Abstract
Anatomy is a major organizing principle for diseases. In the formal definitions provided by SNOMED CT, for example, the role ‘finding site’ relates disorders to anatomical entities. This study investigates SNOMED CT and compares the anatomy-based classification of diseases supported by the role finding site to the anatomy-based classification of diseases provided by subsumption (is-a) relations between diseases. For each of the 3,540 anatomical entities associated with disorders, we compared two sets of disorders: first, the set of disorders associated with any descendant of the anatomical entity under investigation (ANAT); second, the set of disorders corresponding to the union of the descendants of the disorders associated with the anatomical entity under investigation (TAXO). The ANAT and TAXO sets were different for 1,231 anatomical entities (35%). In 607 cases, the overlap between ANAT and TAXO was less than 50%. When a difference was found, the TAXO set was always a subset of the ANAT set. Among the 1,025,904 subsumption relations among disorders generated by the ANAT approach, 40% were not present in TAXO. This approach helps identify missing classes and taxonomic relations and can therefore be used for quality assurance purposes in existing ontologies. It can be generalized to other kinds of partitions of biomedical ontologies.
INTRODUCTION
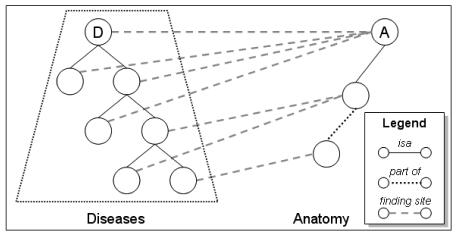
Anatomy is a major organizing principle for diseases. In most medical terminologies, diseases are classified according– at least in part – to the anatomical entity in which they are located. In the International Classification of Diseases (ICD 10), for example, twelve chapters out of twenty correspond to classes of diseases located in a given body system (e.g., Diseases of the nervous system); four other chapters, (Neoplasms, Congenital malformations, Symptoms and signs and Injuries) are subdivided according to anatomical sites (e.g., Injuries to the thorax). In such terminologies, distinctions among anatomical entities are used as implicit classification criteria for diseases. In contrast, in order to support automatic classification and reasoning, formal ontologies represent the properties of entities explicitly. In SNOMED Clinical Terms® (SNOMED CT®)1, the characterization of diseases is based on several roles, including finding site, which relates disorders to anatomical entities. Specialization relations among diseases often parallel partitive and specialization relations among anatomical entities corresponding to their respective locations (e.g. [1-3]). For example, tumors of the brain are tumors of the nervous system because the brain is a part of the nervous system. Analogously, tumors of the mandible are bone tumors because the mandible is a kind of bone. As expected, the specialization relations among diseases (e.g, neoplasm of mandible isa neoplasm of bone) and among anatomical entities (e.g., mandible bone structure isa bone structure) can be found in SNOMED CT, as well as the links between diseases and anatomical entities (e.g., neoplasm of mandible has finding site mandible bone structure and neoplasm of bone has finding site bone structure). Applying this parallel between diseases and anatomical entities to classifying diseases in SNOMED CT, one can assume the following. For a given anatomical entity A and the disease D having A as its finding site, the descendants of A are expected to be finding sites for the descendants of D. More precisely, all diseases having A as their finding site are expected to be descendants of D, and all descendants of D are expected to have A or a descendant of A as their finding site (Fig 1).
Figure 1.
Relation between disorders and anatomical entities
The objective of this study is to evaluate the degree to which, in SNOMED CT, the classification of diseases supported by the role finding site is compatible with the classification of diseases provided by subsumption relations (isa) among diseases. SNOMED CT was selected because it is the most comprehensive biomedical terminology recently developed in native description logics (DL) formalism, which enables this kind of analysis. Moreover, SNOMED CT is expected to play an important role in clinical information systems in the future. As clinical information systems must support accurate retrieval of clinical data, it is important that the disease instances be accurately retrieved, whether searched for by browsing the hierarchy of diseases or that of anatomical entities to which they are related. SNOMED CT has been available as part of the Unified Medical Language System® (UMLS®)2 since 2004. The version of SNOMED CT used in this study was released on January 31, 2004 and corresponds to version 2004AA of the UMLS.
METHODS
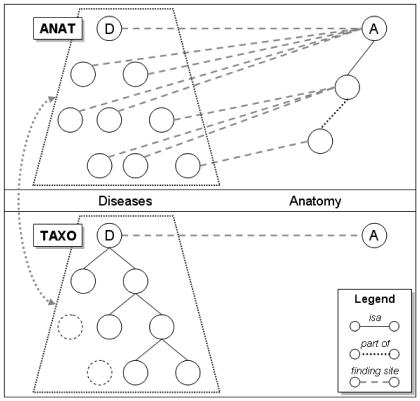
The methods can be summarized as follows. As illustrated in Figure 2, for a given anatomical entity and the disorders associated with it through the role finding site, we computed two sets of disorders: first, the set of disorders associated with any descendant of the anatomical entity under investigation; second, the set of disorders corresponding to the union of the descendants of the disorders associated with the anatomical entity under investigation. Each set of disorders corresponds to a set of SNOMED CT concepts. We then compared these two sets, with a special emphasis on the disorders specific to each set, i.e., not shared by both sets.
Figure 2.
Overview of the methods
Establishing the list of disorder-anatomy associations
Disorders are associated with anatomical entities through the role finding site. All SNOMED CT finding site relations were extracted (i.e., not limited to primitives) and restricted to concepts whose status is "current". 90,702 such associations were extracted, corresponding to 63,190 distinct disorders and 3,540 distinct anatomical entities.
Establishing the set of disorders associated with the descendants of a given anatomical entity
Starting from a given anatomical entity A, the set of all descendants of A can be easily established by traversing recursively the inverse_isa links. Because SNOMED CT uses a representation of anatomical entities based on Structure-Entire-Part (SEP) distinctions [4], traversing the isa link yields both the concepts subsumed by a given anatomical entity and the concepts corresponding to parts of this anatomical entity. For example, the adrenal cortex (Adrenal cortex structure) is a part of the adrenal gland (Adrenal gland structure). In SNOMED CT, Adrenal cortex structure is related to Adrenal gland structure through the following links:
Adrenal cortex structure isa Layer of adrenal gland
Layer of adrenal gland isa Adrenal part
Adrenal part isa Adrenal gland structure
The point here is that both parts and specialized entities can be extracted by exploring solely the isa links. From the set of descendants of a given anatomical entity, we extracted all disorders having any anatomical entity from this set as finding site. This set of disorders, referred to as ANAT, constitutes the set of SNOMED CT disorder classes that are associated with the descendants of the anatomical concept under investigation.
This process yielded 3,540 sets (one for each anatomical entity). The cardinality of these sets ranges from 1 to 63,190 (median = 7). For example, the ANAT set for Structure of peritonsillar tissue (1015003) contains three disorders: Peritonsillar abscess (15033003), Peritonsillar cellulites (102453009), and Peritonsillar cyst (300931007).
Establishing the set of descendants of all disorders associated with a given anatomical entity
We showed in the first subsection how we extracted a set of disorders associated with a given anatomical entity. We then created the union of their descendants. in SNOMED CT (again by traversing recursively the inverse_isa links). The resulting set of disorders, referred to as TAXO, represents the descendants of all SNOMED CT disorders associated with the anatomical entity under investigation.
This process yielded 3,540 sets. The cardinality of these sets ranges from 1 to 63,189 (median = 4). For example, the TAXO set of Structure of peritonsillar tissue (1015003) corresponds to three disorders: Peritonsillar abscess (15033003), Peritonsillar cellulitis (102453009), and Peritonsillar cyst (300931007).
Comparing the two sets of disorders
In order to compare the set of disorders associated with the descendants of a given anatomical entity (ANAT) to the set of descendants of all disorders associated with this anatomical entity (TAXO), we simply computed the intersection of the two sets.
RESULTS
Quantitative results
3,540 pairs of sets of descendants were obtained using the ANAT and TAXO methods respectively. Of these, 2,309 (65%) were identical in both methods. For example, Structure of peritonsillar tissue has the same three diseases in both ANAT and TAXO sets of descendants.
In 1,231 cases (35%), differences were found between the set of descendants obtained by the two methods. Among the cases with differences, the average percentage of descendants common to ANAT and TAXO sets was 48%. For 607 anatomical entities (17% of all the anatomical entities, and 49% of the cases with differences) the overlap between ANAT and TAXO sets was less than 50%. For 250 anatomical entities (7% of all the anatomical entities, 20% of the cases with differences) it was less than 10%.
When a difference is found between the two sets for a given anatomical entity, the TAXO set is always a subset of the ANAT set. In other words, the ANAT approach identifies descendants that are not identified by the TAXO approach. Conversely, the TAXO approach does not retrieve any descendant that would not have been identified by the ANAT approach. The number of disorders extracted specifically by the ANAT method ranges from 1 (e.g., for Fifth metatarsal structure (301000)) to 43,832 (for Body part structure (38866009)). For instance, as illustrated in Table 1, the concept Gastric fundus structure (414003) is associated with nine disorders in TAXO and with twelve disorders in ANAT. The latter group includes the nine disorders present in TAXO.
Table 1.
Disorders corresponding to Gastric fundus structure according to ANAT and TAXO (* indicates the top-level disorder)
| Concept Name | Concept ID | Method |
|---|---|---|
| Neoplasm of fundus of stomach* | 126826009 | ANAT, TAXO |
| Benign neoplasm of fundus of stomach | 92116006 | ANAT, TAXO |
| Carcinoma in situ of fundus of stomach | 92598002 | ANAT, TAXO |
| Carcinoma of fundus of stomach | 254555008 | ANAT, TAXO |
| Fundic gland polyposis of stomach | 235686008 | ANAT, TAXO |
| Malignant tumor of fundus of stomach | 187741001 | ANAT, TAXO |
| Neoplasm of uncertain behavior of fundus of stomach | 94849000 | ANAT, TAXO |
| Primary malignant neo- plasm of fundus of sto- mach | 93809003 | ANAT, TAXO |
| Secondary malignant neoplasm of fundus of stomach | 94311007 | ANAT, TAXO |
| Atrophic fundic gland gastritis | 42740008 | ANAT |
| Hypertrophic glandular gastritis | 80018001 | ANAT |
| Solitary fundic gland polyp | 399468008 | ANAT |
Starting from the initial 3,540 anatomical entities, a total of 1,025,904 subsumption relations among disorders were generated by the ANAT approach. Among these, 613,021 relations were also identified by the TAXO approach and 412,021 relations (40%) were specific to ANAT.
Qualitative results
For many anatomical entities linked to disorders, there exists one high-level entity for all disorders located in this anatomical entity. For example, Disorder of the adrenal cortex (129636003) corresponds to the anatomical entity Adrenal cortex structure (68594002). It was expected that a concept Disorder of X would be found in SNOMED CT for each anatomical entity X. In fact, many of the 3,540 anatomical entities investigated in this study are related to no such high-level class of disorders. This finding seems to explain the differences observed between sets of disorders ANAT and TAXO. Consider for example the anatomical entity Fifth metatarsal structure (301000). Six disorders are associated with it in SNOMED CT, including Closed fracture of fifth metatarsal bone (70204006). The ontology of anatomy indicates that structure of base of fifth metatarsal is a fifth metatarsal structure. Therefore, the ANAT set corresponding to Fifth metatarsal structure includes Fracture of base of fifth metatarsal. In contrast, the TAXO set does not. In fact, because there is no such class as Disorder of the fifth metatarsal in SNOMED CT (corresponding to the anatomical structure Fifth metatarsal structure), Fracture of base of fifth metatarsal is a direct descendant of Metatarsal bone fracture. Therefore, it is not a descendant of any of the six diseases associated with Fifth metatarsal structure and thus not a member of the TAXO set (Fig. 3).
Fig. 3.
Representation of disorders associated to fifth metatarsal structure
In the sets of descendants obtained by the ANAT approach, we were expecting to find a hierarchical organization resulting in one top-level disorder corresponding to the anatomical entity at the origin of the set. For example, the 85 descendants of the ANAT set obtained for Adrenal cortex structure (68594002) were all expected to be descendants of the concept Disorder of the adrenal cortex (129636003). In practice, the number of top-level disorders in the sets ranges from 1 to 113 (median = 2). More precisely, we found that only 1,698 sets (48%) exhibited the single top-level disorder property. In the remaining cases, there were multiple top-level disorders associated with the anatomical entity. The presence of multiple top-level disorders in a set suggests that no single class of disorders encompassing these top-level disorders has been represented. For example, Structure of anterior naris (1797002) is associated with seven disorders in ANAT (Atresia of the anterior nares (204511005), Congenital malposition of nares (93337001), Congenital stenosis of nares (2828008), Congenital stenosis of the anterior nares (204513008), Folliculitis nares perforans (54865008), Foreign body in nostril (33890007), Single naris (95266003)). The presence of a unique top-level disorder subsuming these three disorders was expected (e.g., Disorder of anterior naris). Instead, these seven disorders are organized according to three top-level disorders:
Congenital malposition of nares (93337001)
Congenital stenosis of nares (2828008)
Folliculitis nares perforans (54865008)
DISCUSSION
Ontological features of SNOMED CT
By exploiting not only the explicit subsumption relations between disorders, but also the finding site role in disorder definition, the present study has verified that, in SNOMED CT, for any A, all “diseases of A” are members of the set {disorders whose finding site is A or a descendant of A}. In other terms, the taxonomy of disorders is consistent with the classification of disorders associated with the anatomical entities ontology. The formal properties of SNOMED CT certainly contribute to its consistency. Moreover, we demonstrated that the ontology of anatomy included in SNOMED CT supports the identification of additional relations among disorder entities and therefore contributes to enrich the taxonomy of disorders that is explicitly represented in SNOMED CT. What has been rather unexpected is the proportion of new relations in the ANAT set compared with the intial taxonomy of disorders. Starting from the initial 3,540 anatomical entities, a total of 1,025,904 subsumption relations were generated by the ANAT approach. Among these, 412,021 (40%) were specific to ANAT. These results confirm the benefit of providing complete formal definitions of disorders, linked to a reference ontology of anatomy.
Concepts in taxonomies
Beyond anatomy, the general principles used for taxonomy design are questionable. Rosch argued that categories within taxonomies were structured in such a way that there is generally one level of abstraction at which the most basic category cuts can be made [5]. A basic level of abstraction can be formalized in terms of cue validity or in terms of the set theoretic representation of similarity provided by Tversky [6]. A category with high cue validity (e.g. chair) is more differentiated from other categories than one of lower cue validity (e.g. furniture, or kitchen chair). Category resemblance corresponds to the weighted sum of the measures of all the common features within a category minus the sum of the measures of all of the distinctive features. Their hypothesis is that basic categories (e.g. chair) in taxonomies maximize both cue validity and category resemblance.
In constrast to general taxonomies studied by Rosch and Tversky, SNOMED CT represents a specialized scientific domain. Our findings in SNOMED CT suggest that, as in general taxonomies of concrete objects, disorder categories in TAXO have higher cue validity and better category resemblance than classes inferred from ANAT. For example, Disorder of bone (76069003) exhibits high cue validity and category resemblance (e.g., it is clearly differentiated from Disorder of kidney), but Disorder of fifth metatarsal structure has low cue validity and category resemblance (e.g. it would share a lot of features with Disorder of fourth metatarsal structure). The first one is present in SNOMED CT while Disorder of fifth metatarsal structure is not. It must be noted that the latter is found neither in ICD 10 nor in the UMLS. Similarly, while both right kidney and left kidney may be represented in an ontology of anatomy, there is generally no need for distinguishing between Disorder of right kidney and Disorder of left kidney in a Disease ontology. While right kidney exists in SNOMED CT, Disease of right kidney does not exist in the UMLS or in SNOMED CT.
Generalization
This study has focused on SNOMED CT and classification of diseases with respect to anatomy. SNOMED CT represents the partonomic hierarchy of body parts by a taxonomy of reified part-of relations, i.e. X-struture is the reification of part-of X. Applied to another ontology, our approach would take into account subsumption relations and partitive relations between anatomical concepts. This approach can be applied to other large biomedical ontologies and to other kinds of partitions of the biomedical domain. For example, it may be used to check consistency between hierarchical relations and other relations in the UMLS Metathesaurus. Knowing that a disease D is related to a body part B, it is possible to infer that D may be related by a hierarchical relation to the concept corresponding to disease of B. Furthermore, several kinds of partitions of the biomedical domain can be created (e.g. [7]). A partition of a domain consists in a view on reality with a specific type of focus. For example the classification of disorders with respect to anatomy corresponds to a locative partition. Beyond anatomy, other reference ontologies may be used to organize partitions of a domain. For example an ontology of chemical entities may be used to classify molecular functions with respect to the chemicals involved. Given a reference ontology and a set of rules that connects it to an ontology of more complex entities, the reference ontology can help identify new relations between the more complex entities. These new relations are not limited to subsumption relations. For example, we have used ChEBI, an ontology of chemicals, to identify associative relations within the Gene Ontology [8].
CONCLUSION
The discrepancies observed in SNOMED CT between the hierarchy of diseases and the classification of diseases with respect to anatomy can be attributed to missing classes: a class of diseases is not systematically defined for each anatomical structure. While we are not necessarily suggesting that such classes be defined in SNOMED CT, we argue that the approach presented in this study can be used for quality assurance purposes, for example, by focusing the attention of SNOMED CT editors on these cases, which could help identify missing classes.
Acknowledgments
F. Mougin received a grant from the Région Bretagne (PRIR).
Footnotes
References
- 1.Hahn U, Schulz S, Romacker M. Part-whole reasoning: a case-study in medical ontology engineering. IEEE Intelligent Systems & their applications. 1999;14(5):59–67. [Google Scholar]
- 2.Rector AL. Analysis of propagation along transitive roles: Formalisation of the GALEN experience with Medical Ontologies, 2002 International Workshop on Description Logics DL2002, Tou-louse, France, April 19-21, 2002
- 3.Ceusters W, Smith B, Flanagan J. Ontology and medical terminology: why description logics are not enough. TEPR 2003, Conf. Towards an Electronic Patient Record, San Antonio, 10-14 May 2003
- 4.Schulz S, Hahn U, Romacker M. Modeling anatomical spatial relations with description logics. Proc AMIA Symp. 2000:779–83. [PMC free article] [PubMed] [Google Scholar]
- 5.Rosch E. Principles of catagorization, in Cognition and Categorization, Rosch E and B.B. Lloyd edts, L. Erlbaum, Hillsdale, NJ, 1978, pp 28-46
- 6.Tversky A, Gati I. Studies of similarity, in Cognition and Categorization, Rosch E and B.B. Lloyd edts, L. Erlbaum, Hillsdale, NJ, 1978, pp 81-95
- 7.Smith B, Williams J, Schulze-Kremer S. The ontology of the gene ontology. AMIA Annu Symp Proc. 2003:609–13. [PMC free article] [PubMed] [Google Scholar]
- 8.Burgun A, Bodenreider O. An ontology of chemical entities helps identify dependence relations among Gene Ontology terms. SMBM 2005 workshop, Cambridge, 11-13 Apr 2005.