Abstract
Resting cells experience mutations without apparent external mutagenic influences. Such DNA replication-independent mutations are suspected to be a consequence of processing of spontaneous DNA lesions. Using experimental systems based on reversions of frameshift alleles in Saccharomyces cerevisiae, we evaluated the impact of defects in DNA double-strand break (DSB) repair on the frequency of replication-independent mutations. The deletion of the genes coding for Ku70 or DNA ligase IV, which are both obligatory constituents of the non-homologous end joining (NHEJ) pathway, each resulted in a 50% reduction of replication-independent mutation frequency in haploid cells. Sequencing indicated that typical NHEJ-dependent reversion events are small deletions within mononucleotide repeats, with a remarkable resemblance to DNA polymerase slippage errors. Experiments with diploid and RAD52- or RAD54-deficient strains confirmed that among DSB repair pathways only NHEJ accounts for a considerable fraction of replication-independent frameshift mutations in haploid and diploid NHEJ non-repressed cells. Thus our results provide evidence that G0 cells with unrepressed NHEJ capacity pay for a large-scale chromosomal stability with an increased frequency of small-scale mutations, a finding of potential relevance for carcinogenesis.
Keywords: adaptive mutation/homologous recombination/mutational spectrum/selection-induced
Introduction
Most types of cell, whether they are unicellular organisms or members of a tissue of a multicellular organism, spend large parts of their life in a resting state. Thus, mutations originating in cell cycle-arrested cells may make a considerable contribution to the overall mutational benefit or burden of a population or tissue. In unicellular organisms, rare mutations that relieve the strong pressure of growth arrest may be regarded as beneficial since they allow the new genotype to thrive and spread while non-mutated cells continue to rest and eventually die. However, with respect to mammals, spontaneous mutations in genes responsible for signalling or execution of cell cycle arrest may lead to tumour initiation. As mutations like that present an advantage to the individual cell and, in several experimental setups, give the impression of being a response to selective pressure, the term ‘adaptive mutation’ was repeatedly used to describe them. Without bias, we shall refer to this type of mutation as ‘replication-independent mutation’. Mechanistically, the turnover and processing of damaged DNA by repair processes is suspected as the cause of replication-independent mutations. In contrast, spontaneous mutations in proliferating cells are mainly the consequence of DNA polymerase errors during replication, and, for better linguistic distinction, we shall not refer to them as replication-dependent mutations, but as ‘proliferation-dependent mutations’.
Initiated by observations made in the bacterium Escherichia coli (Cairns et al., 1988; reviewed by Foster, 2000; Rosenberg, 2001), replication-independent mutations in an eukaryotic organism were first reported for the budding yeast Saccharomyces cerevisiae in 1992 (Hall, 1992; Steele and Jinks-Robertson, 1992). Since then, different research groups have tried to elucidate the mechanisms underlying this special kind of spontaneous mutagenesis (Baranowska et al., 1995; Heidenreich and Wintersberger, 1997, 1998, 2001; Babudri et al., 2001; Cejka et al., 2001). During our studies of DNA sequence spectra resulting from reversions of frameshift alleles in yeast, we noticed a typical signature of reversions taking place during cell cycle arrest (Heidenreich and Wintersberger, 2001). In contrast to a highly variable spectrum of reversion sites found for proliferation-dependent reversions, we observed a prevalence of simple deletions in mononucleotide repeats for replication-independent reverse mutations of the same allele. Such a type of mutation is usually assigned to polymerase slippage errors that escaped mismatch repair or were corrected wrongly. These findings suggested the hypothesis that most replication-independent frameshift mutations are produced by error-prone DNA (re-)synthesis, probably induced by the processing of spontaneous DNA damage. Yet another possibility for the generation of frameshift-causing small deletions proceeding without DNA synthesis would be double-strand break (DSB) repair by non-homologous end joining (NHEJ).
The repair of DSBs is essential for the maintenance of genomic integrity. We can distinguish between three major processes of DSB repair: homologous recombination (HR), single-strand annealing (SSA) and NHEJ (reviewed by Pfeiffer et al., 2000; Pastink et al., 2001). HR and SSA (sometimes considered as a subpathway of HR) both rely on the existence of homologous sequences, 5′ to 3′ resection of DNA ends and the activity of the Rad52 protein, but differ considerably in other properties. HR is a mainly accurate mechanism that depends on an undamaged homologous donor sequence, normally located on the sister chromatid or a homologous chromosome. The hallmarks of HR are Rad50/Mre11/Xrs2-mediated end processing, Rad52-, Rad54-, Rad55- and Rad57-assisted formation of Rad51-complexed nucleoprotein filaments and strand invasion into the homologous donor duplex, DNA synthesis and, finally, resolution of the intermediate (reviewed by Paques and Haber, 1999). Unlike HR, SSA does not require a homologous chromosome for break repair, but tandem repeats on either side of the DSB. When these homologies are exposed by 5′ to 3′ resection of the broken ends, the Rad52 protein can catalyse annealing, subsequently leading to the covalent ligation of the two fragments. SSA does not involve Rad51, Rad54, Rad55 and Rad57 and, obligatorily, leads to the deletion of the region between the two repeats.
Alternatively, NHEJ restores the continuity of the broken chromosome by a joining mechanism without the need for a homologous sequence. NHEJ in S.cerevisiae is mediated by the DNA double-stranded end-recognizing and end-binding activities of the Yku70/Yku80 heterodimer (Doherty and Jackson, 2001) followed by the end-bridging function of the Rad50/Mre11/Xrs2 protein complex (Chen et al., 2001) and finalized by the action of DNA ligase IV, which consists of the catalytic subunit Dnl4p and the regulatory subunit Lif1p (for a review see Lewis and Resnick, 2000). Homologues of all these proteins are known in mammals. Recently, a pathway of regulation of NHEJ activity and its dependence on homozygosity versus heterozygosity of the mating type locus was described (Frank-Vaillant and Marcand, 2001; Kegel et al., 2001). This mechanism ensures a modulation of the different DSB repair pathways in accordance with the ploidy of the cell. It was shown that intracellular localization of Lif1p is regulated in response to mating type, resulting in a downregulation of NHEJ in diploid cells (Valencia et al., 2001).
The relative contributions of the alternative DSB repair pathways change with ploidy and cell cycle stage, but also differ between yeast and mammals. NHEJ and SSA, as well as gene conversion by HR, can make significant contributions to mammalian DSB repair (Liang et al., 1998). However, owing to a strong preference for the sister chromatid as a template for DSB repair by HR (Johnson and Jasin, 2000), the importance of HR is expected to diminish in G0 cells where sister chromatids are not available. In contrast, DSB repair by homologous recombination mechanisms predominates in S.cerevisiae. Nevertheless, NHEJ in yeast is important in special situations. It has recently been reported that NHEJ efficiency increases in the G1 stage of the cell cycle and after transition to the stationary phase (Karathanasis and Wilson, 2002). A study of DSB repair pathways other than HR in resting G1/G0 yeast cells is facilitated experimentally by the use of haploid strains.
In this study, we investigated whether DNA break repair mechanisms contribute to frameshift mutagenesis in cell cycle-arrested yeast cells. By using diploid strains and strains deficient in DSB repair due to either defects in homology-dependent repair pathways or in non-homologous end joining, we were able to identify an important role for NHEJ in creation of mutations in resting cells. The implications of this novel mechanism, which is probably DNA polymerase independent, of spontaneous mutagenesis in non-replicating cells are discussed.
Results
Assay for the observation of replication-independent mutations
A system that relies on growth-permitting reversions of frameshift alleles in the budding yeast S.cerevisiae was set up for the study of mutations arising in non-replicating cell populations. The allele mainly used, lys2ΔBglII, was constructed by filling-in a BglII restriction site of the LYS2 gene; the resulting +4 shift in the open reading frame causing an auxotrophy for lysine. A second frameshift detection system was based on the hom3-10 allele in which a +1 mutation in the HOM3 coding sequence causes auxotrophy for threonine and methionine. Following the transfer from the growth medium to the selective medium without the essential amino acid, cells typically enter cell cycle arrest as unbudded G1/G0 cells (see Supplementary figure 1 available at The EMBO Journal Online). Only if the correct reading frame of the lys2 or hom3 allele is restored by a compensating frameshift mutation, is relief of the selective pressure (lack of an essential amino acid) possible by means of the production of the functional enzyme. Subsequently, such events allow DNA replication and give rise to colonies that are available for further analysis. In a first wave, 2–3 days after the exposure to the selective medium, revertant colonies originating from reversion events during the preculture phase become visible, as has been proved previously by reconstruction growth tests (Heidenreich and Wintersberger, 2001). Some pre-existing hom3-10 revertants need 4 days to form readily visible colonies (Heidenreich and Wintersberger, 1998).
In the present study, we included colonies newly arising on day 4 in the proliferation-dependent mutant count to account for the possibility of slowly growing revertant clones, even in the case of the lysine starvation system. These cumulative counts were used to calculate the mutation rates of all strains (Table I) by using the method of the median (Lea and Coulson, 1949). Following the appearance of these colonies formed by pre-existing mutants, further colonies continued to arise as a result of replication-independent reversion events that occurred during starvation-induced cell cycle arrest. The origin of these latter clones is the matter of our interest. To make a clear distinction between replication-independent and replication-based reversion events, we show the increase of colony numbers from day 4 onwards (cf. Figures 1, 2 and 5).
Table I. Mutation rates of proliferation-dependent reversions.
| Test allele and relevant genotype | Reversion rate | Relation to respective wild types |
|---|---|---|
| lys2ΔBglII, haploid strains | ||
| Wild type | 1.10 ± 0.21 × 10–8 | |
| DNL4-deficient | 1.15 ± 0.12 × 10–8 | 1.0× |
| YKU70-deficient | 1.20 ± 0.27 × 10–8 | 1.1× |
| RAD52-deficient | 6.21 ± 1.42 × 10–8 | 5.6× |
| RAD54-deficient | 1.46 ± 0.16 × 10–8 | 1.3× |
| lys2ΔBglII, diploid strains | ||
| Wild type | 0.99 ± 0.05 × 10–8 | |
| DNL4-deficient | 0.87 ± 0.18 × 10–8 | 0.9× |
| YKU70-deficient | 1.35 ± 0.13 × 10–8 | 1.4× |
| RAD52-deficient | 5.78 ± 0.61 × 10–8 | 5.9× |
| RAD54-deficient | 1.49 ± 0.30 × 10–8 | 1.5× |
| MATa/a wild type | 1.75 ± 0.36 × 10–8 | 1.8× |
| hom3-10, haploid strains | ||
| Wild type | 3.45 ± 1.04 × 10–9 | |
| DNL4-deficient | 3.54 ± 0.34 × 10–9 | 1.0× |
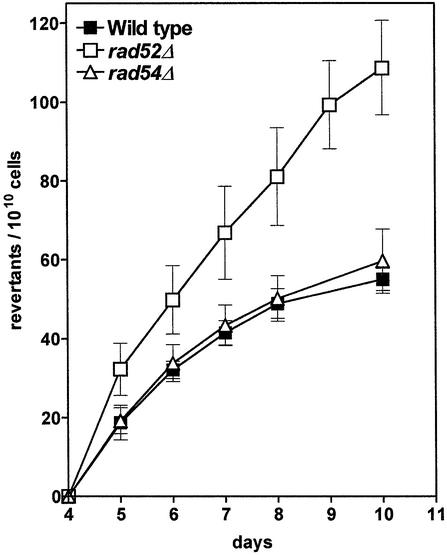
Fig. 1. Frequencies of replication-independent Lys+ revertant colonies in a haploid repair-proficient (wild type) and two DSB repair-deficient strains with deletions of RAD52 or RAD54, respectively. A time course starting with day 4 after plating is shown because earlier arising colonies were considered as originating (surely or potentially) from pre-existing revertants. Cumulative colony counts were normalized to the numbers of cells on the plates. The mean of three to four experiments is shown together with standard error bars.
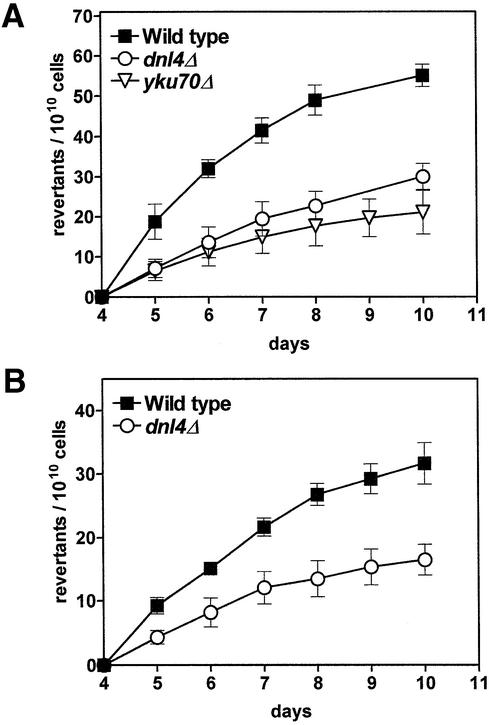
Fig. 2. Frequencies of replication-independent revertant colonies in haploid NHEJ-deficient strains. Two different frameshift alleles were analysed. (A) Reversion of lys2ΔBglII-caused lysine auxotrophy. (B) Reversion of hom3-10-caused methionine auxotrophy. Graph style as described for Figure 1.
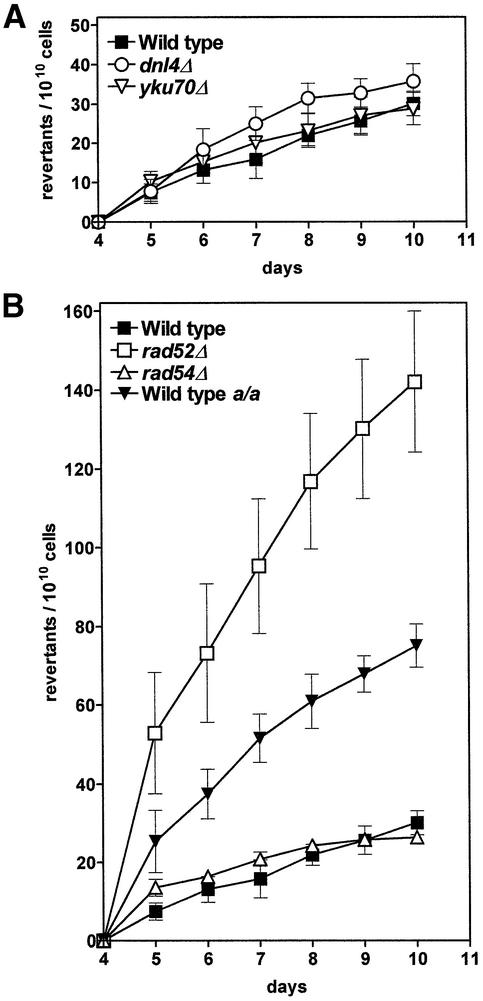
Fig. 5. Frequencies of replication-independent Lys+ revertant colonies in diploid strains. (A) NHEJ-deficient strains. (B) Strains deficient in homology-dependent DSB repair pathways, together with wild-type strains differing in mating type properties. Wild-type a/a is repair proficient and homozygous for the mating type allele MATa. All other strains are MATa/α. Graphs are drawn to the same scale; style as described for Figure 1.
Effects of homology-dependent DSB repair pathways on replication-independent mutation frequencies
To study the influence of DSB repair pathways on replication-independent mutations we constructed a set of haploid lys2ΔBglII strains defective in the major pathways of DSB repair. For the generation of a deficiency in HR, a RAD54-deletion strain was constructed. Rad54p is a DNA-dependent ATPase that stimulates Rad51p-mediated strand exchange during recombination (Petukhova et al., 1998; Solinger and Heyer, 2001). To address a possible role of SSA, we constructed a RAD52-deficient strain. In contrast to RAD54, which is involved solely in HR, RAD52 has a dual function in both HR and SSA. Therefore an effect of SSA should become apparent in the phenotypic differences between a RAD54-deleted (rad54Δ) and a RAD52-deleted (rad52Δ) strain. The replication-independent revertant frequencies of the haploid rad54Δ and rad52Δ strains were compared with that of the isogenic DSB repair-proficient strain (Figure 1). Whereas the deletion of RAD54 did not cause any change, the incidence of replication-independent revertants was increased significantly in the rad52Δ strain, indicating a RAD52-dependent frameshift-antagonizing process.
The frequency of replication-independent frameshift reversions is reduced to ∼50% in NHEJ-deficient strains
To evaluate a possible contribution of NHEJ to the mutation formation in cell cycle-arrested cells, we employed lys2ΔBglII strains deficient for DNL4 (dnl4Δ) or YKU70 (yku70Δ). DNL4 codes for the catalytic subunit of DNA ligase IV, the only activity in yeast cells for sealing DSBs (Schär et al., 1997; Teo and Jackson, 1997; Wilson et al., 1997). No additional function has yet been reported for this protein, which made it a suitable candidate for our purpose. The deletion of the YKU70 gene abolishes the function of the Yku70/Yku80 heterodimer. As, unlike DNA ligase IV, this heterodimer is involved in additional cellular processes (e.g. telomere maintenance), NHEJ-independent side-effects might influence the result.
In both haploid NHEJ-deficient strains (dnl4Δ and yku70Δ), the lys2ΔBglII reversion assay resulted in significantly decreased replication-independent mutation frequencies compared with the repair-proficient wild type (Figure 2A). Approximately half of the reversion events were abolished in both the dnl4Δ and the yku70Δ cells, a remarkable finding pointing to a mutagenic role of the NHEJ pathway in resting yeast cells. To examine whether this effect is also true for another frameshift allele, we made use of the hom3-10 allele. An isogenic pair of hom3-10 strains, either repair proficient (wild type) or NHEJ deficient by deletion of the DNL4 gene, was prepared and analysed with regard to the occurrence of revertant colonies during starvation for methionine (Figure 2B). Analogously to the lys2ΔBglII system, a decrease of ∼50% in the replication-independent revertant frequency was observed after the inactivation of DNA ligase IV.
Sequence analysis of frameshift reversions in the DNA ligase IV-deficient strain
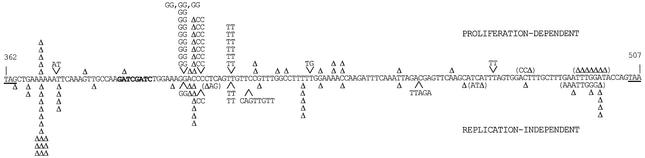
In previous work, we reported a difference between the mutational spectra of proliferation-dependent and replication-independent reversions of the lys2ΔBglII allele (Heidenreich and Wintersberger, 2001). Mutations arising during proliferation resulted in a wide-ranging spectrum of different mutation types. However, replication-independent mutations showed a significant bias for simple deletions in mononucleotide runs. As we now observed a reduced replication-independent mutation frequency in a DNL4-deficient strain, we were interested in the resulting effect on the mutational spectrum. Therefore the relevant region of the LYS2 gene derived from the genomic DNA of 58 proliferation-dependent and 67 late-arising revertant colonies was sequenced (Figure 3). In addition to the reversion events indicated in Figure 3, two proliferation-dependent and eight replication-independent revertant clones showed no alteration in this reversion window of the LYS2 gene. As we were primarily interested in locus reversion at the LYS2 gene, we did not characterize the extragenic origin of these frameshift suppressions further and thus all the following statistical comparisons of mutation types are based on intragenic reversions only.
Fig. 3. Sequence analysis of lys2ΔBglII reversions in the haploid DNA ligase IV-deficient strain. Only the relevant part of the LYS2 allele from nucleotides 362 to 507 is shown (numbered relatively to the translation start). Reverting frameshifts of the lys2ΔBglII-specific GATC duplication (bold) must take place in the represented sequence stretch between two out-of-frame stop codons (underlined) with respect to the wild-type reading frame. Fifty-six intragenic proliferation-dependent reversions are indicated above the sequence, and 59 intragenic replication-independently generated reversions are displayed below the sequence. Symbols: –1 deletions are shown by Δ; inserted nucleotides are shown together with arrow heads; individual complex sequence alterations (e.g. deletions accompanied by base substitutions) are displayed in parentheses.
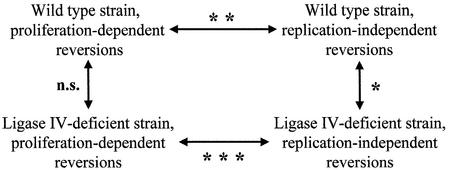
At first sight, the spectra of proliferation-dependent and replication-independent DNL4-deficient revertants differ in their distribution of ‘hotspots’ over the sequence range, where reverting frameshifts can take place. An analysis of the overall similarity between the two spectra (Figure 4, lower part) found them to be significantly different (P << 0.001). Comparison with previously published reversion spectra of the isogenic wild-type strain revealed that the differences between the two genotypes (Figure 4, vertical comparisons) were not as prominent as those between the two modes of mutagenesis, proliferation dependent or replication independent (Figure 4, horizontal comparisons). The similarity of the two proliferation-dependent reversion spectra (Figure 4, left-hand side) and the calculated mutation rates (Table I) document that a lack of Dnl4p leads to a change in neither quality nor quantity of reversion events in proliferating haploid cells. In contrast, the two sets of replication-independent reversion sequences differ not only in quantity (Figure 2A), but also in their composition (Figure 4, right-hand side).
Fig. 4. Statistical analysis of reversion sequence spectra. The similarities of pairs of sequence datasets of haploid wild-type or DNA ligase IV-deficient revertants were calculated using the algorithm of Adams and Skopek, available as a computer program (Cariello et al., 1994). Significance levels are indicated as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., no significant difference. Wild-type sequence data are derived from Heidenreich and Wintersberger (2001).
More information can be gained if we group the mutation events into types for a comparative evaluation (Table II). Whereas in the DNL4-deficient strain the percentage of insertions and deletions is reasonably balanced in the proliferation-dependent subset of reversion events, there is a highly significant shift (P << 0.001) to a prevalence of deletions among the replication-independent reversion events. This extreme difference is partially due to three ‘hotspots’ for insertions (Figure 3, positions 402–403, 405–408 and 413–414) present in the proliferation-dependent subset but unobtrusive in the replication-independent subset. Considering the sequence context of deletions and insertions, the following was noticed. In general, similarly to the reversion spectra of wild-type cells (Heidenreich and Wintersberger, 2001), mononucleotide repeats are more prone to frameshift-reverting mutations, whether they are nucleotide gains or losses. Insertions in runs of three or more identical nucleotides (≥3 runs in Table II) are typical of proliferation-dependent reversion events (P = 0.012), whereas deletions in ≥3 runs predominate in replication-independent reversions (P = 0.022). If all alterations in mononucleotide runs of three or more are pooled (i.e. insertions plus deletions), the fractions of the total for proliferation-dependent and replication-independent subsets are similar (41.1% versus 49.2%). This is in contrast to the NHEJ-competent strain (Table II, lower part), where such ‘DNA polymerase slippage error resembling’ mutations are significantly allocated to the replication-independent mutants (26.3% versus 60.5%). In a direct comparison of replication-independent reversions in dnl4Δ and DNL4+ strains, alterations in mononucleotide runs are slightly reduced in the DNA ligase IV-deficient strain (49.2% versus 60.5%), whereas deletions of non-iterated bases are increased (Table II, last column).
Table II. Incidence of selected mutation types.
| Data sets of lys2ΔBglII revertant sequences | Incidence (%) |
||||
|---|---|---|---|---|---|
| Insertions total | Insertions in ≥3 runs | Deletions total | Deletions in ≥3 runs | Deletions of non-iterated nucleotides | |
| dnl4Δ, proliferation-dependent | 48.2 | 14.3 | 51.8 | 26.8 | 5.4 |
| dnl4Δ, replication-independent | 10.2 | 1.7 | 89.8 | 47.5 | 20.3 |
| DNL4+, proliferation-dependenta | 36.8 | 10.5 | 63.2 | 15.8 | 13.2 |
| DNL4+, replication-independenta | 10.5 | 0.0 | 89.5 | 60.5 | 7.9 |
aData derived from Heidenreich and Wintersberger (2001).
Generally, the frequency of replication-independent frameshift reversions in diploid strains is reduced to ∼50% of the level found for haploid NHEJ-proficient strains
As the experiments with haploid strains indicated that DSB repair pathways play a role in replication-independent mutagenesis, we proceeded with diploid strains in order to study the influence of homologous recombination or ploidy-specific regulation. A diploid repair-proficient strain and strains homozygous for deletions of DNL4, YKU70, RAD52 or RAD54, respectively, were constructed, all of them containing the lys2ΔBglII frameshift detection allele. Figure 5 shows the results of assays for replication-independent mutagenesis in resting cells. In contrast to the haploid strain set, no significant difference was observed between the revertant frequencies of the repair-proficient wild type and both NHEJ-deficient strains (Figure 5A). Instead, the curves cluster around the same basal level (∼29 revertant colonies/1010 cells by day 10) observed with the haploid NHEJ-deficient strains (cf. Figure 2A). This concurrence could indicate that NHEJ is not active in diploid wild-type cells (see below). The replication-independent revertant frequencies of the diploid rad54Δ strain also decreased to the same basal level, thus showing no significant difference from the wild type. The rad52Δ strain was the only repair-deficient strain to show a deviating mutation frequency. Analogously to data gained from of the haploid strain set, the deletion of the RAD52 gene resulted in a substantially increased replication-independent reversion frequency (Figure 5B).
The frequency of replication-independent frameshift reversions in a MATa/a diploid strain resembles those of haploid NHEJ-proficient strains
If the conclusion derivable from Figure 2, namely that NHEJ is responsible for ∼50% of replication-independent mutations in the haploid wild-type strain, is valid, we have to postulate that for the diploid strains NHEJ should be inactive to explain the low level of revertants, as reported above. In fact, NHEJ is known to be downregulated in MATa/α diploid cells (for details see the Discussion and references cited therein). To investigate whether diploid strains also acquire higher frequencies of replication-independent mutants if NHEJ is active, we provoked NHEJ activity in diploid cells by constructing a MATa/a strain. This strain is homozygous for the a allele of the mating type locus and therefore is non-repressed in its NHEJ activity (E.Heidenreich, V.Holzman and F.Steinböck, unpublished data), although the genome exists in duplicate. Indeed, the frequency of replication-independent revertants was found to be elevated in the MATa/a strain compared with the MATa/α strain (Figure 5B); it rose to a level even higher than that found for the haploid strain (cf. Figure 2A).
Discussion
There are two main ways of mutation generation: (i) DNA polymerase errors that escape repair and undergo fixation as mutations; and (ii) DNA lesion-triggered mutation formation. In principle, both could be active in cell cycle-arrested cells but, owing to the lack of gross DNA replication, type (i) is limited to errors introduced by repair DNA synthesis, which again is likely to be evoked by DNA lesions. The integrity of the genome is constantly endangered by a wide range of spontaneously occurring lesions (Lindahl, 1993; Gupta and Lutz, 1999). The stressful cell cycle-arresting situation of nutrient limitation may well enhance levels of endogenous lesions. Support for this assumption comes from the observation that mitochondrial genomes were progressively degenerating when yeast cells were starved for essential amino acids (Heidenreich and Wintersberger, 1997).
In this study, we wanted to evaluate the importance of DSB repair pathways for spontaneous mutagenesis in arrested cells. The experimental setup used relies on the detection of frameshift mutations in either the lys2ΔBglII or the hom3-10 test allele. In both cases, the reading frame can be restored by compensating small deletions or insertions of nucleotides within a distinct region of the gene surrounding the original frameshift site. Base-substitution mutations are not observable in our systems, except when they accompany a reversion-causing frameshift. Frameshift mutations are highly relevant to human pathogenesis, as they can lead to protein truncations and thereby to a complete loss of protein function. A high incidence of frameshift mutations may become apparent in microsatellite instability.
Using the lys2ΔBglII reversion assay, we observed that strains deficient for the core NHEJ factors (DNA ligase IV or Ku70) exhibited only 38–55% of the revertant frequency of the wild type, while at the same time their viability showed no significant differences (see Supplementary figure 2). These results led us to the conclusion that NHEJ activity was essential for the production of approximately half of all replication-independent frameshift mutations observed in the wild type. In contrast, the rate of proliferation-dependent frameshift reversions was not found to be altered by deletion of DNL4 or YKU70 (Table I). A study of the influence of a DNA ligase IV deficiency on hom3-10 reversions confirmed this NHEJ dependence of ∼50% of replication-independent frameshifts in another genetic system. Together, these findings enabled us to identify the NHEJ pathway as an important factor for the production of frameshift mutations in cell cycle-arrested cells.
The most obvious hypothesis for an explanation of this 50% reduction is that DSBs that have arisen spontaneously may have experienced a small degradation or trimming of DNA ends, followed by DNA ligase IV-mediated religation of the broken ends. In such a way, error-prone NHEJ may convert endogenously induced DSBs to frameshift mutations. As a consequence of the ligation of truncated ends, NHEJ would be expected to produce primarily deletions. Accordingly, in a comparison of dnl4Δ and DNL4+ replication-independent reversion sequence spectra, some deletions should be missing in the dnl4Δ spectrum while insertions should be overrepresented. Surprisingly, this was not the case (Table II), as the proportion of insertions was ∼10% in both spectra. Therefore, we have to postulate that not only deletions but also a small number of insertions originated from NHEJ. Insertions may be infrequently produced by NHEJ if the substrate lesion is a staggered break (Moore and Haber, 1996). Microhomologies on the protruding ends may trigger annealing, and a fill-in synthesis of one or a few nucleotides would be required to fill the resulting gaps. Nevertheless, deletions represented the major reversion type with or without functional DNA ligase IV. Previously, we reported that replication-independent reversion of the lys2ΔBglII allele in the repair-proficient parent strain had shown a strong bias for deletions of one nucleotide within a mononucleotide run (Heidenreich and Wintersberger, 2001). This effect was somewhat diminished by the deletion of DNL4 (Table II). Deletions in mononucleotide repeats still predominated over other mutation types in the replication-independent reversion spectrum, but not as strongly as in the repair-proficient strain. On the other hand, deletions of non-iterated nucleotides were found more frequently in the replication-independent mutation spectrum of the dnl4Δ strain. This shift in the mutational spectrum, together with the quantitative reduction of replication-independent reversions to ∼50% in the dnl4Δ strain, allows the following conclusion: most of the reversion events that were abolished in the DNL4-deleted strain and therefore probably produced by NHEJ in the DNL4+ strain were –1 deletions within mononucleotide repeats. Thus NHEJ, without participation of DNA polymerases, is able to produce a spectrum of mutations closely resembling typical DNA polymerase slippage errors that escaped correction by proofreading or mismatch repair. In other words, ∼50% of the microsatellite instability-like mutations found in our replication-independent NHEJ-competent revertants may actually have been produced by NHEJ and not by inaccurate DNA synthesis.
It is not yet known whether homopolymeric runs are more prone to DSBs during cell cycle-arresting limitation for a certain nutrient component than non-iterative sequences, and whether for this reason mutations in such monotonous sequences predominate. Alternatively, if DSBs arise in a random distribution, they might be repaired most efficiently in mononucleotide repeats, perhaps because of the existence of short complementarity even after a shift in alignment of the two DNA strands.
Since the first reports on adaptive mutations in yeast (Hall, 1992; Steele and Jinks-Robertson, 1992), only haploid strains have been used for analyses. We proceeded with diploid strains and obtained results that complemented our data from haploid strains. The replication-independent revertant frequency of the diploid repair-proficient wild type was already found in the range of 50% of the frequency of the haploid wild type and thus correlated with the frequencies of the haploid NHEJ-deficient strains. Diploid strains homozygously deleted for DNL4 or YKU70 did not show a further decrease in replication-independent revertants. If our hypothesis that NHEJ is responsible for half of the replication-independent frameshift mutations is correct, these results would imply that the NHEJ pathway is not active in diploid cell cycle-arrested cells. Indeed, NHEJ activity in S.cerevisiae is known to be regulated in response to mating type. Heterozygosity at the mating type locus triggers suppression of NHEJ by means of a mechanism proposed recently (Frank-Vaillant and Marcand, 2001; Kegel et al., 2001; Valencia et al., 2001). Haploid cells or MAT-homozygous diploid cells express either MATa or MATα genes and are not prone to suppression of DNA ligase IV activity by downregulation of the cofactor, Nej1p/Lif2p, which regulates the subcellular distribution of Lif1p, the regulatory subunit of DNA ligase IV. This mechanism ensures that HR is the preferred pathway for DSB repair in S.cerevisiae except when no duplicate of the genome is available, and therefore NHEJ is a better choice for preserving the integrity of the genome. To evaluate the influence of NHEJ regulation on replication-independent mutation, we constructed and analysed a diploid strain with two MATa alleles. Such strains show various characteristics of haploid strains; for example, they are unable to sporulate but are able to mate with MATα partners. Unlike the situation in MATa/α diploids, NHEJ is not suppressed in diploid strains that express only one type of the two alter native mating type alleles (Åström et al., 1999; Lee et al., 1999). We observed that the replication-independent mutation frequency in the MATa/a strain was increased considerably compared with the MATa/α diploid, which further supports our hypothesis that a substantial part of replication-independent frameshift mutations in arrested haploid cells was produced by NHEJ. Intriguingly, the mutation frequency was even higher than that of the corresponding haploid strain (Figures 2A and 5B). As a reversion of one of the two lys2ΔBglII alleles present in diploids is sufficient for lysine prototrophy, diploid cells should in principle have up to a 2-fold chance of haploid cells for such reversion events. Obviously, such an increase is only identifiable in the reversion frequencies of NHEJ-derepressed strains like the MATa/a strain.
A lack of NHEJ activity abolished only ∼50% of replication-independent frameshift reversions in our experimental system. The remaining level of mutations still observable in all NHEJ-deficient or NHEJ-repressed strains was fairly constant with regard to strains that differed in ploidy and DSB repair capacity and therefore was probably independent of DSB processing. The molecular mechanisms responsible for the formation of this remaining NHEJ-independent portion of replication-independent frameshift mutations await elucidation. It is conceivable that these mechanisms, in contrast to NHEJ, are connected to error-prone DNA synthesis in the course of repair of spontaneous DNA lesions other than DSBs.
To make a comprehensive survey of all major DSB repair pathways, we also studied replication-independent mutations in strains deficient for HR or SSA. Clearly, the mere incidence of replication-independent mutants in haploid wild-type strains indicates that HR cannot be essential for these mutations since a homologous second copy of the genome is missing in these cell cycle-arrested cells. As expected in the haploid strain set, a deficiency for RAD54 neither reduced nor increased the frequency of replication-independent mutations compared with the wild type. Owing to the preference of S.cerevisiae for DSB repair by HR, we expected a significant protective effect of the mainly accurate HR pathway in diploid strains. Surprisingly, this was not observed. The revertant frequencies of the RAD54+ and the rad54Δ strain pair differed only marginally. On the one hand, this could indicate that mutagenic NHEJ is downregulated in diploids so rigorously that the remaining level of reversions is not elicited by DSBs, so that HR has no influence. On the other hand, we cannot rule out that early steps of HR, especially the production of long 3′ single-stranded tails, are accomplished in rad54Δ strains in almost the same manner as in wild-type strains. Such resection of the DNA ends might irreversibly prevent NHEJ, irrespective of further processing, and could thus lead to an underestimation of an antimutagenic role of HR in diploid G0 cells.
Additional information was gained by the use of RAD52-deleted strains. In contrast to the rad54Δ strains, the revertant frequencies of the rad52Δ strains, both haploid and diploid, were significantly increased compared with wild-type strains of the same ploidy. This frameshift-promoting effect of a RAD52 deficiency was more pronounced in the diploid strain (5-fold wild type) than in the haploid strain (2-fold wild type), pointing to an increased importance of RAD52 in frameshift prevention in diploid cells. As NHEJ is the principal remaining DSB repair pathway in rad52Δ strains, it is most plausible that the increased incidence of frameshifts in rad52Δ strains is produced by error-prone NHEJ. Alternatively, the RAD52 deficiency may elicit an additional mutagenic response by either the unknown mechanism that contributed to the NHEJ-independent ‘basal level’ discussed above or by yet another process that did not contribute to the wild-type reversion level.
What actually is the RAD52-dependent process that counteracts frameshift-producing pathways? In haploid cells, it might predominantly be the SSA pathway, since there is no homologous template for HR. However, a dominance of SSA seems to be a paradox because SSA obligatorily leads to large deletions, an even more severe type of mutation. In diploid cells, the mechanism in question might be a mixture of SSA and HR, especially with respect to the larger extent of the RAD52-dependent effect on frameshift reversions. The apparent discrepancy with the wild-type-like reversion frequency of the rad54Δ strain could be explained by a redundancy between SSA and HR based on common early steps. As stated above, resection of double-stranded ends could suppress NHEJ but, in diploid cells, still provide the choice between HR and SSA.
Our experimental system does not allow a distinction between error-free repair, repair producing large deletions or a failure to repair DSBs. In all cases, cell cycle arrest is maintained and these events escape detection. Therefore the presumptive common outcome of HR and SSA hampers the study of a possible competition between the different DSB repair pathways for their common substrate DSBs, and it is evident that the revelation of pathway channelling and interactions among different DSB repair pathways in cell cycle-arrested cells will require a different approach. Our results clearly demonstrate that, among the DSB repair pathways tested, only NHEJ contributes substantially to the formation of spontaneously occurring frameshift mutations in resting cells and documents that HR and SSA counteract rather than promote this type of mutation.
Prokaryotic ‘adaptive mutation’ is a phenomenon that shares some similarities with replication-independent mutation in yeast. Research on adaptive mutation in prokaryotes is dominated by one particular system, the E.coli FC40 assay of reversions of a lac frameshift allele located on an F′ episome (reviewed by Foster, 2000; Rosenberg, 2001). The proposed mechanism underlying these reversions is complex, but basically is described as error-prone DNA synthesis primed by recombinational DSB repair. The strict requirement for recombination proteins (and for episome-specific functions) clearly distinguishes this type of mutation from the replication-independent mutations that we characterized in S.cerevisiae. If the same frameshift allele was placed on the E.coli chromosome instead of on the episome [reviewed together with other chromosomal alleles elsewhere (Foster, 1999)], the adaptive reversion frequency dropped 100-fold to a level similar to that observed in yeast (about one reversion per 109 cells per day). The genetic requirements for such non-episomal frameshift mutations are largely uncharacterized. Therefore, especially in the light of the recent discovery of a NHEJ complex in prokaryotes (Weller et al., 2002), it is too early to rule out that some mechanisms of (chromosomal) replication-independent mutagenesis may be conserved between prokaryotic and eukaryotic organisms.
For mammals, growing evidence suggests an important role of DSB repair pathways in tumorigenesis (Pierce et al., 2001). A defect in NHEJ results in a high frequency of genomic instability (Ferguson et al., 2000). With respect to the hereditary breast and ovarian cancer susceptibility genes BRCA1 and BRCA2, evidence for a role in DSB repair by HR has been found (Scully and Livingston, 2000; Pierce et al., 2001). These findings underline that DSB repair mechanisms by HR and NHEJ function as caretakers of the genome. In contrast to HR, NHEJ risks altering the local sequence at the junction. While this may be a minor drawback compared with the benefit of healing a broken chromosome, it can pose a serious threat if tumour suppressor genes or regulatory sequences of proto-oncogenes are affected.
In summary, our data indicate that NHEJ does not obviously contribute to frameshift mutagenesis in proliferating haploid and diploid yeast cells, whereas, on the other hand, we observed a substantial contribution of NHEJ to frameshift mutagenesis in non-proliferating haploid and MATa/a diploid cells. Organism-specific differences in the regulation of NHEJ foster the speculation that diploid NHEJ non-repressed MATa/a cells might particularly mimic mammalian cells in their proneness to NHEJ-mediated DSB repair, which, as shown here, is mutagenic. Based on our experimental system and the present findings, we will be able to study mutagenic processes in non-replicating cells in further detail and reveal mechanisms of potential relevance for evolution, ageing and cancer.
Materials and methods
Strain construction
The genotypes of the S.cerevisiae strains used are listed in Table III. The strains EH100, EH140 and EH150 were described previously (Heidenreich and Wintersberger, 1998, 2001). The strains EHV4, YLB4, YLB52 and YLB70 were constructed by oligo-directed gene disruption of the repair genes indicated. Details of the procedure are described in the Supplementary data. For the generation of a RAD54-deleted strain, EH150 was crossed with the congenic strain BGY106. Haploid segregants with the desired genotype were back-crossed to EH150 background three times to generate the largely isogenic RAD54-deleted strain YLB54. All other haploid strains are absolutely isogenic to EH150 or EH100. Strain YNNLα was obtained from EH140 by inducing mating type switching by expression of the HO endonuclease from plasmid YCp50-HO (Herskowitz and Jensen, 1991). Diploid cells resulting from mating of original cells and switched cells were isolated. Plasmid loss was selected for by a medium containing fluoro-orotic acid (FOA) (Toronto Research Chemicals Inc., North York, Canada), which is a counterselection for the URA3 marker of the plasmid. Subsequently, after sporulation and tetrad dissection, YNNLα was isolated as a meiotic segregant. All diploid strains were produced by a two-step procedure: first, MATα strains corresponding to EH150, YLB4, YLB52, YLB54 and YLB70 were obtained as segregants of diploids formed by mating the respective strains with YNNLα (as a donor of MATα); secondly, the isogenic MATa and MATα pairs were mated individually by micromanipulation. Strain YLBDa/a was produced by inducing mating type switching in strain YLBD. YLBD was transformed with plasmid pGal-HO, which harbours the counterselectable URA3 marker and the HO endonuclease gene under control of the galactose-inducible GAL10 regulatory region (Herskowitz and Jensen, 1991). The HO endonuclease was induced by incubation in a galactose medium for 3 h. Switching was stopped by transfer to a glucose medium and plasmid loss was selected for by FOA medium. A clone that mated with MATα tester strains was isolated as strain YLBDa/a. Correct ploidy was tested by mating with different haploid and diploid strains and subsequent spore viability analysis (Herskowitz and Jensen, 1991), and by checking the mutation rate at the CAN1 locus.
Table III. Saccharomyces cerevisiae strains.
| Name | Genotypea | Source |
|---|---|---|
| EH100 | MATa hom3-10 lys2-801a | Heidenreich and Wintersberger (1998) |
| EHV4 | MATa hom3-10 lys2-801a dnl4::HIS3 | This study |
| EH140 | MATa LYS2 | Heidenreich and Wintersberger (2001) |
| YNNLα | MATα LYS2 | This study |
| BGY106 | MATa lys2-801a rad54::TRP1 | YGSCb |
| EH150 | MATa lys2ΔBglII | Heidenreich and Wintersberger (1998) |
| YLB4 | MATa lys2ΔBglII dnl4::HIS3 | This study |
| YLB52 | MATa lys2ΔBglII rad52::HIS3 | This study |
| YLB54 | MATa lys2ΔBglII rad54::TRP1 | This study |
| YLB70 | MATa lys2ΔBglII yku70::HIS3 | This study |
| YLBD | MATa/α homozygous diploid of EH150 | This study |
| YLB4D | MATa/α homozygous diploid of YLB4 | This study |
| YLB52D | MATa/α homozygous diploid of YLB52 | This study |
| YLB54D | MATa/α homozygous diploid of YLB54 | This study |
| YLB70D | MATa/α homozygous diploid of YLB70 | This study |
| YLBDa/a | MATa/a diploid, derived from YLBD | This study |
aAll strains contain the following markers: trp1-Δ, his3-Δ200, ura3-52 and ade2-1o.
bYGSC, Yeast Genetic Stock Center at American Type Culture Collection, Manassas, VA.
Determination of replication-independent mutation frequencies
The experimental procedure was described previously (Heidenreich and Wintersberger, 1998). Briefly, several independent subpopulations of the respective auxotrophic strains were started from single colonies. A fluctuation assay system (Luria and Delbrück, 1943) was arranged to allow an estimation of the proliferation-dependent mutation rate. Then subpopulations were transferred from liquid complete medium to solid selective medium (synthetic complete medium minus lysine for lys2 strains; modified synthetic complete medium minus methionine for hom3 strains) and the appearance of colonies was monitored for 2 weeks. The plating density was 108 cells/plate for lys2 strains, and 2 × 108 cells/plate for hom3 strains. Each experimental series consisted of 13 subpopulations plated on to four plates each.
Sequence analysis
According to rigorous tests described earlier (Heidenreich and Wintersberger, 2001), colonies originating from cells that had already reverted to lysine prototrophy during the proliferation in complete medium appear until day 3 after plating. Thus, for the determination of sequence spectra of proliferation-dependent lys2ΔBglII revertants, day 3 colonies were used while, for the analysis of replication-independent reversions, colonies that appeared on days 7–9 were collected. Sequencing was performed as described previously (Heidenreich and Wintersberger, 2001). Mutation types were statistically compared using χ2 tests of independence (Sokal and Rohlf, 1995).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
This work was supported by a grant from the Herzfeldersche Familienstiftung to E.H.
References
- Åström S.U., Okamura,S.M. and Rine,J. (1999) Yeast cell-type regulation of DNA repair. Nature, 397, 310. [DOI] [PubMed] [Google Scholar]
- Babudri N., Pavlov,Y.I., Matmati,N., Ludovisi,C. and Achilli,A. (2001) Stationary-phase mutations in proofreading exonuclease-deficient strains of the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics, 265, 362–366. [DOI] [PubMed] [Google Scholar]
- Baranowska H., Policinska,Z. and Jachymczyk,W.J. (1995) Effects of the CDC2 gene on adaptive mutation in the yeast Saccharomyces cerevisiae. Curr. Genet., 28, 521–525. [DOI] [PubMed] [Google Scholar]
- Cairns J., Overbaugh,J. and Miller,S. (1988) The origin of mutants. Nature, 335, 142–145. [DOI] [PubMed] [Google Scholar]
- Cariello N.F., Piegorsch,W.W., Adams,W.T. and Skopek,T.R. (1994) Computer program for the analysis of mutational spectra: application to p53 mutations. Carcinogenesis, 15, 2281–2285. [DOI] [PubMed] [Google Scholar]
- Cejka P., Vondrejs,V. and Storchova,Z. (2001) Dissection of the functions of the Saccharomyces cerevisiae RAD6 postreplicative repair group in mutagenesis and UV sensitivity. Genetics, 159, 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Trujillo,K., Ramos,W., Sung,P. and Tomkinson,A.E. (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell, 8, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Doherty A.J. and Jackson,S.P. (2001) DNA repair: how Ku makes ends meet. Curr. Biol., 11, R920–R924. [DOI] [PubMed] [Google Scholar]
- Ferguson D.O., Sekiguchi,J.M., Chang,S., Frank,K.M., Gao,Y., DePinho,R.A. and Alt,F.W. (2000) The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA, 97, 6630–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P.L. (1999) Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet., 33, 57–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P.L. (2000) Adaptive mutation: implications for evolution. BioEssays, 22, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M. and Marcand,S. (2001) NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev., 15, 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.C. and Lutz,W.K. (1999) Background DNA damage for endogenous and unavoidable exogenous carcinogens: a basis for spontaneous cancer incidence? Mutat. Res., 424, 1–8. [PubMed] [Google Scholar]
- Hall B.G. (1992) Selection-induced mutations occur in yeast. Proc. Natl Acad. Sci. USA, 89, 4300–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich E. and Wintersberger,U. (1997) Starvation for a specific amino acid induces high frequencies of rho– mutants in Saccharomyces cerevisiae. Curr. Genet., 31, 408–413. [DOI] [PubMed] [Google Scholar]
- Heidenreich E. and Wintersberger,U. (1998) Replication-dependent and selection-induced mutations in respiration-competent and respiration-deficient strains of Saccharomyces cerevisiae. Mol. Gen. Genet., 260, 395–400. [DOI] [PubMed] [Google Scholar]
- Heidenreich E. and Wintersberger,U. (2001) Adaptive reversions of a frameshift mutation in arrested Saccharomyces cerevisiae cells by simple deletions in mononucleotide repeats. Mutat. Res., 473, 101–107. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. and Jensen,R.E. (1991) Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol., 194, 132–146. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Jasin,M. (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J., 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis E. and Wilson,T.E. (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics, 161, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A., Sjostrand,J.O. and Åström,S.U. (2001) Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol., 11, 1611–1617. [DOI] [PubMed] [Google Scholar]
- Lea D.E. and Coulson,C.A. (1949) The distribution of the numbers of mutants in bacterial populations. J. Genet., 49, 264–285. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Paques,F., Sylvan,J. and Haber,J.E. (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol., 9, 767–770. [DOI] [PubMed] [Google Scholar]
- Lewis L.K. and Resnick,M.A. (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res., 451, 71–89. [DOI] [PubMed] [Google Scholar]
- Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Luria S.E. and Delbrück,M. (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics, 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K. and Haber,J.E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastink A., Eeken,J.C. and Lohman,P.H. (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res., 480–481, 37–50. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Goedecke,W. and Obe,G. (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis, 15, 289–302. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Stark,J.M., Araujo,F.D., Moynahan,M.E., Berwick,M. and Jasin,M. (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol., 11, S52–S59. [DOI] [PubMed] [Google Scholar]
- Rosenberg S.M. (2001) Evolving responsively: adaptive mutation. Nature Rev. Genet., 2, 504–515. [DOI] [PubMed] [Google Scholar]
- Schär P., Herrmann,G., Daly,G. and Lindahl,T. (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev., 11, 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R. and Livingston,D.M. (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature, 408, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R.R. and Rohlf,F.J. (1995) Biometry: The Principles and Practice of Statistics in Biological Research. W.H. Freeman, New York, NY.
- Solinger J.A. and Heyer,W.D. (2001) Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl Acad. Sci. USA, 98, 8447–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D.F. and Jinks-Robertson,S. (1992) An examination of adaptive reversion in Saccharomyces cerevisiae. Genetics, 132, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.H. and Jackson,S.P. (1997) Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J., 16, 4788–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M., Bentele,M., Vaze,M.B., Herrmann,G., Kraus,E., Lee,S.E., Schär,P. and Haber,J.E. (2001) NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature, 414, 666–669. [DOI] [PubMed] [Google Scholar]
- Weller G.R. et al. (2002) Identification of a DNA nonhomologous end-joining complex in bacteria. Science, 297, 1686–1689. [DOI] [PubMed] [Google Scholar]
- Wilson T.E., Grawunder,U. and Lieber,M.R. (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature, 388, 495–498. [DOI] [PubMed] [Google Scholar]