., 361, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.D., Sato,R., Brown,M.S., Hua,X.X., Goldstein,J.L. (1994) Srebp-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell, 77, 53–62. [DOI] [PubMed] [Google Scholar]
- Weir M.L., Xie,H., Klip,A. and Trimble,W.S. (2001) VAP-A binds promiscuously to both v- and tSNAREs. Biochem. Biophys. Res. Commun., 286, 616–621. [DOI] [PubMed] [Google Scholar]
- Wirtz K.W. (1991) Phospholipid transfer proteins. Annu. Rev. Biochem., 60, 73–99. [DOI] [PubMed] [Google Scholar]
- Wirtz K.W. (1997) Phospholipid transfer proteins revisited. Biochem. J., 324, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles J.P., McMaster,C.R. and Ridgway,N.D. (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem., 277, 29908–29918. [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu,Y., Ridgway,N.D. and McMaster,C.R. (2001) Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J. Biol. Chem., 276, 18407–18414. [DOI] [PubMed] [Google Scholar]
- Yang T., Espenshade,P.J., Wright,M.E., Yabe,D., Gong,Y., Aebersold,R., Goldstein,J.L. and Brown,M.S. (2002) Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell, 110, 1–20. [DOI] [PubMed] [Google Scholar]
- Zweytick D., Leitner,E., Kohlwein,S.D., Yu,C., Rothblatt,J. and Daum,G. (2000) Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem., 267, 1075–1082. [DOI] [PubMed] [Google Scholar]