Abstract
Type 1 myotonic dystrophy is caused by the expansion of an unstable CTG repeat in the DMPK gene. We have investigated the molecular mechanisms underlying the CTG repeat instability by crossing transgenic mice carrying >300 unstable CTG repeats in their human chromatin environment with mice knockout for genes involved in various DNA repair pathways: Msh2 (mismatch repair), Rad52 and Rad54 (homologous recombination) and DNA-PKcs (non-homologous end-joining). Genes of the non-homologous end-joining and homologous recombination pathways did not seem to affect repeat instability. Only lack of Rad52 led to a slight decrease in expansion range. Unexpectedly, the absence of Msh2 did not result in stabilization of the CTG repeats in our model. Instead, it shifted the instability towards contractions rather than expansions, both in tissues and through generations. Furthermore, we carefully analyzed repeat transmissions with different Msh2 genotypes to determine the timing of intergenerational instability. We found that instability over generations depends not only on parental germinal instability, but also on a second event taking place after fertilization.
Keywords: CTG instability/double-strand break repair/mismatch repair/transgenic mice
Introduction
Type 1 myotonic dystrophy (DM1) is a neuromuscular condition involving myotonia and progressive muscle wasting. Digestive and respiratory functions are also affected, as are the central nervous system and the eyes (Harper, 2001). DM1 results from the expansion of an unstable CTG repeat in the 3′ untranslated region of the DM protein kinase gene (DMPK) (Aslanidis et al., 1992; Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992). In unaffected individuals, there are 5–37 CTG repeats in this gene, and repeat size remains stable over generations. In DM1 patients, a high level of intergenerational instability is observed, with a strong bias towards expansions. The number of CTG repeats is >50 and increases from generation to generation, to several thousand repeats. The sex and CTG repeat size of the transmitting parent affect CTG size in the offspring (Lavedan et al., 1993; Ashizawa et al., 1994). In DM1 families, the size of the CTG repeat is generally correlated with the severity of the symptoms and age at onset, accounting for the anticipation phenomenon observed from generation to generation (Harper et al., 1992). Somatic CTG repeat instability is observed in various tissues from DM1 patients, with a strong bias towards expansions. This instability probably appears early in embryogenesis, increases after 16 weeks of gestation and continues into adulthood (Martorell et al., 1997).
Since the discovery of dynamic mutations, many studies have focused on the molecular mechanisms of instability. The hairpins and slipped-strand structures observed in vitro with trinucleotide arrays are strongly suspected to induce instability through their interference with replication processes (Gacy et al., 1995; Samadashwily et al., 1997; Moore et al., 1999; Pearson et al., 2002). These structures may cause DNA polymerase to pause while replicating a triplet repeat, resulting in its dissociation from the newly synthesized strand (Ohshima and Wells, 1997; Samadashwily et al., 1997; Viguera et al., 2001). This strand would then separate from its template and might reassociate elsewhere within the repeat, leading to expansions on the resumption of replication, a phenomenon known as replication slippage (Pearson and Sinden, 1998; Petruska et al., 1998). In addition, replication-associated DNA hairpins arising at triplet repeat sequences also seem to inhibit the FEN-1 structure-specific endonuclease and may contribute to instability at the triplet repeat sequence (Spiro et al., 1999; Henricksen et al., 2000). The absence of FEN-1 due to the disruption of the yeast RAD27 gene also leads to destabilization of minisatellites, microsatellites and triplet repeats. DNA breakage within the region of CTG repeats may increase the action of DNA repair mechanisms at this particular site (Freudenreich et al., 1998; Kokoska et al., 1998; Schumacher et al., 1998). Furthermore, the direction of replication and the distance between the origin of replication and the repeat tract determine whether instability tends towards expansions or contractions in vitro and in primate cells (Cleary et al., 2002; Panigrahi et al., 2002). However, analyses of transgenic mice reproducing CTG somatic instability have shown that there is no correlation between somatic mosaicism and tissue division rate, suggesting the involvement of replication-independent mechanisms in non-dividing cells (Lia et al., 1998). Therefore, one or several DNA repair pathways, linked or not to replication, may also be involved in triplet repeat instability.
Double-strand break (DSB) repair involving homologous recombination (DSBR-HR) affects the stability of CAG and CTG repeats in yeast, leading to both expansions and contractions (Richard et al., 1999; Jankowski et al., 2000; Karran, 2000). HR is mediated by the Rad52 group of proteins, including Rad52 and Rad54 (van Gent et al., 2001). DSB repair can also occur via the non-homologous end-joining mechanism (NHEJ), which requires little, if any, sequence homology (Haber, 2000). Mismatch repair (MMR) has been demonstrated to be involved in triplet repeat instability, based on studies of the main component of this pathway, MutS in bacteria and its homolog, Msh2 in yeast and mammals. The absence of MutS in Escherichia coli results in stabilization of 64, 130 or 180 CTGs, and prevention of the large deletions that occur in the presence of MutS (Jaworski et al., 1995; Schumacher et al., 1998). In the yeast Saccharomyces cerevisiae, CTG and CAG repeats with 50 units display large natural deletions and do not seem to be affected by Msh2 mutations, indicating that Msh2 is not involved in such large deletions (Miret et al., 1997). Human Msh2 is known to bind tightly to the secondary structures formed by trinucleotide repeats in vitro and this binding may activate repair mechanisms (Pearson et al., 1997). In a transgenic mouse model for Huntington’s disease (HD) with 130 CAG units, somatic expansions disappear if Msh2 is deleted, indicating that Msh2 is involved in the generation of somatic expansions (Mangiarini et al., 1997; Manley et al., 1999). Further more, in these mice, Msh2 has been proposed to be involved in gap repair generating instability of CAG repeats in spermatozoa (Kovtun and McMurray, 2001). Studies of the partners of Msh2 in MMR: Msh3 and Msh6, in a DM1 knock-in mouse model carrying 84 CTGs have shown that the absence of Msh3 abolishes somatic expansions whereas the absence of Msh6 leads only to a slight increase in somatic expansions (van den Broek et al., 2002).
We decided to investigate the involvement of various DNA repair pathways in CTG repeat instability, using our DM1 transgenic mouse model, which carries a large repeat (>300 CTGs) embedded in its human chromatin environment of >45 kb. The use of long flanking sequences from the human genome and of a large expansion have facilitated faithful reproduction of the dynamics of the CTG repeat observed in DM1 patients, both intergenerationally and somatically (Gourdon et al., 1997b; Seznec et al., 2000). As observed in DM1 families, these mice display >90% CTG repeat length variation over generations, with a strong bias towards expansions (>85%). Parental age, repeat size and sex affect size variation. These mice display a high level of tissue-specific mosaicism biased towards expansions, which increases with age. Our model is therefore perfectly suitable for studying the influence of various DNA repair genes on intergenerational and somatic instability that cannot be approached in unicellular organisms. Analyses of CTG repeat size over generations and in tissues, in various DNA repair-deficient backgrounds, showed that a lack of expression of the Rad54 (DSBR-HR) and DNA-dependent protein kinase catalytic subunit or DNA-PKcs (NHEJ) genes had no major effect on CTG instability. However, the absence of Rad52 (DSBR-HR) had no effect on the frequency of expansions and contractions, but did slightly decrease the size of expansions. Furthermore, in sharp contrast to the stabilization of a CAG trinucleotide repeat observed in transgenic mice deficient in Msh2 (Manley et al., 1999; Kovtun and McMurray, 2001), the absence of Msh2 in our transgenic mice did not result in the stabilization of CTG repeats. Instead, it changed instability from expansions to contractions. In Msh2-deficient transgenic mice, we observed almost exclusively contractions, both somatically and over successive generations. Moreover, analysis of transmissions in parents and offspring of different Msh2 genotypes made it possible to determine the timing of intergenerational instability. We present data showing that intergenerational instability results not only from parental germinal instability, but also from a ‘second event’ taking place just after fertilization.
Results
Experimental strategy
Mice from the DM300–328 line, carrying >300 CTG repeats, were crossed with knockout mice for genes belonging to various DNA repair pathways. We assessed intergenerational instability over successive generations and the somatic instability of the CTG repeat in several tissues, in the various DNA repair-deficient backgrounds. After a series of matings, we obtained generations of transgenic DM300–328 mice lacking both alleles of the assayed gene. We determined the length of the CTG repeat of the transgenic transmitting parent and of the transgenic offspring by PCR, using tail DNA obtained at weaning (Seznec et al., 2000). The change in CTG repeat length from generation to generation in the mutant lines was compared with that of controls wild-type for both alleles of the tested gene, and/or with mice +/– for the studied DNA repair genes.
Effect of the Rad52 and Rad54 genes on intergenerational CTG instability
For DSB repair via HR, it was possible to investigate the potential roles of two genes, Rad52 and Rad54. Rad52 promotes HR by assisting Rad51, the key player in HR which is responsible for recognition of homologous DNA sequences and DNA strand exchange (Paques and Haber, 1999; Sugiyama and Kowalczykowski, 2002). The Rad52 knockout mice are viable and fertile. Rad52 knockout embryonic stem (ES) cells display a slightly reduced frequency of HR as measured by targeted homologous integration, but are not hypersensitive to DSB-inducing agents (Rijkers et al., 1998; Whitehouse et al., 1999). Rad54 also stimulates Rad51-mediated HR, most likely at a step after Rad52 function (Petukhova et al., 1998). Rad54 predominantly promotes HR with the use of the sister chromatid as repair template (Arbel et al., 1999; Dronkert et al., 2000). Rad54 knockout ES cells are sensitive to ionizing radiation and interstrand DNA crosslinking agents, display a reduced frequency of targeted homologous integration and are defective in DSB repair (Essers et al., 1997; Dronkert et al., 2000). Furthermore, Rad54 knockout mice are sensitive to interstrand DNA crosslinking agents and ionizing radiation sensitive in combination with a DNA-PKcs mutation (Essers et al., 1997, 2000).
As the Rad52 and Rad54 knockout mice were bred from mice with a genetic background identical to that of our transgenic DM300–328 mice (C57BL/6), we compared transgenic Rad52 and Rad54 knockout mice with DM300–328 mice. Table I presents the data of the various DNA repair-deficient transgenic lines, for male and female transmissions combined. As reported previously, CTG repeat in the DM300–328 mice showed a high level of intergenerational instability, with >90% size variation in the offspring, and a bias towards expansions (86.3%), with a mean size of +14.5 CTG. Contractions accounted for only 8.7% of total transmissions. In Rad52 or Rad54-deficient mice, the frequency of mutations was unaffected and a strong bias towards expansions was observed (89.7% for Rad52 –/–, 85.5% for Rad54 –/–). Although the mean size of expansions was unchanged by the absence of Rad54 (+13.3 CTG), a significant (P <0.001) decrease was observed for Rad52 –/– to –/– transmissions, with a mean of +9.3 CTG, as opposed to +14.5 for DM300–328 and +14.4 for Rad52 +/– to +/– (not shown). We showed previously that variations of the CTG repeat length from generation to generation depend on the age and CTG repeat size of the transmitting parent. This could eventually introduce a bias in the results if transmissions are obtained from mice with different age and CTG repeat (Seznec et al., 2000). We therefore also analyzed groups of transmitting mice with similar CTG repeat sizes and ages and obtained the same results (data not shown). Overall, our results show that the complete loss of Rad52 and Rad54 does not affect the mutability of the CTG repeat, or the frequency of expansions and contractions in our transgenic mice. However, loss of Rad52 resulted in a significant decrease in the mean size of expansions.
Table I. Intergenerational instability in transgenic mice with different DNA repair-deficient backgrounds.
| Parent to offspring transmission | Transmissionsanalyzed | Expansionfrequency (%) | Mean expansionlength (CTG) | Contraction frequency (%) | Mean contraction length (CTG) | Unchanged repeat size frequency (%) |
|---|---|---|---|---|---|---|
| DM300–328 | 299 | 86.3 | +14.5 | 8.7 | –10.1 | 5.0 |
| Rad52 –/– to –/– | 68 | 89.7 | +9.3 | 4.4 | –12.7 | 5.9 |
| Rad54 –/– to –/– | 55 | 85.5 | +13.3 | 10.9 | –14.5 | 3.6 |
| DNA-PKcs –/– to –/– | 91 | 64.8 | +7.6 | 27.5 | –10.7 | 7.7 |
| DNA-PKcs +/– to +/– | 37 | 62.2 | +10.0 | 27.0 | –10.6 | 10.8 |
| Msh2 –/– to –/– | 82 | 4.9 | +4.0 | 90.2 | –14.5 | 4.9 |
| Msh2 +/– to +/– | 82 | 78.0 | +12.8 | 19.5 | –9.9 | 2.5 |
+/–, –/– represent specific genotypes at the murine locus for the assayed gene: +, wild-type allele; –, mutant allele.
DNA-PKcs has no effect on intergenerational CTG instability
We investigated the effect of the NHEJ repair pathway on CTG repeat instability with the use of a DNA-PKcs mutant. Together with the heterodimer of the Ku70 and Ku80 proteins, DNA-PKcs forms the DNA–PK complex. The complex can juxtapose broken ends and the DNA–PK complex bound to one end may trans-phosphorylate its counterpart on the other end. This could induce a change in the overall conformation of the complex, providing free access for repair proteins such as XRCC4 and ligase IV for repair termination (Haber, 2000). DNA-PKcs is also involved in the V(D)J recombination process, and mice with mutations in the gene encoding DNA-PKcs display a SCID (severe combined immunodeficiency) phenotype (Taccioli et al., 1998). These mice are viable and fertile despite also displaying radiosensitivity and defects in the NHEJ repair process.
The comparison of DNA-PKcs +/– to +/– and –/– to –/– transmissions showed no significant difference in mutation frequency or range (Table I). There was a bias towards expansions for both transmissions (62.2 and 64.8%, respectively), showing that loss of the remaining functional DNA-PKcs allele did not change intergenerational instability in our model. It has to be noted that both the frequency and mean size of expansions were lower in DNA-PKcs +/– to +/– transmissions than in DM300–328 (P = 0.0007, P = 0.04). As the absence of the last DNA-PKcs allele did not affect intergenerational instability (same level of instability in DNA-PKcs +/– and DNA-PKcs –/– transmissions), the differences observed between the DNA-PKcs and DM300–328 mice may be accounted for by the different genetic backgrounds of the two lines (CB17 and C57BL/6). Therefore, DNA-PKcs is probably not involved in the repeat instability mechanisms.
The loss of Msh2 does not affect the mutability of the repeat but drives instability from expansions towards contractions
For MMR, we studied the influence of Msh2 because of its central role in this process. It recognizes both true base–base mismatches (when complexed with Msh6, giving rise to the MutSα complex) and insertion/deletion loops of up to 12 bp (when complexed with Msh3, giving rise to the MutSβ complex) (Marti et al., 2002). In both complexes, Msh2 is thought to act as a scaffold, with its two partners directly involved in DNA defect recognition, possibly in association with the PCNA sliding clamp (Bellacosa, 2001; Bowers et al., 2001). This recognition induces a change in the conformation of the DNA helix at this site, resulting in activation of the MutLα and MutLβ complexes, and ATP/ADP exchanges trigger a cascade of events leading to repair (Marti et al., 2002). In yeast, Msh2, in association with Msh3, ensures that there is a high level of sequence similarity between the invading 3′ ssDNA to be repaired and the matrix chosen as a template, but this protein is not involved in Holliday junction resolution. The knockout mice used in our study had very low MMR efficiency, with microsatellite instability, hyper-recombination and a predisposition to lymphoma. They were, however, viable and fertile (de Wind et al., 1995).
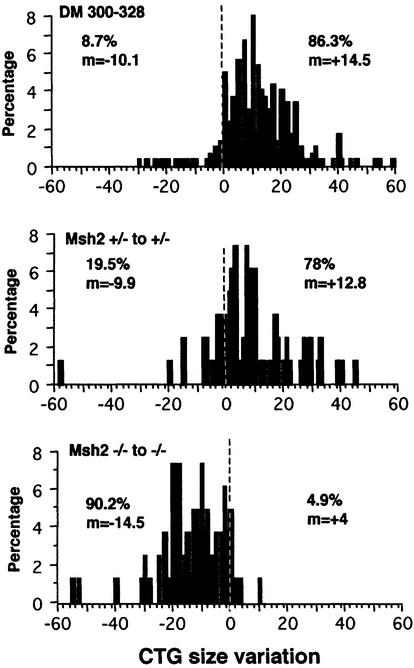
The results obtained with mice transgenic for the repeat and carrying different Msh2 genotypes were more striking than those obtained for the other DNA repair genes (Table I). In the absence of a functional Msh2 in both the transgenic parent and the offspring, there was a bias towards contractions (90.2%) rather than expansions of the CTG repeat over generations, for both male and female transmissions. The overall mutability (expansions + contractions) was unchanged in Msh2-deficient background, 95.1% in Msh2 –/– to –/– versus 97.5% in +/– to +/– and 95% in +/+ to +/+ DM300–328 mice, but the direction of instability (towards expansions or contractions) was reversed (Figure 1). Not only was the frequency of mutation similar with and without Msh2, but also the range of size variations, with +14.5 CTG in DM300–328, +12.8 CTG in Msh2 +/– to +/– and –14.5 CTG in Msh2 –/– to –/– transmissions. Although Msh2 knockout mice had a different genetic background compared with DM300–328 mice, Msh2 +/– to +/– transmissions displayed mostly expansions (78%, +12.8 CTG). A single Msh2 allele is therefore sufficient to drive instability towards expansions.
Fig. 1. Changes in CTG repeat size between transgenic parent and offspring for the various transmissions. The x-axis shows CTG repeat length change and the y-axis shows the percentage corresponding to each size change for all transmissions in each line. The frequency (%) and mean size (m) of expansions are indicated on the right. The frequency (%) and mean size (m) of contractions are indicated on the left. The number of transmissions assayed for each genotype was as follows: DM300–328, 299; Msh2 +/– to +/–, 82; Msh2 –/– to –/–, 82.
Intergenerational contractions and expansions occur not only in the germline, but also after fertilization
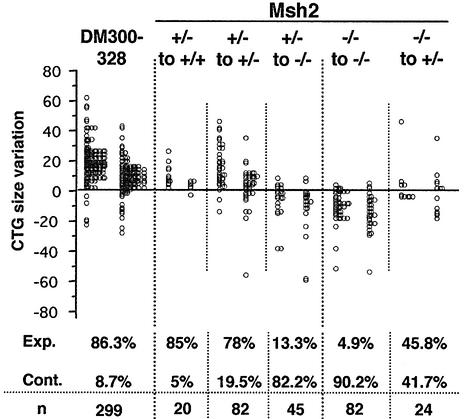
Intergenerational changes in CTG size were analyzed for all possible parental and offspring Msh2 genotypes, to elucidate the role of Msh2 in instability (Figure 2). When the transmitting parent carried one functional Msh2 allele, we observed a bias towards expansions (>78%) in Msh2 +/+ and +/– offspring, for both males and females. However, when these same Msh2 +/– transgenic parents gave rise to Msh2 –/– offspring, we observed mostly contractions (82.2%). These Msh2 +/– to –/– transmissions had similar instability characteristics to the Msh2 –/– to –/– transmissions presented above. Both the frequency of contractions (82.2 and 90.2%) and the mean size of these contractions (–13.75 and –14.5) were very similar.
Fig. 2. CTG repeat size variations following transgene transmission between parent and offspring carrying different Msh2 genotypes. The upper genotype indicates parental Msh2 status and the lower genotype indicates that of the offspring. + is the wild-type allele and – is the mutant allele. In each column, the left bar corresponds to male transmissions and the right bar to female transmissions. The frequencies of expansions (Exp.) and contractions (Cont.) are indicated for each transmission. n is the number of transmissions analyzed.
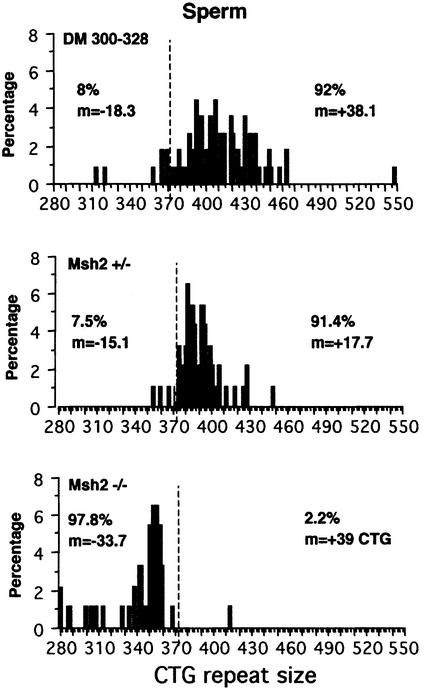
To obtain further insight into intergenerational instability mechanisms, we used single-cell PCR to investigate the distribution of alleles carrying different CTG repeat sizes in individual spermatozoa from 10-month-old males with different Msh2 genotypes. Figure 3 shows CTG repeat sizes in spermatozoa from DM300–328, Msh2 +/– and Msh2 –/– males. The length of the CTG repeat sequence in DNA from the tail of the analyzed male was taken as a reference for expansions or contractions, as this measure was used to calculate intergenerational size changes. Furthermore, the tail CTG repeat showed no mosaicism at weaning (3 weeks). A large majority of expansions was obtained in spermatozoa from the DM300–328 (Msh2 +/+) male (92%, +38.1 CTG) and from the Msh2 +/– male (91.4%, +17.7 CTG). Expansions were smaller on average in Msh2 +/– than in DM300–328 mice, probably due to differences in genetic background, or to a decrease in Msh2 expression. For the Msh2 –/– male, the CTG repeat mutability in spermatozoa was similar to that in the controls, but the size variations were contractions in almost all cases (97.8%, –33.7 CTG). The possible influence of the inherited CTG repeat size on instability dynamics was abolished by choosing mice carrying CTG repeats identical in length. Single-cell PCR profiles for spermatozoa from the DM300–328 male and Msh2 –/– male were consistent with the percentage of expansions and contractions observed in the respective +/+ and –/– offspring. Previously, it was thought that intergenerational instability results mainly from parental germinal instability. This would result in a strong bias towards expansions in all Msh2 +/– transmissions, irrespective of the Msh2 genotype of the offspring. The distribution of expansions and contractions in the Msh2 +/– male matches those in its +/– offspring, but not in its –/– offspring. Regardless of the sex of the transmitting parent and the Msh2 genotype (+/– or –/–) of the non-transgenic parent, there was a strong bias in the Msh2 –/– offspring towards contractions rather than expansions (see Figure 2). These results demonstrate that the offspring Msh2 genotype affects intergenerational instability and that the contraction event in the Msh2 –/– offspring of an Msh2 +/– transgenic parent occurs after fertilization. Interestingly, Msh2 –/– to +/– transmissions displayed an equilibrium between expansions and contractions, suggesting that the presence of a single Msh2 allele in the offspring is not sufficient to counteract entirely the effect of germline contractions.
Fig. 3. Distribution of alleles with different CTG repeat sizes in spermatozoa from 10-month-old male transgenic DM300–328, Msh2 +/– or Msh2 –/– mice, as determined by single-cell PCR. The y axis shows the percentage of each size allele with respect to all single genomes analyzed. The dotted line indicates the CTG repeat size in tail DNA at weaning: 372 CTG for DM300–328, 373 CTG for Msh2 +/– and 372 CTG for Msh2 –/–. The frequency and mean size of expansions are indicated on the right and the frequency and mean size of contractions are indicated on the left. The number of single genomes analyzed for each tissue was as follows: DM300–328, 113; Msh2 +/–, 93; Msh2 –/–, 92.
We investigated the timing of this event by assessing somatic instability in various tissues in 1-day-old pups of different Msh2 genotypes born from parents of various Msh2 genotypes. Weak intratissue somatic mosaicism was present in most tissues at this stage, with a tendency towards expansions in Msh2 +/– newborns with Msh2 +/– parents, and towards contractions in Msh2 –/– pups from transgenic Msh2 +/– or Msh2 –/– parents (data not shown). However, the predominant size of the CTG repeat was similar in the various tissues and we observed no major intertissue mosaicism, suggesting that the second instability event occurred very early in development, probably at the one-cell stage, just after fertilization.
Influence of the various DNA repair genes on somatic instability of the CTG repeat
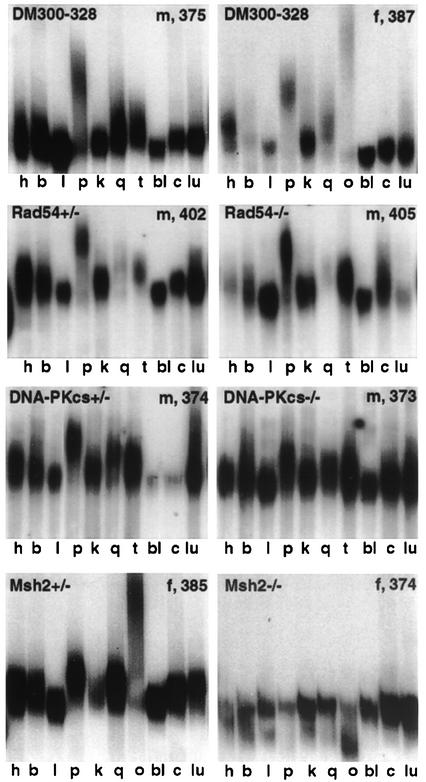
We assayed somatic instability in 11 tissues in 10-month-old male and female mice, –/– or +/– for each DNA repair gene and/or +/+ control mice (Figure 4). We analyzed transgenic mice carrying CTG repeats of identical size in their tail DNA to avoid effects of basal repeat length on instability. Single-cell PCR was also used to resolve the smears and to determine the distribution of alleles with different CTG repeat sizes in several tissues (Figures 5 and 6). CTG repeat length in blood was considered to represent the inherited size in each mouse, because this tissue remains stable throughout the animal’s life.
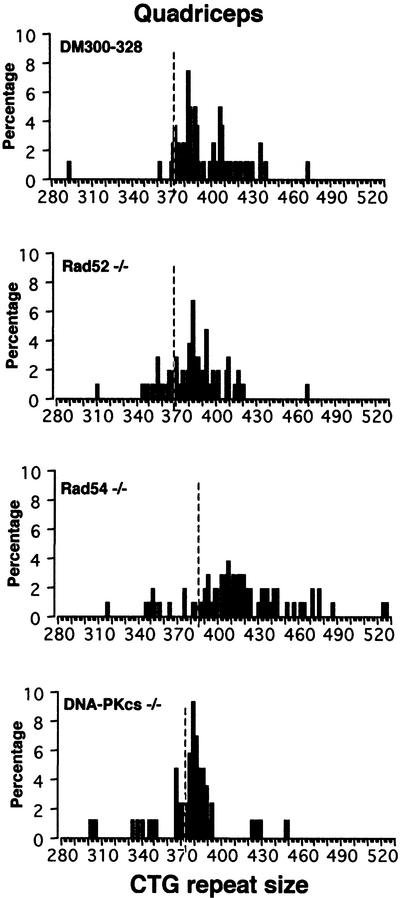
Fig. 4. Somatic instability in 10-month-old male and female DM300–328, Rad54 +/–, Rad54 –/–, DNA-PKcs +/–, DNA-PKcs –/–, Msh2 +/– and Msh2 –/– transgenic mice. The same instability profiles were observed between Rad52 +/– and Rad52 –/– transgenic mice (data not shown). h, heart; b, brain; l, liver; p, pancreas; k, kidney; q, quadriceps; o, ovary; t, testis; bl, blood; c, cerebellum; lu, lung. CTG repeat length in blood is indicated for each mouse. m, male; f, female.
Fig. 5. Distribution of alleles with different CTG repeat sizes in quadriceps from 10-month-old male transgenic DM300–328, Rad52 –/–, Rad54 –/– and DNA-PKcs –/– mice, as determined by single-cell PCR. The dotted line is the CTG repeat size in blood: 372 CTG for DM300–328, 369 CTG for Rad52 –/–, 387 CTG for Rad54 –/– and 373 CTG for DNA-PKcs –/–. The number of single molecules analyzed for each male was as follows: DM300–328: 87; Rad52 –/–, 105; Rad54 –/–, 106; DNA-PKcs –/–, 86.
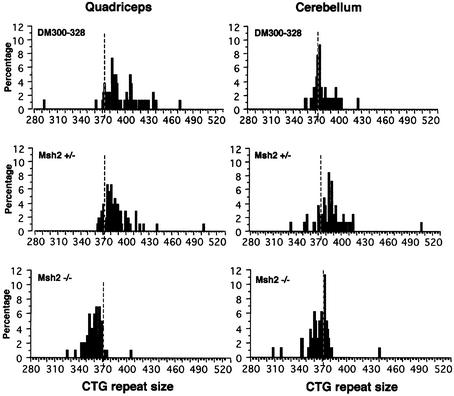
Fig. 6. Distribution of alleles with different CTG repeat sizes in quadriceps and cerebellum from 10-month-old transgenic male DM300–328, Msh2 +/– and Msh2 –/– mice, as determined by single-cell PCR. Dotted lines represent the CTG repeat size measured in blood: 372 CTG for DM300–328, 373 CTG for Msh2 +/– and 372 CTG for Msh2 –/–. The number of single molecules analyzed for each tissue was as follows, DM300–328: quadriceps, 87; cerebellum, 80; Msh2 +/–: quadriceps, 106; cerebellum, 83; Msh2 –/–: quadriceps, 101; cerebellum, 88.
Male and female Rad52 +/–, Rad52 –/–, Rad54 +/– and Rad54 –/– transgenic mice displayed similar patterns of somatic instability to control +/+ mice, with a strong bias towards expansions in tissues presenting mosaicism (Figure 4). We also observed mostly expansions when assessing somatic mosaicism in DNA-PKcs +/– and –/– mice, with no differences between mice that did and did not carry a functional DNA-PKcs allele (Figure 4). The absence of functional Rad52, Rad54 or DNA-PKcs genes in our transgenic mice had no effect on somatic instability, indicating that these genes do not play a critical role in this process. Single-cell PCR performed with DNA from quadriceps, an affected tissue that displays relatively high mosaicism both in DM1 patients and transgenic mice, showed that there was a strong bias towards expansions in DM300–328, Rad52 –/–, Rad54 –/– and DNA-PKcs –/– mice (Figure 5). The mean size of expansions was greatest in Rad54 –/– quadriceps, probably due to the larger basal CTG repeat size in this mouse. However, no major differences were observed in the mean size of expansions between DM300–328 and Rad52 –/– mice, in contrast to that observed in intergenerational analyses. The lower level of mosaicism observed in DNA-PKcs –/– quadriceps may result from the different genetic background carried by DNA-PKcs knockout mice.
Somatic instability dynamics were strongly modified in Msh2-deficient mice (both in female and male, Figure 4 and data not shown). In control and Msh2 +/– mice, a high level of tissue-specific mosaicism was observed, with a strong bias towards expansions, especially in the ovaries and pancreas. In contrast, in Msh2-deficient transgenic mice, mosaicism was observed but there was a shift towards contractions.
The results of single-cell PCR studies performed in 10-month-old male mice, with quadriceps (displaying mosaicism in control mice) and cerebellum (displaying little or no mosaicism in control mice) are shown in Figure 6 for Msh2 +/–, Msh2 –/– and DM300–328 (Msh2 +/+) males. In the DM300–328 male, we observed mostly expansions in the quadriceps, with weaker mosaicism in the cerebellum. The Msh2 +/– male also displayed mosaicism in both tissues, with a strong bias towards expansions, as observed for females (Figure 4). However, lower levels of mosaicism were observed in the quadriceps of Msh2 +/– than in DM300–328, probably due to lower levels of Msh2 expression or differences in genetic background between DM300–328 and Msh2 knockout mice. Interestingly, Msh2 +/– mice displayed higher levels of instability in the cerebellum than did DM300–328 mice. In sharp contrast, in the Msh2 –/– mice, we observed a strong bias towards contractions, with greater size variation in the quadriceps than in the cerebellum. These results show that Msh2 is also necessary to drive instability towards expansions in tissues, and that instability shifts towards contractions in the absence of this gene.
Discussion
To identify the type of DNA repair pathway involved in triplet repeat instability mechanisms, we determined the influence of various DNA repair genes on CTG instability in our transgenic mouse model by monitoring intergenerational and somatic instability in knockout mice for these genes. As CTG repeats have been shown to induce length-dependent breakage in yeast (Freudenreich et al., 1998), we investigated the role of two genes involved in DSBR-HR, Rad52 and Rad54. The absence of Rad52 or Rad54 in our transgenic mice had no major effect on intergenerational and somatic instability. The mutability was high (>90%), and a strong bias towards expansions was observed from generation to generation, in all the tissues tested. Thus, these genes are probably not directly responsible for the CTG repeat instability in our mice and Rad52/Rad54-mediated HR is probably not involved. However, a slight but significant decrease in the mean size of expansions was observed in Rad52 –/– to –/– transmissions (but not in tissues). Our results with Rad54 knockout mice suggest that instability is unlikely to be produced through the DSBR-HR mechanism. Thus, if Rad52 is involved in CTG instability, this is probably not via this pathway. Rad52 could possibly act through its activity in single-strand annealing (SSA), which is an error-prone DSB repair pathway that does not depend on other Rad52 group proteins (Paques and Haber, 1999). Interestingly, in yeast, Msh2 has been shown to participate in SSA as well (Pastink and Lohman, 1999). DSBs arising in CTG tracts, due to the formation of aberrant secondary DNA structures during replication, could also be repaired by the NHEJ. The results obtained with DNA-PKcs knockout mice showed that instability, and expansions in particular, continue to occur in the absence of DNA-PKcs expression, suggesting that the NHEJ may not affect CTG repeats, at least as far as the action of DNA-PKcs is concerned.
Contrasting results were obtained for mice deficient in Msh2. In an Msh2-deficient background, high frequencies of mutation were observed in transgenic mice but, in sharp contrast to what was observed in Msh2-expressing mice, we observed a large majority of contractions in transgenes transmitted between parent and offspring and in tissues. These data show that Msh2 is a key factor driving the instability of the CTG repeat towards expansions from generation to generation and in tissues. Msh2 was reported previously to be involved in the generation of somatic expansions in CAG repeats in transgenic mice carrying the first exon of the huntingtin (HD) gene (with ∼130 CAG), and the CAG tract was found to be stable in Msh2-deficient mouse tissues (Manley et al., 1999). Surprisingly, in our Msh2 –/– transgenic mice, the CTG repeat was not stabilized but there was a shift in instability from expansions towards contractions in tissues, and during transgene transmissions over several generations. The sequences surrounding the trinucleotide repeat affect the mutability of the repeat, and comparison of the various mice models with trinucleotide repeats suggests that flanking sequences also determine whether instability tends towards expansions or contractions (Gourdon et al., 1997a; Brock et al., 1999; Seznec et al., 2000; Cleary et al., 2002). The presence of large GC-rich human flanking sequences around the CTG repeat, recreating the human genomic DNA environment in our mice, may account for the differences observed in the absence of Msh2 in the different mice models. The majority of contractions observed in transgenic Msh2-deficient mice may result from another mechanism, substituting for the action of Msh2 and generating deletions when processing a CTG repeat.
Until now, despite the suggestion of a post-zygotic instability event observed in fragile X-affected fetuses (Malter et al., 1997), it has been thought that intergenerational instability is mainly the result of germinal instability from the transmitting parent. This germinal instability could take place during stem cell divisions, or at diploid or haploid stages of gametogenesis. It has been suggested that in transgenic mice carrying CAG repeats, expansions occur in spermatozoa by a mechanism involving Msh2 and gap repair (Kovtun and McMurray, 2001). However, in our transgenic DM300–328 mice we observed, using cell sorting and single-cell PCR, that CTG repeat expansion occurs already in spermatogonia (C.Savouret, C.Garcia-Cordier, C.Junien and G.Gourdon, in preparation). We also did not observe any effect of the offspring gender on the CTG repeat intergenerational instability in the DM300–328 mice as has been described in the CAG repeat mouse model (Kovtun et al., 2000). This discrepancy in the behavior of the repeats between these two models is probably related to the different genomic context of the repeats. However, in our transgenic mice, taking into account that the Msh2 genotype of the offspring is critical in driving the intergenerational instability towards expansions or contraction, instability in the germline is not sufficient to explain changes in CTG repeat size from generation to generation. Detailed analysis of CTG repeat size variation between transgenic parents and offspring carrying different Msh2 genotypes demonstrated that another event must take place, in addition to gonadal CTG instability. Analysis of somatic CTG instability in tissues from 1-day-old pups carrying various Msh2 genotypes revealed weak intratissue mosaicism, but no strong intertissue mosaicism, indicating that the second instability event must take place very early during development, probably just after fertilization as suggested for CGG repeat in Fragile X syndrome (Malter et al., 1997). The frequencies of expansions and contractions were similar in Msh2 –/– to +/– transmissions, showing that contractions in Msh2 –/– germinal cells were only partially rescued by the presence of a functional Msh2 allele in the offspring, giving rise to both expansions and contractions. Logically, if the effects of the two events were additional, then the mean size of contractions in Msh2 –/– to –/– transmissions would be expected to be higher than that in Msh2 +/– to –/– transmissions. However, the frequency and observed mean size of contractions appear to be almost equal for both these transmissions. Factors such as maternal Msh2 genotype may be critical. If Msh2 is produced during ovogenesis, then a residual fraction of maternal Msh2 proteins produced by a diploid Msh2 +/– ovocyte may be present in the Msh2– haploid ovocyte used for fertilization, potentially modifying CTG repeat instability just after fertilization. This influence of the maternal Msh2 genotype on stability will be monitored by analyzing a larger number of transmissions in our mice.
The data presented here clearly implicate Msh2 not only in the somatic but also in intergenerational instability of the CTG repeat. As reported previously in another study, Msh3 is also involved in CTG repeat somatic instability which disappears on a Msh3-deficient background (van den Broek et al., 2002). Msh3 probably acts in association with Msh2 forming a Msh2–Msh3 complex, involved in MMR but also in other repair pathways. The competition between Msh3 and Msh6 to form complexes with Msh2 could explain why in Msh6-deficient mice, in which only Msh2–Msh3 complexes are formed, there is an increase of somatic instability (van den Broek et al., 2002). This would imply that only the Msh2–Msh3 complex is involved to generate CTG expansions. The implication of Msh3 in intergenerational instability remains to be demonstrated. However, the mechanism by which Msh2 and Msh3 cause expansions is still unknown, particularly since the Msh proteins may also be involved in other types of DNA repair such as DSBR-HR or SSA. Rad52 may also be involved in the mechanism generating expansions, but probably via a Rad54-independent pathway. Taking these results together, we propose that the Msh2–Msh3 complex generates somatic triplet repeat expansions while repairing DSBs through the SSA pathway. This complex would also generate intergenerational expansions via the SSA, in association with Rad52. Others factors involved in SSA need to be assayed to confirm this hypothesis. In the absence of Msh2, other repair mechanism(s) that remain(s) to be identified, could process the CTG repeat generating contractions. As far as the timing of intergenerational instability is concerned, we suggest that Msh2 and the associated mechanism exert their expansion-producing effects not only in the parental germline, but also just after fertilization, and that the combination of these two distinct events accounts for variations in CTG repeat size over generations. Identification of the precise time at which instability occurs in these two steps should help us to determine the molecular mechanism involved.
Materials and methods
Transgenic mice
Transgenic status was determined by PCR with the DMHR4 and DMHR5 primers. Tail DNA was amplified by PCR, using primers 101 and 102, and repeat length was measured by running CTG amplification products on a 4% acrylamide denaturing gel (Seznec et al., 2000). Rad52 +/– (C57BL/6 background), Rad54 +/– (C57BL/6 background) and Msh2 +/– (129/OLA, FVB background) mutant mice have been described elsewhere (de Wind et al., 1995; Essers et al., 1997; Rijkers et al., 1998). DNA-PKcs mutant SCID mice (CB17 background) were purchased from IFFACREDO (France). Rad52, Rad54, Msh2 and DNA-PKcs genotypes were assayed by multiplex PCR, using primers that anneal to either the wild-type or the mutant allele, as reported previously (de Wind et al., 1995; Blunt et al., 1996; Essers et al., 1997; Rijkers et al., 1998).
Somatic instability analyses
Ten-month-old mice were killed and dissected, and DNA was phenol-extracted from tissues (Lia et al., 1998). Semen was taken from the vas deferens and DNA from spermatozoa was extracted with care to ensure that it was not contaminated by DNA from other cells (Seznec et al., 2000). This extraction step is very important, as semen may contain seminal leukocytes and epithelial cells from the epididymis that display a high level of instability, potentially complicating the interpretation of the results. We carried out PCR with 15 ng of DNA from each tissue (DNA from ∼2500 cells in total), with 1.4 µM of primers 101 and 102 surrounding the CTG repeat, in a buffer described elsewhere (Jeffreys et al., 1994; Seznec et al., 2000) with 1 unit of Thermoperfect DNA Polymerase (Integro BV, The Netherlands). After initial denaturation for 5 min at 96°C, we carried out 30 cycles of 45 s at 96°C, 30 s at 68°C and 3 min at 72°C followed by a chase of 1 min at 68°C and 10 min at 72°C. PCR products were mixed with formamide loading buffer, heated for 5 min at 100°C and subjected to electrophoresis in 3.5% acrylamide denaturing gels at 55 W for 30 min and 25 W for 13 h 30 min in 1× TBE buffer (0.09 M Tris, 0.09 M boric acid and 2 mM EDTA). DNA was transferred onto a nylon membrane and hybridized with a (CAG)17 radiolabeled probe.
Single-cell PCR analyses
Single-cell PCR was performed as described previously (Monckton et al., 1995). Briefly, 10 µg of quadriceps, cerebellum or sperm DNA was digested with HindIII. We carried out PCR with 6 pg of DNA, corresponding to the amount of DNA present in a single cell, hence the equivalent of one genome, digested with HindIII. Amplification conditions were as described above. After initial denaturation at 96°C for 5 min, reactions involved 28 cycles (96°C for 45 s, 68°C for 30 s, 72°C for 3 min), with a chase of 1 min at 68°C and 10 min at 72°C. Single-cell PCR products were subjected to electrophoresis in acrylamide gels, as described above. CTG repeat length was determined by comparison with a radiolabeled 100 bp DNA ladder. Experiments were repeated until a significant number of single alleles were obtained, fixed at between 80 and 120 single molecules (Monckton et al., 1995; Lia et al., 1998).
Statistical analyses
Statistical analyses were performed with StatView software using t-test and chi-square test (SAS Institute Inc.).
Acknowledgments
Acknowledgements
We thank E.Lodato for attentively caring for the mice. This work was supported by grants from INSERM, the Association Française contre les Myopathies (AFM) and the Université René-Descartes. C.S. was supported by a grant from the Ministère de la Recherche et de la Technologie.
References
- Arbel A., Zenvirth,D. and Simchen,G. (1999) Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J., 18, 2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa T., Dunne,P.W., Ward,P.A., Seltzer,W.K. and Richards,C.S. (1994) Effects of the sex of myotonic dystrophy patients on the unstable triplet repeat in their affected offspring. Neurology, 44, 120–122. [DOI] [PubMed] [Google Scholar]
- Aslanidis C. et al. (1992) Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature, 355, 548–551. [DOI] [PubMed] [Google Scholar]
- Bellacosa A. (2001) Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ., 8, 1076–1092. [DOI] [PubMed] [Google Scholar]
- Blunt T., Gell,D., Fox,M., Taccioli,G.E., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1996) Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl Acad. Sci. USA, 93, 10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J., Tran,P.T., Joshi,A., Liskay,R.M. and Alani,E. (2001) MSH–MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol., 306, 957–968. [DOI] [PubMed] [Google Scholar]
- Brock G.J.R., Niall,H.A. and Monckton,D.G. (1999) Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet., 8, 1061–1067. [DOI] [PubMed] [Google Scholar]
- Brook J.D. et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell, 68, 799–808. [DOI] [PubMed] [Google Scholar]
- Cleary J.D., Nichol,K., Wang,Y.H. and Pearson,C.E. (2002) Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet., 31, 37–46. [DOI] [PubMed] [Google Scholar]
- de Wind N., Dekker,M., Berns,A., Radman,M. and te Riele,H. (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination and predisposition to cancer. Cell, 82, 321–330. [DOI] [PubMed] [Google Scholar]
- Dronkert M.L., Beverloo,H.B., Johnson,R.D., Hoeijmakers,J.H., Jasin,M. and Kanaar,R. (2000) Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C.H., Kantrow,S.M. and Zakian,V.A. (1998) Expansion and length-dependent fragility of CTG repeats in yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- Fu Y.H. et al. (1992) An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science, 255, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Gacy A.M., Goellner,G., Juranic,N., Macura,S. and McMurray,C.T. (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- Gourdon G., Dessen,P., Lia,A.S., Junien,C. and Hoffman-Radvanyi,H. (1997a) Intriguing association between disease associated unstable trinucleotide repeat and CpG island. Ann. Génét., 40, 73–77. [PubMed] [Google Scholar]
- Gourdon G., Radvanyi,F., Lia,A.S., Duros,C., Blanche,M., Abitbol,M., Junien,C. and Hofmann-Radvanyi,H. (1997b) Moderate instability of a 55 CTG repeat in transgenic mice carrying a 45 kb genomic region from an affected DM patient. Nat. Genet., 15, 190–192. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (2000) Partners and pathways: repairing a DSB. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- Harper P.S. (2001) Myotonic Dystrophy. 3rd edn. Saunders, London.
- Harper P.S., Harley,H.G., Reardon,W. and Shaw,D.J. (1992) Anticipation in myotonic dystrophy: new light on an old problem. Am. J. Hum. Genet., 51, 10–16. [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Tom,S., Liu,Y. and Bambara,R.A. (2000) Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem., 275, 16420–16427. [DOI] [PubMed] [Google Scholar]
- Jankowski C., Nasar,F. and Nag,D.K. (2000) Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA, 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski A., Rosche,W.A., Gellibolian,R., Kang,S., Shimizu,M., Bowater,R.P., Sinden,R.R. and Wells,R.D. (1995) Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl Acad. Sci. USA, 92, 11019–11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A.J., Tamaki,K., MacLeod,A., Monckton,D.G., Neil,D.L. and Armour,J.A. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- Kokoska R.J., Stefanovic,L., Tran,H.T., Resnick,M.A., Gordenin,D.A. and Petes,T.D. (1998) Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol., 18, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun I.V. and McMurray,C.T. (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet., 27, 407–411. [DOI] [PubMed] [Google Scholar]
- Kovtun I.V., Therneau,T.M. and McMurray,C.T. (2000) Gender of the embryo contributes to CAG instability in transgenic mice containing a Huntington’s disease gene. Hum. Mol. Genet., 9, 2767–2775. [DOI] [PubMed] [Google Scholar]
- Lavedan C. et al. (1993) Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability and somatic mosaicism. Am. J. Hum. Genet., 52, 875–883. [PMC free article] [PubMed] [Google Scholar]
- Lia A., Seznec,H., Hofmann-Radvanyi,H., Radvanyi,F., Duros,C., Saquet,C., Blanche,M., Junien,C. and Gourdon,G. (1998) Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age-dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet., 7, 1285–1291. [DOI] [PubMed] [Google Scholar]
- Mahadevan M. et al. (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science, 255, 1253–1255. [DOI] [PubMed] [Google Scholar]
- Malter H.E., Iber,J.C., Willemsen,R., de Graaff,E., Tarleton,J.C., Leisti,J., Warren,S.T. and Oostra,B.A. (1997) Characterization of the full fragile X syndrome mutation in fetal gametes. Nat. Genet., 15, 165–169. [DOI] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam,K., Mahal,A., Mott,R., Seller,M. and Bates,G.P. (1997) Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat. Genet., 15, 197–200. [DOI] [PubMed] [Google Scholar]
- Manley K., Shirley,T.L., Flaherty,L. and Messer,A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet., 23, 471–473. [DOI] [PubMed] [Google Scholar]
- Marti T.M., Kunz,C. and Fleck,O. (2002) DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol., 191, 28–41. [DOI] [PubMed] [Google Scholar]
- Martorell L., Johnson,K. and Boucher,C.A. (1997) Somatic instability of the myotonic dystrophy (CTG)n repeat during human fetal development. Hum. Mol. Genet., 6, 877–880. [DOI] [PubMed] [Google Scholar]
- Miret J.J., Pessoa-Brandao,L. and Lahue,R.S. (1997) Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 3382–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monckton D.G., Wong,L.J.C., Ashizawa,T. and Caskey,C.T. (1995) Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet., 4, 1–8. [DOI] [PubMed] [Google Scholar]
- Moore H., Greenwell,P.W., Liu,C.P., Arnheim,M. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K. and Wells,R.D. (1997) Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem., 272, 16798–16806. [DOI] [PubMed] [Google Scholar]
- Panigrahi G.B., Cleary,J.D. and Pearson,C.E. (2002) In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. J. Biol. Chem., 277, 13926–13934. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastink A. and Lohman,P.H. (1999) Repair and consequences of double-strand breaks in DNA. Mutat. Res., 428, 141–156. [DOI] [PubMed] [Google Scholar]
- Pearson C.E. and Sinden,R.R. (1998) Slipped strand DNA, dynamic, mutations and human disease. In Wells,R.D. and Warren,S.T. (eds), Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego, CA, pp. 585–623.
- Pearson C.E., Ewel,A., Acharya,S., Fishel,R.A. and Sinden,R.R. (1997) Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet., 7, 1117–1123. [DOI] [PubMed] [Google Scholar]
- Pearson C.E., Tam,M., Wang,Y.H., Montgomery,S.E., Dar,A.C., Cleary,J.D. and Nichol,K. (2002) Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res., 30, 4534–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska J., Hartenstine,M.J. and Goodman,M.F. (1998) Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem., 273, 5204–5210. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- Richard G.F., Dujon,B. and Haber,J.E. (1999) Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Mol. Genet., 261, 871–882. [DOI] [PubMed] [Google Scholar]
- Rijkers T., Van Den Ouweland,J., Morolli,B., Rolink,A.G., Baarends,W.M., Van Sloun,P.P., Lohman,P.H. and Pastink,A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily G.M., Raca,G. and Mirkin,S.M. (1997) Trinucleotide repeats affect DNA replication in vivo. Nat. Genet., 17, 298–304. [DOI] [PubMed] [Google Scholar]
- Schumacher S., Fuchs,R.P. and Bichara,M. (1998) Expansion of CTG repeats from human disease genes is dependent upon replication mechanisms in Escherichia coli: the effect of long patch mismatch repair revisited. J. Mol. Biol., 279, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Seznec H., Lia-Baldini,A., Duros,C., Fouquet,C., Lacroix,C., Hofmann-Radvanyi,H., Junien,C. and Gourdon,G. (2000) Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet., 9, 1185–1194. [DOI] [PubMed] [Google Scholar]
- Spiro C. et al. (1999) Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell, 4, 1079–1085. [DOI] [PubMed] [Google Scholar]
- Sugiyama T. and Kowalczykowski,S.C. (2002) Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem., 277, 31663–31672. [DOI] [PubMed] [Google Scholar]
- Taccioli G.E. et al. (1998) Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity, 9, 355–366. [DOI] [PubMed] [Google Scholar]
- van den Broek W.J., Nelen,M.R., Wansink,D.G., Coerwinkel,M.M., te Riele,H., Groenen,P.J. and Wieringa,B. (2002) Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 11, 191–198. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Hoeijmakers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- Viguera E., Canceill,D. and Ehrlich,S.D. (2001) In vitro replication slippage by DNA polymerases from thermophilic organisms. J. Mol. Biol., 312, 323–333. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Flaus,A., Cairns,B.R., White,M.F., Workman,J.L. and Owen-Hughes,T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]