Abstract
Sumoylation is involved in mediating protein–protein interactions, subcellular compartmentalization and protein stability. Our analysis of various Wnt signaling molecules revealed that one of them, Tcf-4, is sumoylated at the endogenous level. At least one sumoylation site, Lys297, of Tcf-4 was identified. The sumoylation of Tcf-4 was enhanced by PIASy, a SUMO E3 enzyme, and inhibited by Axam, a desumoylation enzyme. Although PIASy did not affect the interaction of Tcf-4 with β-catenin or DNA, Tcf-4, SUMO-1 and PIASy were co-localized in the nucleus and present in a complex in the PML body. PIASy enhanced β-catenin-dependent transcriptional activity of Tcf-4, whereas Axam inhibited it. Reduction of the protein level of Axam by RNA interference led to an increase in sumoylation of Tcf-4 and activation of Tcf-4. Furthermore, β-catenin and PIASy activated Tcf-4K297R, in which Lys297 was changed to arginine, less than wild-type Tcf-4. These results suggest that sumoylation of Tcf-4 is involved in β-catenin-dependent and Tcf-4-mediated gene expression in the Wnt signaling pathway.
Keywords: Axam/gene expression/PIAS/sumoylation/Tcf-4
Introduction
Wnt proteins constitute a large family of cysteine-rich secreted ligands that control development in organisms ranging from nematode worms to mammals (Wodarz and Nusse, 1998). The intracellular signaling pathway of Wnt is also evolutionarily conserved and regulates cellular proliferation and differetiation (Wodarz and Nusse, 1998; Bienz and Clevers, 2000; Seidensticker and Behrens, 2000). According to the most widely accepted current model, casein kinase Iα (CKIα) and glycogen synthase kinase-3β (GSK-3β) target cytoplasmic β-catenin for degradation in the absence of Wnt (Yost et al., 1996; Ikeda et al., 1998; Liu et al., 2002). Axin has been shown to form a complex with GSK-3β, CKIα, β-catenin and adenomatous polyposis coli (APC) protein, and to stimulate CKIα- and GSK-3β-dependent phosphorylation of β-catenin (Hart et al., 1998; Ikeda et al., 1998; Kishida et al., 1998; Kikuchi, 1999; Liu et al., 2002). Phosphorylated β-catenin forms a complex with Fbw1, a member of the F-box protein family, resulting in the degradation of β-catenin by the ubiquitin and proteasome pathways (Kitagawa et al., 1999).
When Wnt acts on its cell surface receptor consisting of Frizzled and lipoprotein receptor-related protein (LRP) 5/6, Dvl induces the accumulation of β-catenin in the cytoplasm by inhibiting the phosphorylation of β-catenin. Accumulated β-catenin is translocated to the nucleus, where it binds to the transcription factor T-cell factor (Tcf)/lymphoid-enhancer factor (Lef) and thereby stimulates the expression of various genes (Bienz and Clevers, 2000; Seidensticker and Behrens, 2000; Hurlstone and Clevers, 2002). Thus, post-translational modifications of β-catenin such as phosphorylation and ubiquitylation are important for the regulation of β-catenin stability.
In addition to the post-translational modifications of β-catenin in the cytoplasm, Tcf is also modified post-translationally in the nucleus. Drosophila CREB-binding protein (CBP) interacts with Drosophila Tcf and acetylates a conserved lysine in the Armadillo (Drosophila β-catenin)-binding domain of Drosophila Tcf (Waltzer and Bienz, 1998). This acetylation lowers the affinity of Tcf for Armadillo. NEMO-like kinase binds directly to and phosphorylates Tcf, which then inhibits the binding of the β-catenin–Tcf complex to DNA (Ishitani et al., 1999). Thus, it is likely that post-translational modifications of Tcf-4 such as acetylation and phosphorylation are important for its transcriptional activity.
The small ubiquitin-related modifier (SUMO) modification (sumoylation) pathway resembles the ubiquitin conjugation pathway, but the enzymes involved in these two processes are distinct (Hochstrasser, 2000; Yeh et al., 2000; Müller et al., 2001). There are three mammalian SUMOs, SUMO-1, SUMO-2 and SUMO-3, and one budding yeast homolog, Smt3. SUMO-1 is activated for conjugation by the E1 enzyme Aos1/Uba2, subsequently transferred to the E2 conjugation enzyme Ubc9, and finally conjugated to target proteins by either of the E3 ligases: protein inhibitor of activated STAT (PIAS) (Johnson et al., 1997; Saitoh et al., 1998; Kahyo et al., 2001; Sachdev et al., 2001) or RanBP2 (Kirsh et al., 2002; Pichler et al., 2002). Sumoylation is reversible, and there are at least seven mammalian SUMO-specific proteases, which are designated the SENP family proteins (Yeh et al., 2000). Among them, SENP1, SENP2 (Axam), SENP3 (SMT3IP1) and SENP6 (SUSP1) have been characterized biochemically (Gong et al., 2000; Kim et al., 2000; Nishida et al., 2000; Kadoya et al., 2002). Modification of target proteins by sumoylation changes their subcellular localization, transcriptional activation and protein stability.
It has been suggested that PIASy and Axam (SENP2) are involved in the Wnt signaling pathway. Lef-1 is a member of the Tcf family proteins. PIASy stimulates the sumoylation of Lef-1 and inhibits its transcriptional activity (Sachdev et al., 2001). Axam has been identified as an Axin-binding protein and induces the degradation of β-catenin. This action of Axam requires its desumoylation activity (Kadoya et al., 2002). Although the precise molecular mechanism by which sumoylation and desumoylation regulate the Wnt signaling pathway is not known, it is likely that sumoylation is involved in the Wnt signaling pathway. During the course of analyzing the physiological significance of sumoylation in the Wnt signaling pathway, we found that Tcf-4 is sumoylated. Here we show that sumoylation stimulates β-catenin-dependent and Tcf-4-mediated gene expression.
Results
Modification of Tcf-4 with SUMO
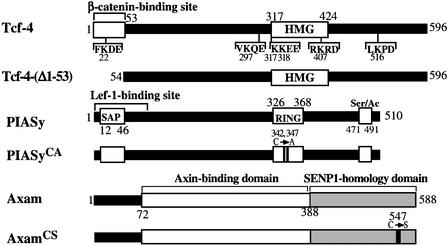
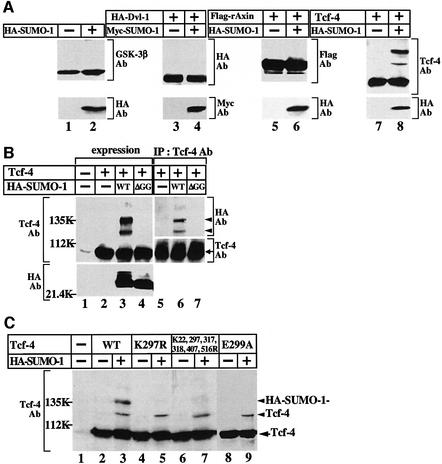
The constructs of Tcf-4, PIASy and Axam used in this study are depicted in Figure 1. It is known that sumoylation reduces the migration of substrates on an SDS–polyacrylamide gel (Matunis et al., 1996; Mahajan et al., 1997). To identify the substrates of sumoylation in the Wnt signaling pathway, we expressed SUMO-1 in 293 cells. Although the expression of hemagglutinin (HA)-SUMO-1 or Myc-SUMO-1 did not influence the migration of endogenous GSK-3β and expressed HA-Dvl-1 or Flag-rAxin on an SDS–polyacrylamide gel, Tcf-4 exhibited a mobility shift (Figure 2A). At least two slowly migrating forms of Tcf-4 were observed. SUMO-1 precursor is processed by a C-terminal hydrolase to produce the mature form, and exposure of the C-terminal Gly–Gly residues is required for conjugation of SUMO-1 to substrates (Li and Hochstrasser, 1999; Gong et al., 2000). When we expressed HA-SUMO-1(ΔGG), in which the C-terminal six amino acids including Gly–Gly were deleted, in 293 cells, the higher molecular mass Tcf-4 bands were not observed (Figure 2B, lane 4). Furthermore, when the lysates of cells expressing Tcf-4 together with HA-SUMO-1 were immunoprecipitated with the anti-Tcf-4 antibody, the slowly migrating forms of Tcf-4 were detected by the anti-HA antibody (Figure 2B, lanes 5–7).
Fig. 1. Schematic representation of Tcf-4, PIASy and Axam constructs used in this study. HMG, high mobility group; Ser/Ac, serine-rich and acidic domain.
Fig. 2. Modification of Tcf-4 with SUMO. (A) Modification of the Wnt signal components with SUMO. 293 cells expressing the indicated proteins were incubated with TCA to precipitate the proteins. The resulting precipitates were probed with the indicated antibodies. (B) Modification of Tcf-4 with SUMO. The precipitates of 293 cells expressing the indicated proteins produced by TCA were probed with the indicated antibodies (lanes 1–4). The lysates (200 µg of protein) prepared by extraction with RIPA buffer were immunoprecipitated with the anti-Tcf-4 antibody, and the immunoprecipitates were probed with the indicated antibodies (lanes 5–7). (C) Sumoylation sites of Tcf-4. The precipitates of 293 cells expressing the indicated proteins produced by TCA were probed with the anti-Tcf-4 antibody. Ab, antibody; IP, immunoprecipitation; WT, wild-type; ΔGG, SUMO-1-(1–95). The results shown are representative of three independent experiments.
It has been proposed that ψKXD/E (ψ is a hydrophobic residue and X is an any amino acid) is a consensus sequence for sumoylation (Melchior, 2000; Yeh et al., 2000; Müller et al., 2001). Tcf-4 contains three of these typical consensus sequences of sumoylation: FK22DE, VK297QE and LK516PD (Figure 1). Furthermore, K317K318EE and RK407RD are similar sequences. We mutated the lysine residues at amino acid positions 22, 297, 317, 318, 407 and 516 to arginine, and expressed the lysine mutants of Tcf-4 with HA-SUMO-1. One of the slowly migrating forms of Tcf-4 disappeared when K297 was mutated (Tcf-4K297R) (Figure 2C, lanes 2–5). However, when all six of the lysine residues were mutated (Tcf-4K22, 297, 317, 318, 407, 516R), one sumoylation band was still detected (Figure 2C, lanes 6 and 7). A lysine residue is also a target site for other post-translational modifications such as ubiquitylation and acetylation. When Glu299 in Tcf-4 was mutated to alanine (Tcf-4E299A), one slowly migrating band disappeared, indicating that K297 is modified with SUMO specifically (Figure 2C, lanes 8 and 9). These results imply that Tcf-4 is sumoylated and that at least K297 is a sumoylation site, and suggest that a lysine residue which is not located in the consensus sequence is sumoylated.
Enhancement of sumoylation of Tcf-4 by PIASy
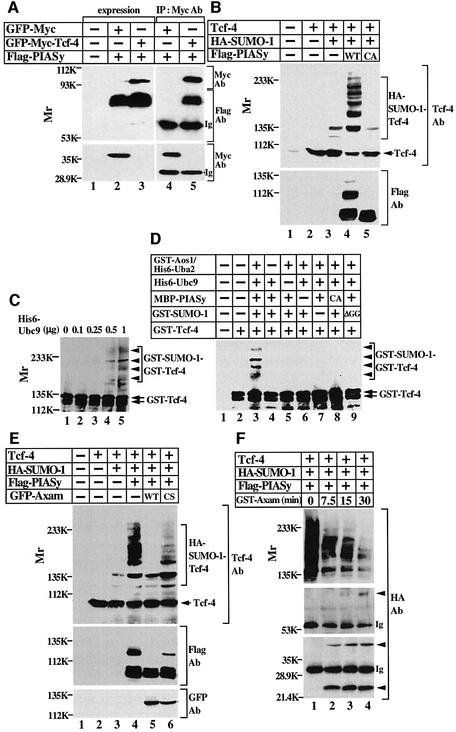
As PIASy acts as a SUMO E3 ligase for Lef-1 (Sachdev et al., 2001), Flag-PIASy formed a complex with green fluorescent protein (GFP)–Myc-Tcf-4 (Figure 3A) and enhanced the sumoylation of Tcf-4 (Figure 3B, lanes 2–4). PIASy has the RING motif, which has been identified as an important functional determinant of ubiquitin and SUMO E3 ligases (Kahyo et al., 2001; Sachdev et al., 2001). A PIASy RING mutant (PIASyCA), in which Cys342 and Cys347 were mutated to alanine, did not interact with Ubc9 in the yeast two-hybrid assay (data not shown). Expression of Flag-PIASyCA did not enhance the sumoylation of Tcf-4, but rather slightly inhibited it, suggesting that PIASyCA acts as a dominant-negative form (Figure 3B, lane 5). These findings suggest that endogenous PIASy is involved in the process of sumoylation of Tcf-4 in intact cells.
Fig. 3. Enhancement of sumoylation of Tcf-4 by PIASy. (A) Interaction of Tcf-4 with PIASy in intact cells. The lysates (20 µg of protein) of 293 cells expressing the indicated proteins were probed with the anti-Myc and anti-Flag antibodies (lanes 1–3). The same lysates (200 µg of protein) were immunoprecipitated with the anti-Myc antibody, and the immunoprecipitates were probed with the indicated antibodies (lanes 4 and 5). (B) Effect of PIASy on the sumoylation of Tcf-4 in intact cells. The precipitates of 293 cells expressing the indicated proteins were probed with the indicated antibodies. (C) Sumoylation of Tcf-4 in vitro. GST–Tcf-4 was incubated with GST–Aos1/His6-Uba2, GST–SUMO-1(GG) and the indicated amounts of His6-Ubc9 for 1 h at 30°C. After the incubation, the mixtures were probed with the anti-Tcf-4 antibody. (D) Enhancement of sumoylation of Tcf-4 by PIASy in vitro. MBP–PIASy and His6-Ubc9 were incubated with the proteins described in (C) for 15 min at 30°C. (E) Effect of Axam on the sumoylation of Tcf-4 in intact cells. The precipitates of 293 cells expressing the indicated proteins were probed with the indicated antibodies. (F) SUMO chain formation. The lysates (200 µg of protein) of 293 cells expressing the indicated proteins were immunoprecipitated with the anti-Tcf-4 antibody, and the immunoprecipitates were incubated with 100 ng of GST–Axam for the indicated times at 30°C. After the incubation, the mixtures were probed with the anti-HA antibody. Arrowheads indicate the positions of monomer, dimer and trimer of HA-SUMO-1. Ig, immunoglobulin; CA, PIASy RING mutant; CS, Axam catalytically inactive mutant. The results shown are representative of three independent experiments.
To examine whether Tcf-4 is sumoylated directly, we used an in vitro reconstituted system with purified recombinant proteins. Incubation with GST–SUMO-1(GG), GST–Aos1/His6-Uba2 and His6-Ubc9 enhanced SUMO conjugation to Tcf-4 in a manner dependent on the dose of His6-Ubc9 (Figure 3C). GST–SUMO-1(GG) is the mature form of SUMO-1. Under conditions that are not appropriate for the sumoylation of Tcf-4, addition of maltose-binding protein (MBP)-fused PIASy (MBP– PIASy) allowed efficient and multiple conjugation of SUMO-1 to Tcf-4, but MBP–PIASyCA did not (Figure 3D, lanes 3, 6 and 8). Conjugation of SUMO-1 to Tcf-4 was dependent on all the other components (Figure 3D, lanes 4, 5, 7 and 9). Taken together, these results indicate that PIASy can function as a SUMO E3 ligase for Tcf-4.
In mammals, four PIAS family proteins have been identified (Liu et al., 1998). Expression of Flag-PIAS3, HA-PIAS1 or HA-PIASxα also enhanced the sumoylation of Tcf-4 in 293 cells (data not shown), suggesting that the PIAS family proteins act as E3 ligases for Tcf-4, at least under conditions in which they are overexpressed. There are at least seven SENP family proteins in mammals (Yeh et al., 2000), and Axam is the same protein as SENP2. Axam completely abolished the sumoylation of Tcf-4 by PIASy in 293 cells, but sumoylation of Tcf-4 was still observed in the presence of a catalytically inactive Axam mutant (AxamCS) (Figure 3E). These findings suggest that the sumoylation of Tcf-4 is regulated by the PIAS family proteins and Axam.
To determine whether highly sumoylated forms of Tcf-4 induced by PIASy contain polysumoylation chains, multiple sites of monosumoylation or a combination of these two possibilities, SUMO-1-modified Tcf-4 prepared from 293 cells was incubated with GST–Axam. As is apparent from the cleavage pattern, HA-SUMO-1 generated species with molecular mass of 23, 45 and 70 kDa, suggesting that SUMO-1 dimers and trimers are formed (Figure 3F). Therefore, PIASy can form polysumoylation chains on Tcf-4 under overexpression conditions.
Sumoylation of Tcf-4 at the endogenous level
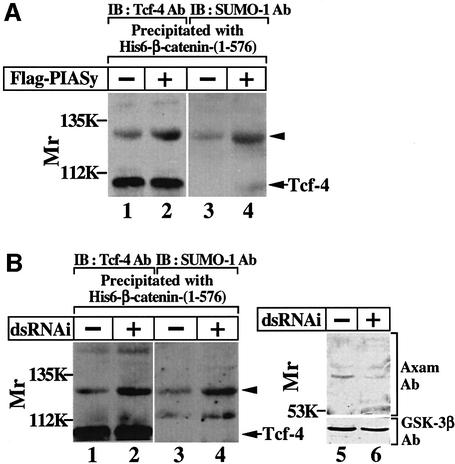
We examined sumoylation of Tcf-4 at the endogenous level. To concentrate Tcf-4, Tcf-4 was precipitated by His6-β-catenin-(1–576) from the nuclear extracts of 293 cells. A slowly migrating band recognized by the anti-Tcf-4 antibody was observed with the unmodified form of Tcf-4 (Figure 4A, lane 1). The band was also recognized by the anti-SUMO-1 antibody (Figure 4A, lane 3). Furthermore, the expression level of this band was increased by PIASy (Figure 4A, lanes 2 and 4). These results indicate that endogenous Tcf-4 is modified with endogenous SUMO-1.
Fig. 4. Sumoylation of Tcf-4 at the endogenous level. (A) Modification of endogenous Tcf-4 with endogenous SUMO-1. The nuclear extracts (200 µg of protein) from 293 cells or 293 cells expressing Flag-PIASy were incubated with His6-β-catenin-(1–576) (50 pmol) immobilized on nickel–agarose. After His6-β-catenin-(1–576) was precipitated by centrifugation, the precipitates were probed with the anti-Tcf-4 (lanes 1 and 2) or anti-SUMO-1 (lanes 3 and 4) antibody. (B) Effect of Axam RNAi on the sumoylation of Tcf-4. HeLa S3 cells were transfected with (lanes 2, 4 and 6) or without (lanes 1, 3 and 5) a dsRNAi oligo for Axam. The total cell lysates (20 µg of protein) were probed with the anti-Axam and anti-GSK-3β antibodies (lanes 5 and 6), and the nuclear extracts (200 µg of protein) were incubated with His6-β-catenin-(1–576) (50 pmol) immobilized on nickel–agarose (lanes 1–4). After His6-β-catenin-(1–576) was precipitated by centrifugation, the precipitates were probed with the anti-Tcf-4 (lanes 1 and 2) or anti-SUMO-1 (lanes 3 and 4) antibody. Arrowheads indicate the position of the endogenous Tcf-4 modified with endogenous SUMO-1. The results shown are representative of five independent experiments.
To examine whether Axam is involved in desumoylation of Tcf-4 at the endogenous level, we tried to deplete Axam via double-stranded (ds) RNA-mediated interference (RNAi) in HeLa S3 cells, because we did not succeed in reducing the protein level of Axam in 293 cells. A dsRNAi oligo for Axam reduced the protein level of Axam (Figure 4B, lanes 5 and 6) and increased the amount of a slowly migrating band that is recognized by both the anti-Tcf-4 and the anti-SUMO-1 antibodies (Figure 4B, lanes 1–4). Under the same conditions, Axam RNAi did not alter the protein level of GSK-3β (Figure 4B, lanes 5 and 6). These results strongly suggest that the sumoylation of Tcf-4 is regulated by Axam at least partly.
Effects of sumoylation on the binding of Tcf-4 to β-catenin and DNA
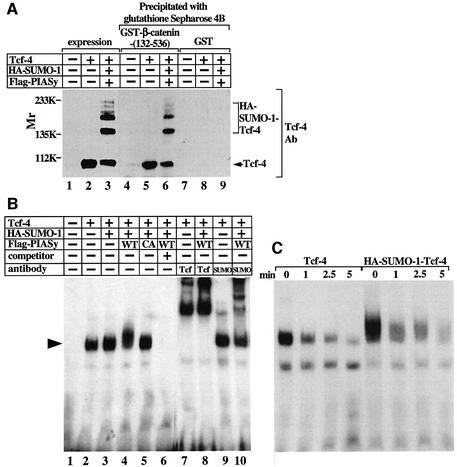
The nuclear extracts expressing Tcf-4, HA-SUMO-1 and Flag-PIASy contained sumoylated Tcf-4 and non-sumoylated Tcf-4 (Figure 5A, lanes 1–3). When the nuclear extracts were incubated with GST–β-catenin-(132–536), which contains Armadillo repeats, both forms of Tcf-4 associated with GST–β-catenin-(132–536), but not with GST alone, with similar efficiency (Figure 5A, lanes 4–9).
Fig. 5. Effects of PIASy on the binding of Tcf-4 to β-catenin and DNA. (A) Binding of sumoylated Tcf-4 to β-catenin. Nuclear extracts (10 µg of protein) from 293 cells expressing the indicated proteins were probed with the anti-Tcf-4 antibody (lanes 1–3). The same nuclear extracts (100 µg of protein) were incubated with GST–β-catenin-(132–536) (lanes 4–6) or GST (lanes 7–9), and the precipitates were probed with the anti-Tcf-4 antibody. (B) Electrophoretic mobility shift assay. Nuclear extracts (2 µg of protein) from 293 cells expressing the indicated proteins were incubated with a 32P-labeled oligonucleotide containing the Tcf-binding sequence in the presence (lane 6) or absence (lanes 1–5) of a 100-fold molar excess of unlabeled oligonucleotide, or in the presence of the indicated antibodies (0.5 µg) (lanes 7–10). An arrowhead indicates the position of the Tcf-4 specific complex. WT, wild-type; CA, PIASy RING mutant. (C) Dissociation rate assay. A 100-fold molar excess of unlabeled oligonucleotide was added to measure the dissociation rates of DNA binding. The time after the addition of excess unlabeled oligonucleotide is indicated. The results shown are representative of three independent experiments.
Tcf-4 contains the high mobility group (HMG) domain, and this domain binds to the Tcf-responsive element directly (Hurlstone and Clevers, 2002). The DNA-binding activity was observed in the nuclear extracts expressing Tcf-4 alone (Figure 5B, lane 2). Co-expression with HA-SUMO-1 or HA-SUMO-1 and Flag-PIASy did not affect the DNA-binding activity of Tcf-4, although a broad band was observed in the nuclear extracts of cells expressing both HA-SUMO-1 and Flag-PIASy (Figure 5B, lanes 3 and 4). Co-expression with HA-SUMO-1 and Flag-PIASyCA did not affect the DNA-binding activity of Tcf-4 (Figure 5B, lane 5). Formation of this band was competed efficiently against an excess of unlabeled oligonucleotide (Figure 5B, lane 6) and the band showed a supershift by the addition of the anti-Tcf-4 or anti-SUMO-1 antibody (Figure 5B, lanes 7–10). The effects of PIASy on the relative DNA-binding affinity of Tcf-4 were examined by determining the dissociation rate. Tcf-4 and sumoylated Tcf-4 were found thereby to exhibit similar affinities for DNA (Figure 5C). These results suggest that sumoylation does not affect the ability of Tcf-4 to bind to β-catenin and DNA.
Localization of Tcf-4 to PML bodies in a PIASy-dependent manner
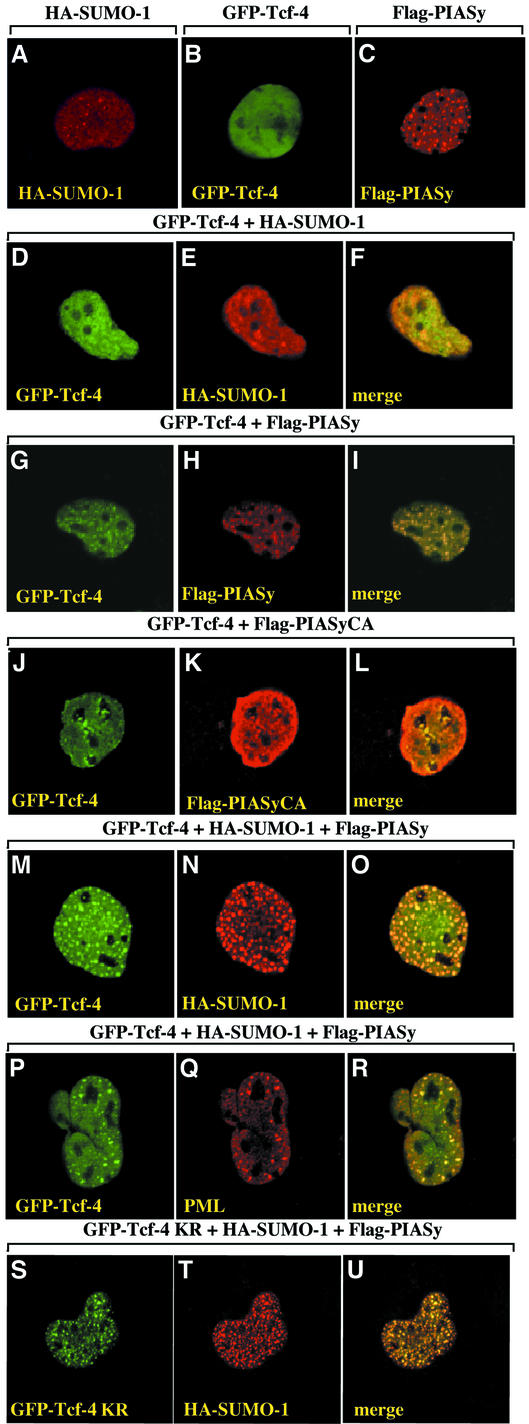
HA-SUMO-1 was largely homogenous in the nucleus, and was also concentrated in nuclear dot-like structures that resemble nuclear bodies (Figure 6A). Nuclear bodies are cell cycle-regulated, matrix-associated subnuclear structures that appear as punctate foci (Zhong et al., 2000). When GFP–Tcf-4 alone was expressed, it showed a diffuse nuclear distribution, whereas Flag-PIASy was localized predominantly to punctate structures in the nucleus (Figure 6B and C). Co-expression with HA-SUMO-1 had little effect on the nuclear distribution of GFP–Tcf-4, but they partly overlapped (Figure 6D–F). In contrast, co-expression with Flag-PIASy but not with Flag-PIASyCA resulted in a remarkable redistribution of GFP–Tcf-4 into punctate structures, which co-localized with Flag-PIASy (Figure 6G–L). Co-expression with both HA-SUMO-1 and Flag-PIASy greatly enhanced the distribution of GFP–Tcf-4 into nuclear bodies, and resulted in overlapping GFP–Tcf-4 and HA-SUMO-1 (Figure 6M–O). Flag-PIASy was also co-localized with them (data not shown). PML is the most prominent component of the nuclear bodies and a substrate for sumoylation (Müller et al., 2001). The nuclear punctate structures of GFP–Tcf-4 induced by HA-SUMO-1 and Flag-PIASy overlapped with the staining with the anti-PML antibody (Figure 6P–R). These results suggest that PIASy targets Tcf-4 to a subset of PML nuclear bodies. When GFP–Tcf-4K297R was co-expressed with HA-SUMO-1 and Flag-PIASy, GFP–Tcf-4K297R localized to nuclear dots, as did GFP–Tcf-4 (Figure 6S-U), suggesting that sumoylation of K297 is not essential for the sequestration of Tcf-4 in nuclear bodies.
Fig. 6. Localization of Tcf-4 to PML bodies in a PIASy-dependent manner. COS cells expressing the indicated proteins were stained with the anti-HA antibody to detect SUMO-1 (A, E, N and T), anti-Flag antibody to detect PIASy (C, H and K), and with anti-PML (Q) antibody. Some cells were viewed directly with a confocal laser-scanning microscope to detect GFP–Tcf-4 (B, D, G, J, M, P and S). The merged image shows the co-localization of GFP–Tcf-4 with HA-SUMO-1 (F, O and U), Flag-PIASy (I) or PML (R), but not with Flag-PIASyCA (L). KR, K297R. The results shown are representative of three independent experiments.
Synergistic activation of Tcf-4 by PIASy and β-catenin
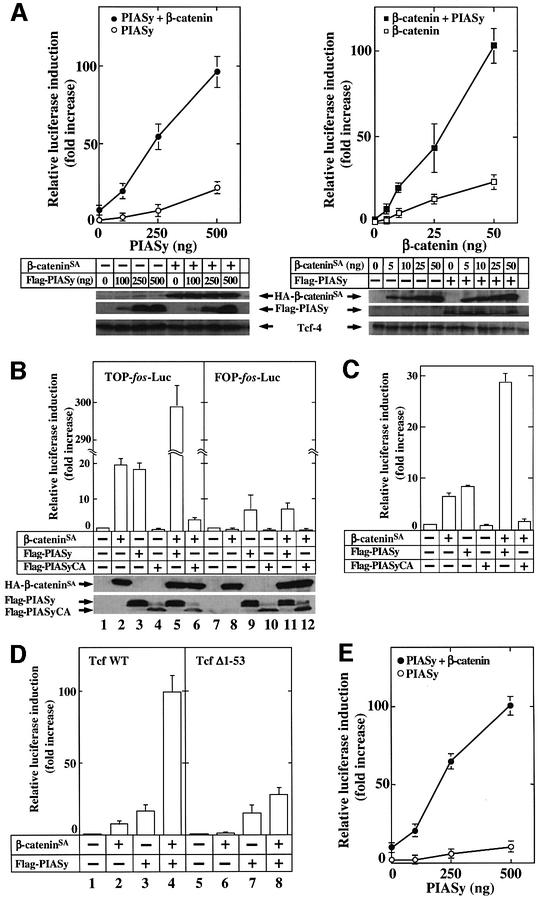
To examine the effects of sumoylation on Tcf-4 transcriptional activity, we used TOP-fos-Luc (Korinek et al., 1997), which contains four Tcf-binding sites and a c-fos promoter sequence ligated to a luciferase gene, as a reporter gene. We also expressed Tcf-4 exogenously to detect a significant increase in luciferase activity. Expression of either PIASy or β-cateninSA, a β-catenin mutant which is not degraded, alone in 293 cells activated Tcf-4 in a dose-dependent manner (Figure 7A). A small amount of β-catenin, which did not by itself activate Tcf-4 efficiently, strongly enhanced PIASy-dependent Tcf-4 activity (Figure 7A, left panel). PIASy also enhanced β-catenin-dependent Tcf-4 activity (Figure 7A, right panel). These results suggest that PIASy and β-catenin activate Tcf-4 synergistically.
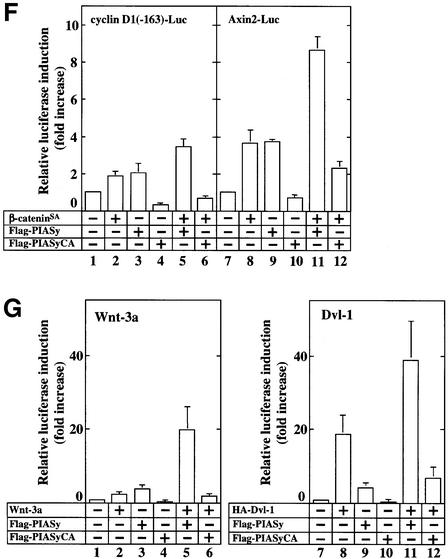
Fig. 7. Synergistic activation of Tcf-4 by β-catenin and PIASy. (A) Activation of Tcf-4 by β-catenin and PIASy. The indicated amounts of pCMV5-Flag/PIASy, pEF-BOS/hTcf-4E (0.1 µg) and TOP-fos-Luc (0.5 µg) were transfected into 293 cells with or without pUC/EF-1α/β-cateninSA (10 ng) (left panel). The indicated amounts of pUC/EF-1α/β-cateninSA, pEF-BOS/hTcf-4E (0.1 µg) and TOP-fos-Luc (0.5 µg) were transfected into 293 cells with or without pCMV5-Flag/PIASy (0.1 µg) (right panel). The luciferase activity was measured and expressed as the fold increase compared with the level observed in cells transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E. Left panel: Flag-PIASy alone (open circles); β-cateninSA and Flag-PIASy (filled circles). Right panel: β-cateninSA alone (open squares); Flag-PIASy and β-cateninSA (filled squares). The lysates (20 µg of protein) of 293 cells expressing the indicated proteins were probed with the anti-HA, anti-Flag and anti-Tcf-4 antibodies (lower panel). (B) pUC/EF-1α/β-cateninSA (50 ng) (lanes 2, 5, 6, 8, 11 and 12), pCMV5-Flag/PIASy (0.5 µg) (lanes 3, 5, 9 and 11) and pCMV5-Flag/PIASyCA (1 µg) (lanes 4, 6, 10 and 12) were transfected into 293 cells with pEF-BOS/hTcf-4E (lanes 1–12) and TOP-fos-Luc (lanes 1–6) or FOP-fos-Luc (lanes 7–12). The lysates (20 µg of protein) of 293 cells expressing the indicated proteins were probed with the anti-HA and anti-Flag antibodies (lower panel). (C) Activation of Lef-1 by β-catenin and PIASy. The assay conditions were the same as those in (B) except that pCMV5-T7/Lef-1 (0.1 µg) was used instead of pEF-BOS/hTcf-4E (0.1 µg). (D) Effects of PIASy on Tcf-4-(Δ1–53). pUC/EF-1α/β-cateninSA (10 ng) (lanes 2, 4, 6 and 8) and/or pCMV5-Flag/PIASy (0.5 µg) (lanes 3, 4, 7 and 8) were transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E (0.1 µg) (lanes 1–4) or pEF-BOS/hTcf-4E-(Δ1–53) (0.5 µg) (lanes 5–8). (E) Synergistic activation of endogenous Tcf family proteins by β-catenin and PIASy. The indicated amounts of pCMV5-Flag/PIASy and TOP-fos-Luc (0.5 µg) were transfected with or without pUC/EF-1α/β-cateninSA (10 ng). Flag-PIASy alone (open circles); β-cateninSA and Flag-PIASy (filled circles). (F) Effects of PIASy and β-catenin on the Tcf activity for natural Tcf-responsive promoters. pUC/EF-1α/β-cateninSA (0.5 µg) (lanes 2, 5, 6, 8, 11 and 12), pCMV5-Flag/PIASy (0.5 µg) (lanes 3, 5, 9 and 11) and pCMV5-Flag/PIASyCA (1 µg) (lanes 4, 6, 10 and 12) were transfected with cyclin D1(-163)-Luc (0.5 µg) (lanes 1–6) or Axin2-Luc (0.5 µg) (lanes 7–12). (G) Effect of PIASy on Wnt-3a- or Dvl-1-dependent Tcf activity. pPGK/Wnt-3a (0.5 µg) (lanes 2, 5 and 6) or pCGN/Dvl-1 (0.5 µg) (lanes 8, 11 and 12) was transfected with TOP-fos-Luc and pCMV5-Flag/PIASy (0.5 µg) (lanes 3, 5, 9 and 11) or pCMV5-Flag/PIASyCA (1 µg) (lanes 4, 6, 10 and 12). All the experiments were performed at least three times, and the results shown are means ± SE.
PIASyCA did not activate Tcf-4, and it inhibited β-catenin-dependent Tcf-4 activity, suggesting that endogenous PIASy is involved in the activation of Tcf-4 (Figure 7B). β-Catenin did not activate Tcf-4 towards FOP-fos-Luc, in which the Tcf-binding sites were mutated (Figure 7B). PIASy showed decreased ability to activate Tcf-4 when FOP-fos-Luc was used, suggesting that PIASy stimulates TOP-fos-Luc both dependently and independently of Tcf-4. β-Catenin and PIASy did not activate Tcf-4 synergistically towards FOP-fos-Luc (Figure 7B). When we used other reporter plasmids (TOP-TK-Luc and FOP-TK-Luc) that contained the Tcf-binding element or its mutant with a thymidine kinase promoter instead of the c-fos promoter, the results obtained were basically the same as those in the experiments using TOP-fos-Luc and FOP-fos-Luc (data not shown). Although it has been shown that PIASy inhibits β-catenin-dependent Lef-1 activation (Sachdev et al., 2001), PIASy and β-catenin activated Lef-1 synergistically and PIASyCA inhibited β-catenin-dependent Lef-1 activation in our assay conditions (Figure 7C). β-Catenin did not activate Tcf-4-(Δ1–53), in which the β-catenin-binding site was deleted, but PIASy still activated it to a similar extent as with wild-type Tcf-4 (Figure 7D). β-catenin did not enhance the PIASy-dependent activation of Tcf-4-(Δ1–53) significantly (Figure 7D).
Since at least Tcf-1 and Tcf-4 were observed to be expressed endogenously in 293 cells, we performed the same experiments without the expression of Tcf-4. PIASy alone slightly activated endogenous Tcf towards TOP-fos-Luc, and β-catenin greatly enhanced PIASy-dependent activation of endogenous Tcf (Figure 7E). Co-expression of PIASy and β-catenin at least additively increased the transcriptional activity of endogenous Tcf towards the cyclin D1 and Axin2 promoters, which are the natural Tcf-responsive promoters (Tetsu and McCormick, 1999; Jho et al., 2002; Lustig et al., 2002), but PIASyCA inhi bited β-catenin-dependent activity of these promoters (Figure 7F). Furthermore, PIASy enhanced Wnt-3a- or Dvl-1-dependent Tcf activity, whereas PIASyCA inhibited it (Figure 7G). Taken together, these results support the idea that β-catenin and PIASy activate Tcf-4 synergistically, and suggest that sumoylation is involved in the β-catenin-dependent and Tcf-4-mediated gene expression.
Involvement of sumoylation in the activation of Tcf-4
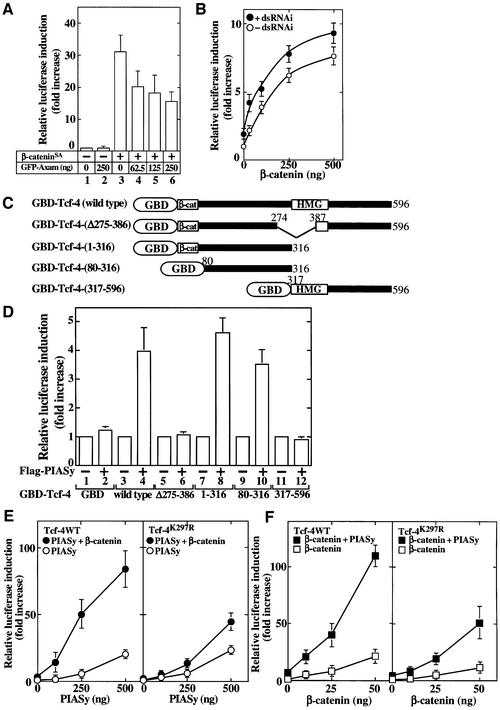
To confirm that sumoylation of Tcf-4 is really involved in the activation of Tcf-4, we asked what were the effects of Axam on the transcriptional activity of Tcf-4. Axam inhibited β-catenin-dependent Tcf-4 activation in 293 cells (Figure 8A). Axam RNAi increased the basal activity of Tcf-4 slightly and enhanced β-catenin-dependent Tcf-4 activation in HeLa S3 cells (Figure 8B). These results strongly suggest that desumoylation by Axam is involved in the regulation of Tcf-4.
Fig. 8. Involvement of sumoylation of Tcf-4 in its activation. (A) Effects of Axam on Tcf-4 activity. pUC/EF-1α/β-cateninSA (0.1 µg) (lanes 3–6) and the indicated amounts of pEGFP-C1/Axam (lanes 2 and 4–6) were transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E. The luciferase activity was measured and expressed as the fold increase compared with the level observed in cells transfected with TOP-fos-Luc and pEF-BOS/hTcf-4E. (B) Effects of Axam RNAi on Tcf-4 activity. After HeLa S3 cells had been transfected with (filled circles) or without (open circles) a dsRNAi oligo for Axam, the cells were transfected further with the indicated amounts of pUC/EF-1α/β-cateninSA, pEF-BOS/hTcf-4E (0.1 µg) and TOP-fos-Luc (0.5 µg). (C) Schematic representation of Tcf-4 deletion mutants fused to the GAL4 DNA-binding domain (GBD). β-cat, β-catenin-binding site; HMG, high mobility group. (D) Transcriptional activity of Tcf-4 deletion mutants fused to the GAL4 DNA-binding domain. pCMV5 (2 µg) (lanes 1, 3, 5, 7, 9 and 11 ) or pCMV5-Flag/PIASy (2 µg) (lanes 2, 4, 6, 8, 10 and 12) and pG4-TK-Luc (0.5 µg) (lanes 1–12) were transfected into 293 cells with pCMXGAL4 (lanes 1 and 2), pCMXGAL4/hTcf-4E (lanes 3 and 4), pCMXGAL4/hTcf-4E-(Δ275–386) (lanes 5 and 6), pCMXGAL4/hTcf-4E-(1–316) (lanes 7 and 8), pCMXGAL4/hTcf-4E-(80–316) (lanes 9 and 10) or pCMXGAL4/hTcf-4E-(317–596) (lanes 11 and 12) (0.5 µg each). (E) The indicated amounts of pCMV5-Flag/PIASy, TOP-fos-Luc (0.5 µg) and pEF-BOS/hTcf-4E (0.1 µg) (left panel) or pEF-BOS/hTcf-4E K297R (0.1 µg) (right panel) were transfected into 293 cells with or without pUC/EF-1α/β-cateninSA (10 ng). Flag-PIASy alone (open circles); Flag-PIASy and β-cateninSA (filled circles). (F) The indicated amounts of pUC/EF-1α/β-cateninSA, TOP-fos-Luc (0.5 µg) and pEF-BOS/hTcf-4E (0.1 µg) (left panel) or pEF-BOS/hTcf-4E K297R (0.1 µg) (right panel) were transfected into 293 cells with or without pCMV5-Flag/PIASy (0.1 µg). β-CateninSA alone (open squares); β-cateninSA and Flag-PIASy (filled squares). All the experiments were performed at least three times, and the results shown are means ± SE.
To examine which region of Tcf-4 is responsible for the PIASy-induced gene expression, Tcf-4 and its deletion mutants were fused to the GAL4 DNA-binding domain (GBD) of their N-termini (Figure 8C). A plasmid that contains three GAL4-binding sites upstream of the thymidine kinase promoter and the luciferase gene was used as the reporter gene (pG4-TK-Luc). PIASy stimulated the luciferase activity 3- to 5-fold in the presence of GBD-Tcf-4 (wild-type) (Figure 8D). PIASy also stimulated the luciferase activity in the presence of GBD-Tcf-4-(1–316) or GBD-Tcf-4-(80–316), but not in the presence of GBD-Tcf-4-(Δ275–386) or GBD-Tcf-4-(317–596). These results suggest that the region encompassed by amino acids 275–316 of Tcf-4 is important for the PIASy-induced gene expression. Since the amino acid region 275–316 includes Lys297, we examined whether sumoylation of Lys297 of Tcf-4 is involved in its activation. PIASy activated Tcf-4K297R to the same extent as wild-type Tcf-4 (Figure 8E), suggesting that sumoylation of Lys297 is not necessary for the PIASy-dependent Tcf-4 activation. Under these assay conditions, PIASy still enhanced sumoylation of Tcf-4K297R, but its extent was decreased in comparison with wild-type Tcf-4 (data not shown). Therefore, sumoylation of other lysine residues may be important. Interestingly, β-catenin activated Tcf-4K297R to a lesser extent than wild-type Tcf-4 (Figure 8F). Furthermore, synergistic activation of Tcf-4K297R by β-catenin and PIASy was greatly reduced compared with that of wild-type Tcf-4 (Figure 8E and F). Therefore, it is likely that the sumoylation of Lys297 of Tcf-4 is involved in the synergistic activation of Tcf-4 by β-catenin and PIASy.
Discussion
Sumoylation of Tcf-4
Endogenous Tcf-4 was modified with endogenous SUMO-1. Among six lysine residues of Tcf-4 we mutated, only Lys297 was a sumoylation site. Since the Tcf-4 mutant, in which all possible sumoylation sites were mutated, was still sumoylated, there must be non-consensus SUMO conjugation sites. Alternatively, these mutations may lead to sumoylation at a secondary site.
PIASy, PIAS1, PIAS3 and PIASxα all enhanced the sumoylation of Tcf-4 (data not shown). In contrast to ubiquitin E3 ligases, the substrate specificity of SUMO E3 ligases and the specific role of the distinct SUMO E3 ligases are not clear. PIAS1 acts as a SUMO E3 ligase for p53 and c-Jun (Schmidt and Müller, 2002). PIASy and PIASxα also enhance sumoylation of Lef-1 and androgen receptor, respectively (Sachdev et al., 2001; Kotaja et al., 2002). Therefore, the PIAS proteins may be less important determinants for the substrate specificity in the SUMO pathway than ubiquitin E3 ligases in the ubiquitin pathway. Our in vitro study using the purified proteins also showed that Aos1/Uba2 and Ubc9 sumoylate Tcf-4 in the absence of PIASy. Hence, the PIAS proteins may play a role in stabilizing the interaction between Ubc9 and substrates.
A number of SUMO-specific proteases have been isolated and shown to carry out SUMO maturation (C-terminal hydrolase) and deconjugation (isopeptidase), and the mammalian enzymes have been designated SENPs (Yeh et al., 2000). PIASy-dependent sumoylation of Tcf-4 was inhibited by Axam. Axam RNAi experiments strongly supported the idea that Axam (SENP2) is involved in the desumoylation of Tcf-4. Therefore, it is conceivable that the sumoylation state of Tcf-4 is regulated by PIAS and Axam.
Regulation of subcellular localization and transcriptional activity of Tcf-4 by sumoylation
It has been demonstrated that the N-terminal SAP domain of Lef-1 is necessary to bind to a matrix attachment region on DNA (Aravind and Koonin, 2000; Sachdev et al., 2001). Consistent with the case of Lef-1, PIASy targeted Tcf-4 to nuclear bodies. Since co-expression of SUMO-1 with Tcf-4 and PIASy further enhanced the targeting of Tcf-4 to nuclear bodies and the PIASy RING mutant was unable to sequester Tcf-4 in nuclear bodies, its SUMO modification and nuclear body sequestration seem to be closely linked.
Although sumoylation did not alter the ability of Tcf-4 to interact with β-catenin or to bind to DNA, PIASy stimulated β-catenin-dependent and Tcf-4-mediated transcriptional activation. These results suggest that sumoylation regulates the function of Tcf-4 positively. Several lines of evidence support this conclusion. (i) Experiments using two synthetic reporter genes (TOP-fos-Luc and TOP-TK-Luc) showed similar results: PIASy enhanced β-catenin-dependent activation of Tcf-4, but the PIASy RING mutant rather inhibited it. (ii) PIASy activated Tcf-4-(Δ1–53), but it did not act synergistically with β-catenin to activate this Tcf-4 mutant. (iii) PIASy cooperated with β-catenin to activate reporter constructs in which the natural cyclin D1 and Axin 2 promoters are linked to the luciferase gene, but the PIASy RING mutant inhibited it. (iv) PIASy enhanced Wnt-3a- or Dvl-1-dependent Tcf-4 activity, but the PIASy RING mutant inhibited it. (v) Axam repressed the β-catenin-dependent activity of Tcf-4, and Axam RNAi resulted in the activation of Tcf-4.
PIASy and β-catenin activated Lef-1 synergistically in our assay conditions. These are not consistent with the previous observations that overexpression of PIASy represses the β-catenin-stimulated transcriptional activity of Lef-1 (Sachdev et al., 2001). Although we do not know the reasons for the differences in the results at present, they may be dependent on the abundance of various substrates and interacting proteins of PIASy. It is also possible to speculate that sumoylation regulates positively or negatively the transcriptional activity of Tcf-4 and Lef-1 for their target genes, depending on their promoter architecture.
The amino acid region 275–316 of Tcf-4 is responsible for PIASy-induced transcription. Direct sumoylation of this region of Tcf-4 or sumoylation of a protein that binds to this region may be important for the activation of Tcf-4. Furthermore, we have demonstrated that β-catenin and PIASy activate Tcf-4K297R to a lesser extent than wild-type Tcf-4. These results indicate that at least sumoylation of Lys297 of Tcf-4 plays a role in the synergistic activation Tcf-4 by β-catenin and PIASy. An unknown co-activator may recognize the sumoylation of Tcf-4 at Lys297 and act synergistically with β-catenin to activate Tcf-4.
Involvement of sumoylation in the regulation of the Wnt signaling pathway
The Wnt signaling pathway is regulated by several post-translational modifications including phosphorylation, ubiquitylation and acetylation. We previously have demonstrated that Axam, an Axin-binding protein, is a desumoylation enzyme and that the desumoylation activity of Axam is necessary for its ability to induce the downregulation of β-catenin (Kadoya et al., 2000, 2002). In this study, we have shown that PIASy and Axam stimulate and inhibit, respectively, β-catenin-dependent Tcf-4 mediated transcription. Taken together, these facts imply that sumoylation and desumoylation may regulate positively and negatively, respectively, the Wnt signaling pathway.
Materials and methods
Materials and chemicals
GST–Aos1 and His6-Uba2 purified from Spodoptera frugiperda Sf9 cells were supplied by Dr H.Yasuda (Tokyo University of Pharmacy and Life Science, Tokyo, Japan). pcDNA3/Flag-rAxin, pCMV5-Flag/PIAS (1, 3, xα and y), pUC/EF-1α/β-cateninSA, pCMV5-T7/Lef-1, Axin2-luciferase (Axin2-Luc), -163 cyclin D1-luciferase [cyclin D1(-163)-Luc], pcDNAI/hTcf-4E, TOP-fos-Luc, FOP-fos-Luc, pPGK/Wnt-3a, pG4-TK-Luc and pCMXGAL4 were provided by Drs K.Miyazono (Tokyo University, Tokyo, Japan), K.Shuai (University of California, Los Angels), A.Nagafuchi (Kumamoto University, Kumamoto, Japan), R.Grosschedl (University of Munich, Munich, Germany), F.Costantini (Columbia University, New York), T.Akiyama (Tokyo University, Tokyo, Japan), H.Clevers (University Medical Center, Utrecht, The Netherlands), S.Takada (Kyoto University, Kyoto, Japan) and K.Igarashi (Hiroshima University, Hiroshima, Japan), respectively. GST–Tcf-4 was purified from Sf9 cells. Other GST or MBP fusion proteins and His6-tagged proteins were purified from Escherichia coli according to the supplier’s instructions. Anti-Myc antibody was prepared from 9E10 cells. Other materials were purchased from commercial sources.
Plasmid construction
pEF-BOS-HA/hTcf-4E, pEGFP-C1/Axam, pEGFP-C2/AxamC547S, pGEX-2T/Axam, pGEX-2TK/SUMO-1(GG), pRSETA/Ubc9 and pCGN/Dvl-1 were constructed as described (Kadoya et al., 2002). Standard recombinant DNA techniques were used to construct the following plasmids: pEF-BOS/hTcf-4E, pEF-BOS-GFP-Myc/hTcf-4E, pEGFP-C1/hTcf-4E, pVIKS/hTcf-4E, pEF-BOS-HA/hTcf-4E-(Δ1–53), pEF-BOS/hTcf-4EK297R, pEF-BOS/hTcf-4EK22/297/317/318/407/516R, pEF-BOS/hTcf-4EE299A, pEGFP-C1/hTcf-4EK297R, pCGN/SUMO-1, pBJ-Myc/SUMO-1, pCGN/SUMO-1(ΔGG), pMAL-c2/PIASy, pGBKT7/PIASy, pMAL-c2/PIASyC342/347A, pCMV5-Flag/PIASyC342/347A, pGBKT7/PIASyC342/347A, pGAD/Ubc9, pEF-BOS-HA/PIAS1, pEF-BOS-HA/PIASxα, pCMXGAL4/hTcf-4E, pCMXGAL4/hTcf-4E-(Δ275–386), pCMXGAL4/hTcf-4E-(317–596), pCMXGAL4/hTcf-4E-(1–316), pCMXGAL4/hTcf-4E-(80–316), pRSETA/β-catenin-(1–576) and pGEX-KG/β-catenin-(132–536). Some of the constructs in these plasmids were made by digesting the original plasmids with restriction enzymes and inserting the fragments of interest into the vectors. The other constructs were made by inserting fragments generated using the Expand High Fidelity PCR system (Roche Diagnostics GmbH, Manheim, Germany) into the vectors.
Sumoylation assay
To examine the sumoylation of Tcf-4 in intact cells, 293 cells (35 mm diameter dishes) were transfected with pEF-BOS-, pCMV5- and pCGN-derived plasmids. At 60 h after transfection, the cells were supplemented with 500 µl of 10% trichloroacetic acid (TCA) containing 2 mM dithiothreitol (DTT) and incubated for 10 min at 4°C. After proteins were precipitated by centrifugation at 20 000 g for 5 min at 4°C, the resulting precipitate was dissolved in 200 µl of Laemmli’s sample buffer, and the samples were probed with the anti-Tcf-4 antibody. For co-immunoprecipitation analysis, the cells were lysed in 100 µl of RIPA buffer (10 mM Na-phosphate buffer pH 7.2, 150 mM NaCl, 1% Na-deoxycholate, 1% Triton X-100 and 0.1% SDS) containing 1 mM DTT, 1 µg/ml aprotinin and leupeptin, 10 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 0.4 mM Na-orthovanadate and 10 mM N-ethylmaleimide. The lysates (200 µg of protein) were immunoprecipitated with the anti-Tcf-4 antibody, and then the precipitates were probed with the anti-Tcf-4, anti-HA, anti-Myc or anti-Flag antibody. To detect SUMO chain formation on Tcf-4, the precipitates were incubated further with 100 ng of GST–Axam in 50 µl of reaction mixture (50 mM Tris–HCl pH 7.5, 1 mM DTT and 150 mM NaCl) for the indicated times at 30°C. After the incubation, the mixtures were probed with the anti-HA antibody.
To observe sumoylation of Tcf-4 at the endogenous level, nuclear extracts were prepared from 293 cells as described (Ishitani et al., 1999). The nuclear extracts (200 µg of protein) were incubated with His6-β-catenin-(1–576) (50 pmol) immobilized on nickel–agarose for 2 h at 4°C. After His6-β-catenin-(1–576) was precipitated by centrifugation, the precipitates were probed with the anti-Tcf-4 or anti-SUMO-1 antibody.
To examine the sumoylation of Tcf-4 in vitro, 0.25 µg of GST–Tcf-4 was incubated with 0.5 µg of GST–Aos1/His6-Uba2, 0.5 µg of His6-Ubc9, 10 µg of GST–SUMO-1(GG) or 0.2 µg of MBP-PIASy in 50 µl of reaction mixture (50 mM Tris–HCl pH 7.5, 2 mM DTT, 1 mM MgCl2 and 5 mM ATP) at 30°C for 15 min. After the incubation, the mixtures were probed with the anti-Tcf-4 antibody.
RNA interference
Oligonucleotides specific for human Axam 5′-GAUCAGAGUGACAG UUACCTT-3′ (sense) were synthesized and a dsRNA oligonucleotide was annealed in vitro before transfection. Transfection was done with Oligofectamine (Invitrogen) on HeLa S3 cells (35 mm diameter dishes). At 96 h after the transfection, the cells were used for experiments.
Electrophoretic mobility shift assay
A double-stranded 56 nucleotide oligonucleotide (5′-atg gat cca aga tca aag ggg gta aga tca aag ggg gta aga tca aag gga tcc at-3′) containing three potential Tcf/Lef-binding sites (underlined) was used as the optimal Tcf-4 probe (Ishitani et al., 1999). The double-stranded oligonucleotide was labeled with [γ-32P]ATP and T4 polynucleotide kinase. Binding reactions were done for 10 min at room temperature by incubating 2 µg of nuclear extracts and 0.15 pmol of labeled oligonucleotide in 20 µl of binding buffer [10 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 1 µg of poly(dI–dC) and 1 µg of salmon sperm DNA]. Competition analyses were performed by adding excess (15 pmol) unlabeled probe. To measure the dissociation rates of Tcf-4–DNA complexes, after binding reactions were performed for 10 min, unlabeled probe (15 pmol) was added. After the stated incubation period at room temperature, the residual Tcf-4–DNA complexes were loaded on a 4% polyacrylamide gel.
Immunofluorescence study
COS cells (35 mm diameter dishes) transfected with pEGFP-, pCMV5- or pCGN-derived plasmid were fixed with 4% paraformaldehyde and incubated with the anti-Flag, anti-HA or anti-PML antibody. After washing with phosphate-buffered saline (PBS), the cells were viewed with a confocal laser-scanning microscope (LSM510, Carl-Zeiss, Jena, Germany) as described (Kadoya et al., 2002).
Assay of Tcf-4 activity
To examine the Tcf-4 activity, the indicated amounts of pUC/EF-1α/β-cateninSA, pEGFP-C1/Axam, pCMV5-Flag/PIASy, pCGN/Dvl-1 or pPGK/Wnt-3a were transfected into 293 cells (35 mm diameter dishes) with TOP-fos-Luc (0.5 µg), pME18S/lacZ (0.5 µg) and pEF-BOS/hTcf-4E (0.1 µg). At 46 h after transfection, the cells were lysed, and the luciferase activity was measured as described (Hino et al., 2001; Yamamoto et al., 2001). In some experiments, FOP-fos-Luc, TOP-TK-Luc, FOP-TK-Luc, Axin2-Luc or cyclin D1(-163)-Luc was used instead of TOP-fos-Luc. To examine Lef-1 activity, pCMV5-T7/Lef-1 (0.1 µg) was used instead of pEF-BOS/hTcf-4E (0.1 µg).
To determine which region of Tcf-4 is responsible for PIASy, pCMXGAL4/hTcf-4E or its deletion mutants (0.5 µg) were transfected into 293 cells (35 mm diameter dishes) with pG4-TK-Luc (0.5 µg), pME18S/lacZ (0.5 µg) and pCMV5 or pCMV5-Flag/PIASy (2 µg). At 46 h after transfection, the cells were lysed and the luciferase activity was measured.
Acknowledgments
Acknowledgements
We are grateful to Drs H.Yasuda, K.Miyazono, K.Shuai, A.Nagafuchi, R.Grosschedl, F.Costatini, T.Akiyama, H.Clevers, S.Takada, and K.Igarashi for donating proteins and plasmids, and Drs K.Tanaka (Tokyo Metropolitan Institute) and K.Igarashi for helpful discussion. This work was supported by Grants-in-Aid for Scientific Research and for Scientific Research on priority areas from the Ministry of Education, Science and Culture, Japan (2001, 2002), and by grants from the Yamanouchi Foundation for Research on Metabolic Disorders (2001).
References
- Aravind L. and Koonin,E.V. (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci., 25, 112–114. [DOI] [PubMed] [Google Scholar]
- Bienz M. and Clevers,H. (2000) Linking colorectal cancer to Wnt signaling. Cell, 103, 311–320. [DOI] [PubMed] [Google Scholar]
- Gong L., Millas,S., Maul,G.G. and Yeh,E.T. (2000) Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem., 275, 3355–3359. [DOI] [PubMed] [Google Scholar]
- Hart M.J., de los Santos,R., Albert,I.N., Rubinfeld,B. and Polakis,P. (1998) Downregulation of β-catenin by human Axin and its association with the APC tumor suppressor, β-catenin and GSK-3β. Curr. Biol., 8, 573–581. [DOI] [PubMed] [Google Scholar]
- Hino S.-I., Kishida,S., Michiue,T., Fukui,A., Sakamoto,I., Takada,S., Asashima,M. and Kikuchi,A. (2001) Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol. Cell. Biol., 21, 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2000) Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol., 2, 153–157. [DOI] [PubMed] [Google Scholar]
- Hurlstone A. and Clevers,H. (2002) T-cell factors: turn-ons and turn-offs. EMBO J., 21, 2303–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida,S., Yamamoto,H., Murai,H., Koyama,S. and Kikuchi,A. (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J., 17, 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T. et al. (1999) The TAK1–NLK–MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature, 399, 798–802. [DOI] [PubMed] [Google Scholar]
- Jho E.-H., Zhang,T., Domon,C., Joo,C.K., Freund,J.N. and Costantini,F. (2002) Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol., 22, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Schwienhorst,I., Dohmen,R.J. and Blobel,G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J., 16, 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya T., Kishida,S., Fukui,A., Hinoi,T., Michiue,T., Asashima,M. and Kikuchi,A. (2000) Inhibition of Wnt signaling pathway by a novel Axin-binding protein. J. Biol. Chem., 275, 37030–37037. [DOI] [PubMed] [Google Scholar]
- Kadoya T. et al. (2002) Desumoylation activity of Axam, a novel Axin-binding protein, is involved in downregulation of β-catenin. Mol. Cell. Biol., 22, 3803–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahyo T., Nishida,T. and Yasuda,H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell, 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kikuchi A. (1999) Roles of Axin in the Wnt signalling pathway. Cell Signal., 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Kim K.I. et al. (2000) A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem., 275, 14102–14106. [DOI] [PubMed] [Google Scholar]
- Kirsh O. et al. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J., 21, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida S., Yamamoto,H., Ikeda,S., Kishida,M., Sakamoto,I., Koyama,S. and Kikuchi,A. (1998) Axin, a negative regulator of the Wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J. Biol. Chem., 273, 10823–10826. [DOI] [PubMed] [Google Scholar]
- Kitagawa M. et al. (1999) An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J., 18, 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker,N., Morin,P.J., van Wichen,D., de Weger,R., Kinzler,K.W., Vogelstein,B. and Clevers,H. (1997) Constitutive transcriptional activation by a β-catenin–Tcf complex in APC–/– colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- Kotaja N., Karvonen,U., Jänne,O.A. and Palvimo,J.J. (2002) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol., 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.J. and Hochstrasser,M. (1999) A new protease required for cell-cycle progression in yeast. Nature, 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Liu B., Liao,J., Rao,X., Kushner,S.A., Chung,C.D., Chang,D.D. and Shuai,K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA, 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li,Y., Semenov,M., Han,C., Baeg,G.-H., Tan,Y., Zhang,Z., Lin,X. and He,X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell, 108, 837–847. [DOI] [PubMed] [Google Scholar]
- Lustig B. et al. (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol., 22, 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin,C., Guan,T., Gerace,L. and Melchior,F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas,E. and Blobel,G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol., 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO—nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Müller S., Hoege,C., Pyrowolakis,G. and Jentsch,S. (2001) SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell. Biol., 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Nishida T., Tanaka,H. and Yasuda,H. (2000) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem., 267, 6423–6427. [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast,A., Seeler,J., Dejean,A. and Melchior,F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell, 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn,L., Sieber,H., Pichler,A., Melchior,F. and Grosschedl,R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev., 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Sparrow,D.B., Shiomi,T., Pu,R.T., Nishimoto,T., Mohun,T.J. and Dasso,M. (1998) Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol., 8, 121–124. [DOI] [PubMed] [Google Scholar]
- Schmidt D. and Müller,S. (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA, 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidensticker M.J. and Behrens,J. (2000) Biochemical interactions in the wnt pathway. Biochim. Biophys. Acta, 1495, 168–182. [DOI] [PubMed] [Google Scholar]
- Tetsu O. and McCormick,F. (1999) β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Wodarz A. and Nusse,R. (1998) Mechanisms of Wnt signaling in development. Annu. Rev. Cell. Dev. Biol., 14, 59–88. [DOI] [PubMed] [Google Scholar]
- Yamamoto H. et al. (2001) Inhibition of the Wnt signaling pathway by the PR61 subunit of protein phosphatase 2A. J. Biol. Chem., 276, 26875–26882. [DOI] [PubMed] [Google Scholar]
- Yeh E.T., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]
- Yost C., Torres,M., Miller,J.R., Huang,E., Kimelman,D. and Moon,R.T. (1996) The axis-inducing activity, stability and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev., 10, 1443–1454. [DOI] [PubMed] [Google Scholar]
- Zhong S., Salomoni,P. and Pandolfi,P.P. (2000) The transcriptional role of PML and the nuclear body. Nat. Cell Biol., 2, 85–90. [DOI] [PubMed] [Google Scholar]