Abstract
The DNA damage-dependent poly(ADP-ribose) polymerases, PARP-1 and PARP-2, homo- and heterodimerize and are both involved in the base excision repair (BER) pathway. Here, we report that mice carrying a targeted disruption of the PARP-2 gene are sensitive to ionizing radiation. Following alkylating agent treatment, parp-2–/–-derived mouse embryonic fibroblasts exhibit increased post-replicative genomic instability, G2/M accumulation and chromosome mis-segregation accompanying kinetochore defects. Moreover, parp-1–/–parp-2–/– double mutant mice are not viable and die at the onset of gastrulation, demonstrating that the expression of both PARP-1 and PARP-2 and/or DNA-dependent poly(ADP-ribosyl) ation is essential during early embryogenesis. Interestingly, specific female embryonic lethality is observed in parp-1+/–parp-2–/– mutants at E9.5. Meta phase analyses of E8.5 embryonic fibroblasts highlight a specific instability of the X chromosome in those females, but not in males. Together, these results support the notion that PARP-1 and PARP-2 possess both overlapping and non-redundant functions in the maintenance of genomic stability.
Keywords: DNA damage/G2/M arrest/mouse development/NAD metabolism/X-chromosome instability
Introduction
DNA damage induced by environmental genotoxic stress or arising as an outcome of cellular metabolism is counteracted by an evolved and intricate network of DNA repair pathways. Central to the pathways that maintain genomic integrity is the modification of histones and nuclear proteins by polymers of ADP-ribose catalyzed by DNA-dependent poly(ADP-ribose) polymerases (PARPs). PARPs constitute a large family of 18 proteins encoded by 18 different genes; they all share the ‘PARP signature’ that contains most of the active site amino acids (J.-C.Amé and G.de Murcia, in preparation). So far, PARP-1 and PARP-2 are the sole members whose catalytic activity is stimulated in vitro and in vivo by DNA strand breaks, suggesting that they are both involved in the cellular response to DNA damage (Amé et al., 1999, 2001; Schreiber et al., 2002).The involvement of PARP-1 and PARP-2 in the base excision repair (BER) pathway has been established in mouse embryonic fibroblasts (MEFs) lacking either PARP-1 (Trucco et al., 1998; Beneke et al., 2000; Dantzer et al., 2000; Masutani et al., 2000) or PARP-2 (Schreiber et al., 2002) which display a severe defect in alkylation-induced DNA strand break repair. PARP-1 knockout mice revealed the dual facets of PARP-1’s physiological role. With low or moderate DNA damage, PARP-1 functions as a survival factor involved in DNA damage detection and signaling (Ménissier de Murcia et al., 1997; Wang et al., 1997; Masutani et al., 2000) in concert with many other DNA damage response checkpoint proteins. On the other hand, in post-mitotic cells, massive DNA damage, as observed in pathophysiological conditions such as cerebellar or cardiac ischemia or septic shock, overactivates PARP-1 that, in turn, causes energy depletion and cell death (for a review, see Szabo and Dawson, 1998).
To analyze the physiological role of PARP-2, we generated parp-2–/– mice by disrupting exon 9 of the mouse gene by homologous recombination. As in the case of parp-1–/– mice, parp-2-deficient mice are sensitive to ionizing radiation. At the cellular level, parp-2 deficiency leads to chromosomal breaks, preferentially in centromeric regions, after 2 Gy irradiation. Exposure to a sublethal dose of N-methyl-N-nitrosourea (MNU) considerably increases the level of sister chromatid exchanges (SCEs) in parp-2–/– primary MEFs and induces a G2/M cell cycle block mirrored by a high level of mitotic chromosomal aberrations.
To gain further insight into the functional importance of DNA-dependent poly(ADP-ribosyl)ation reactions at the animal level, we crossed parp-2+/– mice with parp-1+/– mice. We show that the double null mutant parp-1–/– parp-2–/– embryos die early in development at the onset of gastrulation. Interestingly, a specific female embryonic lethality was observed in the parp-1+/–parp-2–/– background, associated with specific X-chromosome instability. Together, our results provide genetic evidence for a functional interaction between PARP-1 and PARP-2 possessing both overlapping and non-redundant functions in the maintenance of genomic stability. In addition, these findings also reveal the essential role of both PARP-1 and PARP-2 and/or DNA-dependent poly(ADP-ribosyl)ation during embryonic development.
Results
Targeted disruption of the parp-2 gene and generation of parp-2–/– mice
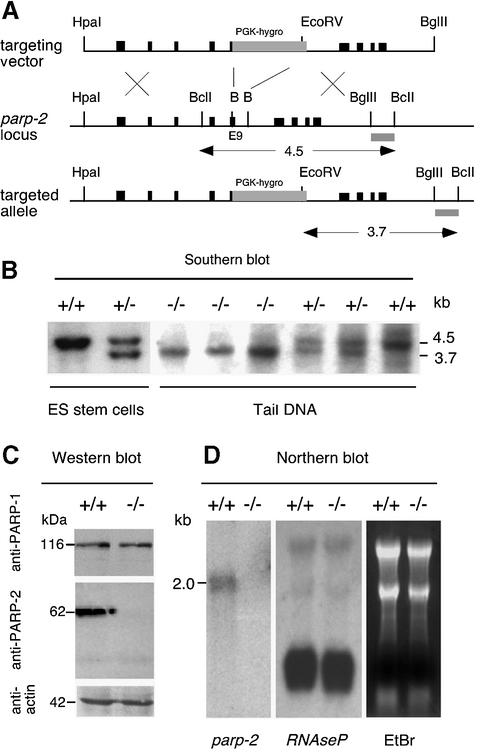
We have shown previously that the parp-2 gene lies head to head with the gene encoding the H1 RNA component of RNase P (Amé et al., 2001; Myslinski et al., 2001). RNase P is a ribonucleoprotein involved in the maturation of the 5′ end of tRNAs; it is an essential ubiquitous enzyme present in all cells and cellular compartments that synthesize tRNA during active proliferation (Xiao et al., 2002). To avoid any influence on the expression of H1 RNA, we inactivated exon 9 at the position corresponding to residue I285 of the parp-2 gene by homologous recombination in embryonic stem (ES) cells from the 129Sv/Pas mouse line by inserting a PGK-hygromycin cassette (Figure 1A). Gene targeting was confirmed by Southern blotting using a 3′ probe (Figure 1B). Two heterozygote ES clones were used to generate chimeric and mutant mice on C57BL/6;129sv/Pas mixed background using standard procedures. The phenotypes of these mice were examined concurrently to rule out genetic background effects. No phenotypic difference has been seen between mice generated from these two ES cell clones. Southern blot analysis of DNA from tail biopsies confirmed the disruption of both parp-2 alleles in homozygous mutants (Figure 1B). Mutant mice were completely devoid of PARP-2 as judged by western blot analysis of cells isolated from testis, using polyclonal antibodies raised against the catalytic domain (Figure 1C) or the N-terminal domain of mouse PARP-2 (not shown). No compensation by PARP-1 upregulation could be observed (Figure 1C). Northern blot analysis showed that the disruption of exon 9 abrogates the expression of the parp-2 gene (Figure 1D) but did not influence the expression of RNase P RNA whose function was preserved in parp-2–/– mice. Mice lacking PARP-2 display no visible abnormal phenotype by 18 months of life and are not tumor prone.
Fig. 1. Targeted disruption of parp-2. (A) Map of the parp-2 murine genomic locus and targeting vector. Exons are indicated by black boxes and the position of the external probe is represented by a gray box. The PGK-hygromycin cassette is inserted in the opposite transcriptional orientation and replaces a BamHI (B) fragment of exon 9 and intron 10. (B) Representative genomic blot probed with the external probe. A wild-type 4.5 kb and a recombined 3.7 kb fragment are shown. Mice were identified as wild-type (+/+), heterozygous (+/–) or homozygous mutant (–/–). (C) PARP-2 is not expressed in cells isolated from the testis of parp-2–/– mice. (D) Northern blot analysis of total RNA from testis using a 820 bp EcoRI fragment of parp-2 cDNA as a probe. Note that the expression of the RNase P RNA is not influenced by the parp-2 disruption.
Radiosensitivity of parp-2–/– mice and cells
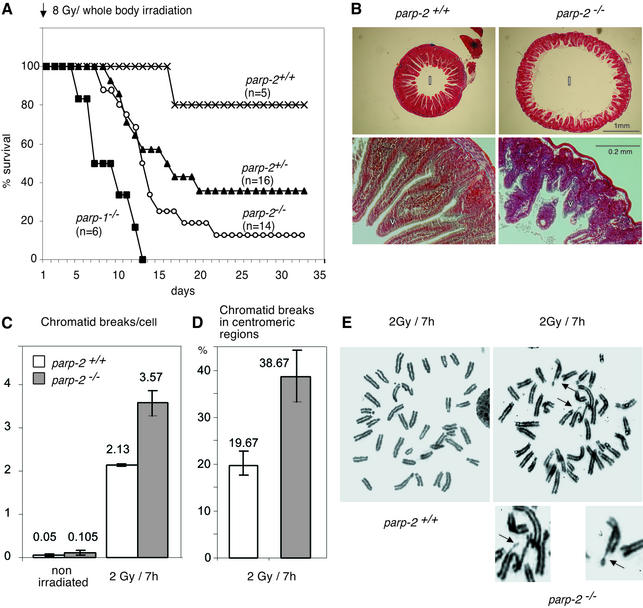
The DNA damage dependence of PARP-2 activity (Amé et al., 1999; Schreiber et al., 2002) suggests its implication in genome surveillance and protection. We determined the sensitivity of parp-2–/– mice to 8 Gy whole-body irradiation. Fourteen parp-2–/–, 16 parp-2+/–, six parp-1–/– and five parp-2+/+ mice 6–8 weeks of age were irradiated and monitored for mortality (Figure 2A). As previously reported, all parp-1–/– mice were dead by 13 days post-irradiation (Ménissier de Murcia et al., 1997). By 32 days post-irradiation, two parp-2–/– (12.5%) mice, five parp-2+/– (35.7%) mice and four wild-type mice (80%) were alive. The gastrointestinal tracts of mutant and wild-type mice were examined 6 days post-irradiation. The duodenum of irradiated parp-2–/– mice appeared dilated, and villi (v in Figure 2B) were more severely shortened than in parp-2+/+ irradiated mice, displaying epithelial crypt degeneration. Therefore, parp-2–/– mice most probably die from acute radiation toxicity to the epithelium of the small intestine.
Fig. 2. parp-2–/– mice are sensitive to ionizing radiation. (A) Kaplan–Meier survival curve after 8 Gy of irradiation. Wilcoxon test: P (parp-2+/+ versus parp-2–/–) < 10–5; P (parp-2–/– versus parp-2+/–) < 0.002. (B) Stained transverse sections of the duodenum from a parp-2+/+ and a parp-2–/– mouse 6 days after a dose of 8 Gy was given. Lumen (l), villi (v). (C) Mean number of chromatid breaks in bone marrow cells taken from mock-irradiated mice or 7 h after 2 Gy whole-body irradiation. (D) Occurrence of chromatid breaks in the centromeric region 7 h after 2 Gy irradiation. (E) Metaphase spreads of bone marrow cells from irradiated parp-2+/+ and parp-2–/– mice. Chromatid break in the centromeric region (top arrow) and tetraradian in metaphase (bottom arrow) from an irradiated parp-2–/– mouse. Magnifications are details pointed to by arrows.
Following 2 Gy irradiation, bone marrow cells taken from irradiated animals showed an increase of chromatid breaks in the parp-2–/– background, whereas this increase was much lower in parp-2+/+ cells (Figure 2C), suggesting a DNA repair deficiency of radiation-induced damage, above all S phase and during G2. Interestingly, chromatid breaks occurred more frequently in centromeric regions in parp-2–/– cells than in parp-2+/+ cells (Figure 2D and E), thus providing evidence for a possible role for PARP-2 in the maintenance of centromeric sequence integrity (Saxena et al., 2002a,b).
G2/M accumulation and sensitivity of parp-2–/– cells to the monofunctional alkylating agent MNU
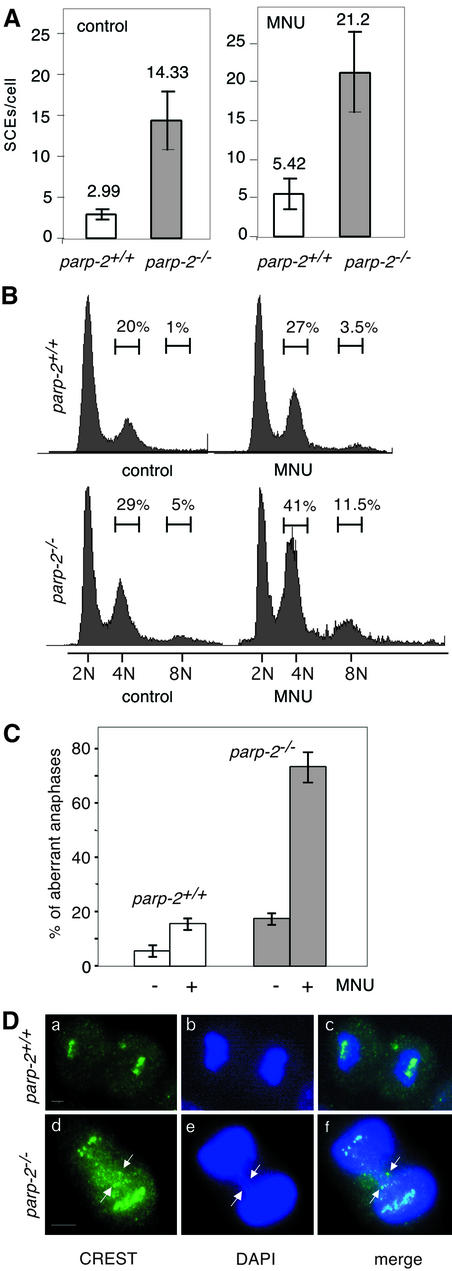
Given the involvement of PARP-2 in the cellular response to DNA strand breaks, we speculated that the lack of PARP-2 might cause chromosomal rearrangements and loss of genomic stability. SCEs were analyzed in bone marrow cells of wild-type and mutant mice, 9 h following MNU (50 mg/kg) treatment. As shown in Figure 3A, the level of SCEs was ∼4-fold higher in parp-2–/– than in parp-2+/+ mice, indicating that DNA repair through homologous recombination between sister chromatids (Sonoda et al., 1999) was strongly stimulated, due to the persistence of DNA damage-induced breaks when PARP-2 is absent (Schreiber et al., 2002).
Fig. 3. (A) Sister chromatid exchanges in bone marrow cells from parp-2+/+ and parp-2–/– mice 9 h after i.p injection with (50 mg/kg) MNU. (B) Cell cycle profile analyzed after propidium iodide staining of parp-2+/+ and parp-2–/– MEFs at passage 2, 24 h following 2 mM MNU treatment. The percentage of cells containing (2N) (4N) and (8N) is indicated. (C) Quantitative analysis of chromosome mis-segregation in parp-2+/+ and parp-2–/– MEFs at passage 2, 24 h following 2 mM MNU treatment. The fraction of mitotic cells that exhibited a defect in segregation, such as lagging chromosomes (see D, panels d–f), is shown. (D) Anaphases in parp-2+/+ and parp-2–/– MEFs following 2 mM MNU treatment. Kinetochores (green) are immunostained with the CREST serum (a and d). Lagging chromosomes forming an anaphase bridge indicated by arrows in parp-2–/– cells. Bars indicate 10 µm.
5-Bromodeoxyuridine (BrdU) incorporation assays revealed no defect in cellular proliferation in parp-2–/– MEFs in the absence of DNA damage (data not shown). We next investigated whether functional PARP-2 is required for genotoxic stress-induced cell cycle checkpoint control. Asynchronously dividing parp-2+/+ and parp-2–/– primary MEFs were exposed to a sublethal dose of the monofunctional alkylating agent MNU (2 mM). In a typical experiment, in the absence of DNA damage, parp-2–/– MEFs undergo a minor G2/M arrest (Figure 3B). At 24 h after MNU treatment, parp-2–/– MEFs exhibited a prominent G2/M arrest (41%) compared with parp-2+/+ MEFs (27%), indicating that parp-2-deficient cells failed to resume their progression in the cell cycle after DNA base damage. This prolonged G2/M accumulation was accompanied by the acquisition of 8N DNA content (11%) (where N is the amount of DNA in a haploid cell) as mutant cells proceed through the next cell cycle. The same batch of control and damaged cells was processed for indirect immunofluorescence to analyze the possible causes of G2/M accumulation. As shown in Figure 3C and D, a high proportion of aberrant anaphases were detected specifically in parp-2–/– MEFs following MNU treatment. A significant fraction of anaphase cells contained one or more lagging (non-segregated) chromosomes and dispersed centromeres in between the two parp-2–/– daughter cell DNA (Figure 3D, panels d–f). The G2/M delay observed in damaged parp-2–/– MEFs together with the elevated rate of chromosome mis-segregation is suggestive of centromere and kinetochore defects (Nasmyth, 2002).
DNA damage-induced apoptosis in parp-2–/– mice
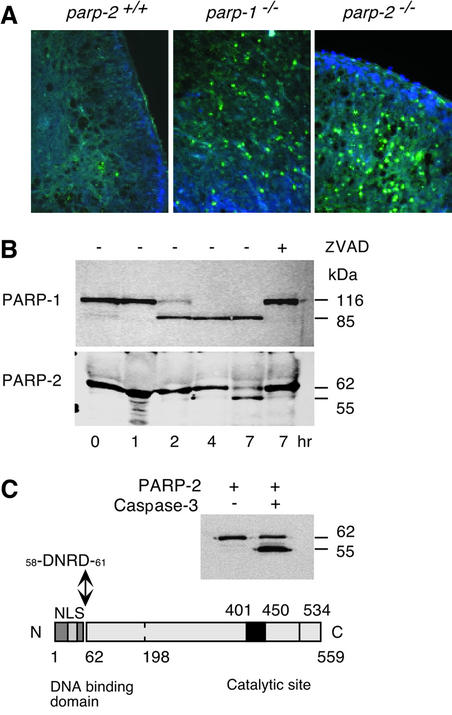
We have shown previously that parp-1–/– cells underwent much more rapid apoptosis than wild-type when exposed to DNA-damaging agents (Ménissier de Murcia et al., 1997). The apoptotic response of thymocytes was therefore studied by TUNEL assay, 48 h following 6 Gy whole-body irradiation in parp-2+/+, parp-1–/– and parp-2–/– mice. As shown in Figure 4A, parp-2+/+ thymus displayed circumscribed foci of apoptotic cells characteristic of a normal response to irradiation. In contrast, parp-2–/– thymus exhibited a high degree of DNA fragmentation (Figure 4A) similar in intensity to that of parp-1–/–, suggesting that both PARP-2 and PARP-1 play a positive role allowing the cell not to undergo apoptosis when it is able to repair the damaged DNA. Thus, we speculated that PARP-2 might be cleaved during the executive phase of apoptosis. To address this issue, we used the model system of HL60 cells treated with VP16, a topoisomerase II inhibitor, to visualize a possible PARP-2 cleavage, concurrently with that of PARP-1. As shown in Figure 4B, PARP-1 cleavage was almost achieved 2 h after treatment, as previously published (Germain et al., 1999), whereas PARP-2 cleavage occurred 5 h later, resulting in a 55 kDa polypeptide. Pre-incubation of cells with the potent caspase inhibitor, benzyloxycarbonyl-VAD-fluoromethylketone, completely prevented both PARP-2 and PARP-1 cleavage. Given the presence of several consensus sequences, we speculated that caspase-3 might be responsible for PARP-2 cleavage in HL60 cells. To identify the site, an in vitro cleavage assay was performed using purified caspase-3 and recombinant mouse PARP-2. As shown in Figure 4C, PARP-2 cleavage in vitro released a fragment of similar size to that obtained in VP16-induced HL60 apoptosis. Microsequencing of the 55 kDa polypeptide N-terminus gave the amino acid sequence 58DNRD61, a caspase-3 consensus motif, strictly conserved between human and mouse PARP-2 and located at the junction between the 62 amino acids forming the DNA-binding domain and the catalytic domain (Amé et al., 1999; Schreiber et al., 2002) (Figure 4C). Therefore, as in the case of PARP-1, the DNA-dependent activation of PARP-2 is abrogated following caspase cleavage that, in both cases, physically separates the DNA-binding function from the C-terminal catalytic function.
Fig. 4. (A) Fluorescent TUNEL stain of thymus 2 days after 6 Gy irradiation. Apoptotic cells are labeled in green, and cell nuclei are stained with DAPI. parp +/+ are sparsely stained whereas parp-1–/– and parp-2–/– show a strong green staining of apoptotic cells. (B) PARP-1 and PARP-2 cleavage kinetics after 100 µM VP16 treatment in HL-60 cells. Z-VAD-fmk inhibitor was used at 0.5 µg/µl and was added 1 h before VP16 treatment. For western blot, 105 cells were harvested at the indicated time, mixed with loading buffer and sonicated. Polyclonal antibodies anti-PARP-1 and anti-PARP-2 were used. (C) In vitro cleavage of purified mPARP-2 (2.5 µg) by active caspase-3 (8 U, Chemicon International) shows the same cleavage pattern in HL-60 cells after VP-16 treatment. The N-terminus of the 55 kDa band was micro sequenced and was found to be 58DNRD61, a consensus cleavage site for caspases 3/7.
parp-1–/–parp-2–/– double mutant embryos are not viable
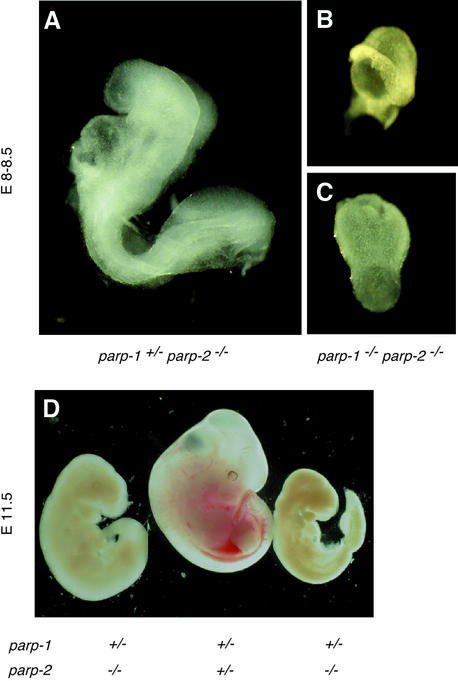
To elucidate potential functional interactions between PARP-1 and PARP-2, we bred parp-1–/– mice with parp-2+/– mice (both in a mixed C57BL/6;129Sv background) to generate parp-1+/–parp-2+/– mice. Double heterozygous parp-1+/–parp-2+/– mice were interbred to generate all possible combinations of parp-1 and parp-2 targeted alleles. Of 221 progeny analyzed, no double homozygous mice (14 expected) and only 10 parp-1+/–parp-2–/– mice (28 expected) (see below) were found, whereas mice of the other genotypes were recovered within experimental variability at Mendelian frequencies. parp-1–/–parp-2+/– mice were intercrossed further to generate doubly null mutant mice, but genotype analysis at term (n = 82) failed to identify any animal of this genotype (20 were expected) while 68% of parp-1–/– parp-2+/– and 32% of parp-1–/–parp-2+/+ were obtained. The stage at which the double null embryos arrested in development was then investigated by isolation of embryos from time-mated parp-1–/–parp-2+/– heterozygous crosses. At E8.0–8.5, six out of 21 embryos were doubly homozygous. These embryos were severely growth retarded (Figure 5B and C) and less developed than wild-type embryos of the same age (Figure 5A). By E10.5, three out of 17 double null embryos were largely resorbed. These studies indicate that the development of the parp-1–/–parp-2–/– embryos was arrested before E8.0, a stage where cellular proliferation increases dramatically (Snow, 1977). Taken together, these results demonstrate that parp-1 and parp-2 gene products are essential during early embryogenesis.
Fig. 5. External views of E8.0–8.5 (A–C) and E11.5 (D) embryos. (A) A normal parp-1+/–parp-2–/– embryo. (B and C) Retarded parp-1–/– parp-2–/– embryos. (D) Example of two parp-1+/–parp-2–/– female embryos that are phenotypically arrested at E9.5, and one normal parp-1+/–parp-2+/– female embryo at E11.5. The same magnifications were used in (A), (B) and (C).
Female lethality in parp-1+/–parp-2–/– embryos
As stated above, live parp-1+/–parp-2–/– mice were generated at a less than expected frequency. Because these mice are poorly fertile (3.5 ± 0.6 offspring per litter compared with 7.4 ± 0.4 offspring per litter in wild-type), we crossed parp-1+/–parp-2–/– or parp-1–/–parp-2+/– males with parp-1+/–parp-2+/– females to enrich for embryos with null alleles. Again, no double null mutant was found. When the sex of the 44 parp-1+/–parp-2–/– offspring was examined, only 10 females were found, which corresponds to a significant reduction (P < 0.01). No sex ratio distortion was found in our parp-1–/– and parp-2–/– colonies. To determine when parp-1+/–parp-2–/– female mutants died, parp-1+/+parp-2–/– mice were crossed with parp-1–/– parp-2+/– mice to obtain a maximum number of parp-1+/–parp-2–/– embryos without any double null mutant. At E8.5 and E11.5, embryos were assessed for developmental defects and their genotype and sex were determined by PCR (Table I). A total of 28 embryos and one resorption were recovered from four females at day 8.5 of gestation. Of these, eight females and six males were found to be parp-1+/–parp-2–/–. None of these embryos showed morphological abnormalities. At day 11.5 of gestation, 31 embryos and three resorption sites were recovered from four females. Of the parp-1+/–parp-2–/– embryos, five out of seven females were retarded and only one (probably representing a spontaneous resorption event) out of nine males was retarded. Based on their gross morphology, the retarded parp-1+/–parp-2–/– females had undergone developmental arrest at around 9.5 days of gestation (Figure 5D). As X-chromosome loss is clearly associated with aging (Surralles et al., 1999), the viable parp-1+/–parp-2–/– females were carefully followed-up, but no symptom of premature aging was detected except that they all display a severe hypofertility (not shown).
Table I. Sex and phenotype of timed post-implantation parp-1+/– parp-2–/– embryos.
| Days of development | Resorption sites | No. of typed embryos | Female parp-1+/–parp-2–/– | Male parp-1+/–parp-2–/– | ||
|---|---|---|---|---|---|---|
| |
|
|
Normal |
Retarded |
Normal |
Retarded |
| 8.5 | 1 | 28 | 8 | 0 | 6 | 0 |
| 11.5 | 3 | 31 | 2 | 5 | 8 | 1 |
| Borna | 10 | 34 | ||||
aRefers to 44 live-born animals after weaning.
parp-1+/–parp-2–/– female embryonic lethality is associated with X-chromosome instability
We sought to determine why this lethality is female specific. Given that equal X-linked gene dosage between the sexes is achieved by random X-chromosome inactivation in the female embryo around gastrulation (for a review, see Avner and Heard, 2001), we reasoned that female-specific embryonic lethality could be due to a non-inactivation or an unstable X chromosome. We took advantage of the fact that inactive X chromosome contained in the Barr body is enriched in histone macro-H2A1.2 (Costanzi and Pehrson, 2001) to examine, by double immunofluorescence, the inactivation status of cells from three different E8.5 parp-1+/–parp-2–/– female embryos. One spot of macro-H2A1.2 was found in almost all the cells (data not shown), indicating that inactivation of the X chromosome was already achieved in the parp-1+/–parp-2–/– females and cannot account for female-specific embryonic lethality.
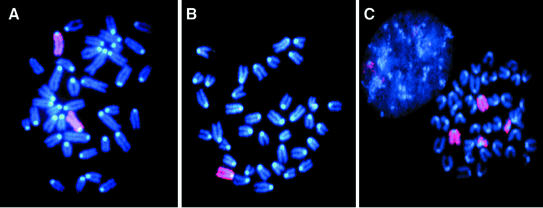
In mammalian females, X chromosomes are the most frequently lost and the most unstable (Tucker et al., 1996; Surralles et al., 1999; Catalan et al., 2000) possibly as a consequence of their late replication in S phase and/or a faulty kinetochore or centromere which could result in lagging of the whole duplicated chromosome during mitosis. We looked for the stability of the X chromosome in embryonic cells derived from E8.5 parp-1+/–parp-2–/– embryos (eight females and two males) and parp-1+/+ parp-2–/– and parp-1–/–parp-2+/+ females. Chromosome spreads revealed a dramatic instability of the X chromosome in parp-1+/–parp-2–/– females only (Figure 6; Table II). Around 40% of the metaphases displayed aneuploidy (1X or 3X) consistent with a severe defect in X-chromosome segregation, and 14% harbored a derivative of one X chromosome fused to an autosomal chromosome (Table II). No Robertsonian fusion and only one dicentric X chromosome were observed, suggesting that no shortening of the telomere length occurred in these cells. As an internal control, we examined the stability of chromosome 14, an autosomal chromosome of comparable length. We found two metaphases harboring only one chromosome 14 among 64 metaphases (not shown). Thus, at E8.5, the defect in the parp-1+/–parp-2–/– female embryonic cells is associated with an increased instability specific to the X chromosome rather than with an overall chromosomal instability.
Fig. 6. parp-1+/–parp-2–/– females display X-chromosome instability. X chromosomes are stained in pink (rhodamine), and chromosomes are counterstained in blue (DAPI). Examples of metaphases with various numbers of X chromosomes in parp-1+/–parp-2–/– females: (A) 2X; (B) 1X; (C) 2X + derX.
Table II. X chromosome instability in females (F) or males (M) of different genotypes.
| Sex | Genotype | No. of cells | (nX) | (nX) + X | (nX) – X | (nX) + derX |
|---|---|---|---|---|---|---|
| Female | parp-1+/+parp-2–/– | 30 | 29 | 0 | 1 | 0 |
| Female | parp-1–/–parp-2+/+ | 21 | 21 | 0 | 0 | 0 |
| Male | parp-1+/–parp-2–/– | 35 | 35 | 0 | 0 | 0 |
| Female | parp-1+/–parp-2–/– | 132 | 66 | 19 | 33 | 14 |
(nX) = metaphases with normal number of X chromosomes: XX, for female, X for male; (nX) + X = metaphases with one X in surplus; (nX) – X = metaphases with one X missing; (nX) + derX = metaphases with any kind of X aneuploidy and a derivative of X chromosome.
Discussion
The disruption of the parp-1 gene allowed the discovery of a residual enzymatic activity (Shieh et al., 1998) catalyzed by a novel DNA-dependent PARP now named PARP-2 (Amé et al., 1999). By disrupting exon 9 of the parp-2 gene in mice, we have created a mouse model deficient in this enzyme, responsible for only 10–15% of the total PARP activity fully stimulated by DNA strand breaks in a cellular extract (Amé et al., 1999; Schreiber et al., 2002). Interestingly, the parp-2 mutant mouse displays some similar phenotypes to that which we and others have described previously for the parp-1–/– mouse (Ménissier de Murcia et al., 1997; Wang et al., 1997; Masutani et al., 2000), in spite of the large difference existing between their respective specific enzymatic activities.
Several defects observed in parp-2 mutant mice and cells strongly support a role for PARP-2 in the cellular response to DNA strand break damage induced by ionizing radiation or monofunctional alkylating agents. The fact that PARP-1 and PARP-2 interact with each other and share common partners, such as XRCC1, DNA polymerase β and DNA ligase III (Schreiber et al., 2002) directly involved in the BER pathway, reinforces the proposal that both are required for genome protection and explains why the deficiency in either PARP-1 or PARP-2 slows down the BER process. At present, we do not know whether cells lacking both PARP-1 and PARP-2 suffer a total deficiency in BER. However, our results demonstrate that the expression of both PARP-1 and PARP-2 and/or DNA-dependent poly(ADP-ribosyl)ation is necessary during early post-implantation development. During this restricted developmental window, undifferentiated stem cells that form the epiblast sustain a high cell division rate (Snow, 1977) and become hypersensitive to DNA damage (Heyer et al., 2000). To ensure survival, cellular responses to DNA damage are required; DNA repair defects could therefore explain the demise of mutated embryos at this stage. Interestingly, lethality occurring shortly after gastrulation was observed in mutant embryos lacking other BER factors, such as mutants lacking XRCC1 (Tebbs et al., 1999) and APE (Xanthoudakis et al., 1996; Ludwig et al., 1998), indicating that no redundancy of these functions exists in the cell. In line with this observation, our results suggest that no other redundant function in vivo overcomes the defect due to both parp-1parp-2 gene disruption.
As a survival factor, PARP-2 is cleaved by caspase-3 at the 58DNRD61 site after saturating DNA damage to avoid unnecessary DNA repair and facilitate nuclear disintegration during apoptosis. Similarly, during middle cerebral artery occlusion, PARP-2 appears to be the preferred nuclear target of caspase-8 (Benchoua et al., 2002); the cleavage at the 183LQMD186 site inactivates PARP-2 in neurons, pointing to the necessity to downregulate its activity for the correct execution of the apoptotic program. It remains to be tested whether the absence of PARP-2 may also lead to substantial protection in inflammatory models of reperfusion injury, as in the case of parp-1 deficiency (for a review, see Virag and Szabo, 2002).
Interestingly, parp-1 haplo-insufficiency in a parp-2–/– background causes female-specific embryonic lethality associated with X-chromosome instability. It has been published recently that a fraction of PARP-1 (Earle et al., 2000; Saxena et al., 2002a) and PARP-2 (Saxena et al., 2002b) molecules are associated with active centromeres and interact with CENP-A, CENP-B and the spindle checkpoint protein BUB3 during mitosis. Indeed, in parp-2-deficient cells at least, kinetochore defects are observed (Figure 3), but there is no obvious explanation for the fact that aneuploidy preferentially affects the X chromosomes in females. A possible explanation of this phenotype might be that half parp-1 dosage in the absence of PARP-2 at centromeres of mutant embryos induces a high frequency of kinetochore defects and aneuploidy during X-chromosome segregation, thus increasing its lagging character. An alternative explanation of our results could be related to the late replication of X chromosomes (Migeon, 1994). We showed that parp-1 and parp-2 deficiency separately lead to DNA damage-induced chromosome instability and G2/M accumulation (Ménissier de Murcia et al., 1997; this study) as a consequence of a BER defect. We propose that the lack of coordination of the mitotic exit combined with a further burst of cellular proliferation occurring during embryo ‘turning’ at E8.5–9.0 could increase X-chromosome instability. Aneuploidy or fragmented X chromosomes at E8.5 (Figure 6) may be the first event that then leads to a global mitotic catastrophe and developmental arrest at around E 9.5 after a number of cell divisions.
In conclusion, we describe here two parp-1 dosage-linked phenotypes in a parp-2 knockout background probably resulting from two different levels of the quality control of DNA integrity, leading either to total lethality or to sex-specific lethality. Interestingly, a similar female-specific embryonic lethality was described previously for mice doubly null for Msh2 and p53 (Cranston et al., 1997; Cranston and Fishel, 1999). Furthermore, human female-specific lethality has been reported associated with a common variant in BRCA2 (Healey et al., 2000). Thus, the sex-dependent fetal death might be a common feature of mutations in caretaker genes. In that case, even if a critical level of the DNA repair capacity is reached, avoiding a complete early lethality, the poor DNA repair efficiency may lead to a severe genomic instability, detrimental first to the females.
Materials and methods
Gene targeting in ES cells and generation of mice
The mouse parp-2 gene was isolated from a 129sv strain genomic library. The parp-2 targeting vector was constructed using a 5.2 kb HpaI–BglII fragment extending from intron 4 to intron 14 by inserting a PGK-hygro cassette in BamHI sites located in exon 9 and intron 9. Following electroporation, ES cells were selected in 200 µg/ml hygromycin. Genomic DNA was restricted by EcoRV and BclI and separated on a 0.8% agarose gel. The targeted alleles were detected by Southern analysis using a 3′ EcoRI–BglII fragment. A single targeted allele was observed in eight of 75 isolated ES cell colonies by Southern analysis. Two heterozygote clones microinjected into C57BL/6 blastocysts gave rise to chimeric offspring, which in turn were mated with C57BL/6. Pups were genotyped by Southern blot analysis of tail DNA for germline transmission. PCR genotyping was performed as a routine assay using a common primer located in intron 8 (5′-GGGACTCTGGTTTGG TGCCT-3′) in combination with primers discriminating wild-type alleles (5′-TGCTGCCGTCCCTTATTCTAAGCT-3′) in intron 9 and knockout alleles encoding the 3′ end of the hygromycin resistance gene (5′-TAA GGGCCAGCTCATTCCTCCCA-3′). Total RNA from parp-2+/+ and parp-2–/– testis was isolated, subjected to northern blot analysis and probed with the gene coding for the RNase P RNA subunit. Protein crude extracts were prepared from testis by gentle laceration, separated by 10% SDS–PAGE and probed with a polyclonal antibody raised against the mouse PARP-2 (Yuc). A parp-2+/– male was mated with two parp-1–/– females (Ménissier de Murcia et al., 1997), and the double heterozygous outcomes were interbred in order to generate parp-1–/–parp-2–/– double null mice. Sex determination of embryos was performed as described by Sah et al. (1995).
Survival curves
parp-2+/+, parp-2+/–, parp-2–/– and parp-1–/– mice were γ-irradiated at 8 Gy (0.5 Gy/min) with a cobalt 60Co source. The percentage of deceased mice from each genotype was compared with that of parp-1–/– mice by Kaplan–Meier analysis, and the log-rank test P-values were calculated. Duodenums were collected in phosphate-buffered saline, then fixed in Bouin’s fluid for 14 h, dehydrated and embedded in paraffin. Serial sections were cut at 6 µm and stained with hematoxylin and eosin.
Cell culture and cell cycle analysis
MEFs were isolated by standard procedures. Individual embryos were grown in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% fetal calf serum (Eurobio), 0.05 mg/ml gentamicin at 37°C with 5% CO2. Cell cycle analysis was performed as previously described (Ménissier de Murcia et al., 1997).
Chromosomal stability measurements
BrdU tablets were implanted subcutaneously in mice 32 h (two cell cycles of bone marrow cells) and colchicine (0.6 mg/kg) was injected 2 h before sacrifice (Allen et al., 1977). SCEs and chromatid breaks measurements were performed as previously described (Ménissier de Murcia et al., 1997). Chromosome paintings were performed according to the manufacturer (Adgenix, Oncor).
TUNEL assay and indirect immunofluorescence
Six-week-old mice were γ-irradiated with 6 Gy. Two days later, thymuses were removed, fixed in Bouin’s fluid for 14 h, dehydrated and embedded in paraffin. Tissue sections were (paraffined) assayed with the ApoAlert DNA Fragmentation Kit (Clontech, Palo-Alto, CA) for apoptosis. Indirect immunofluorescence was performed as described (Amé et al., 1999) using CREST antiserum for immunostaining of kinetochores.
Acknowledgments
Acknowledgements
We thank Dr J.Pehrson for the gift of macro H2A1.2 antibody, Dr M.Mark for comments and advice on histological sections, Dr G.Prévot for all the irradiation experiments, A.Staub for microsequencing, and Dr J.Surrallés for critical reading of the manuscript. This work was supported by funds from the Centre National de la Recherche Scientifique, INSERM, the Association pour la Recherche Contre le Cancer, Electricité de France, the Ligue Nationale contre le Cancer, the Commissariat à l’Energie Atomique and BASF-AG. The work in the L.S. laboratory was supported by contract number FIGH-CT-1999-00009 from the CEC.
References
- Allen J.W., Shuler,C.F., Mendes,R.W. and Latt,S.A. (1977) A simplified technique for in vivo analysis of sister-chromatid exchanges using 5-bromodeoxyuridine tablets. Cytogenet. Cell Genet., 18, 231–237. [DOI] [PubMed] [Google Scholar]
- Amé J.C. et al. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- Amé J.C., Schreiber,V., Fraulob,V., Dollé,P., de Murcia,G. and Niedergang,C.P. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J. Biol. Chem., 276, 11092–11099. [DOI] [PubMed] [Google Scholar]
- Avner P. and Heard,E. (2001) X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet., 2, 59–67. [DOI] [PubMed] [Google Scholar]
- Benchoua A., Couriaud,C., Guegan,C., Tartier,L., Couvert,P., Friocourt,G., Chelly,J., Menissier-de Murcia,J. and Onteniente,B. (2002) Active caspase-8 translocates into the nucleus of apoptotic cells to inactivate poly(ADP-ribose) polymerase-2. J. Biol. Chem., 277, 34217–34222. [DOI] [PubMed] [Google Scholar]
- Beneke R., Geisen,C., Zevnik,B., Bauch,T., Muller,W.U., Kupper,J.H. and Moroy,T. (2000) DNA excision repair and DNA damage-induced apoptosis are linked to poly(ADP-ribosyl)ation but have different requirements for p53. Mol. Cell. Biol., 20, 6695–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan J., Falck,G.C. and Norppa,H. (2000) The X chromosome frequently lags behind in female lymphocyte anaphase. Am. J. Hum. Genet., 66, 687–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C. and Pehrson,J.R. (2001) MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem., 276, 21776–21784. [DOI] [PubMed] [Google Scholar]
- Cranston A. and Fishel,R. (1999) Female embryonic lethality in Msh2-Trp53 nullizygous mice is strain dependent. Mamm. Genome, 10, 1020–1022. [DOI] [PubMed] [Google Scholar]
- Cranston A., Bocker,T., Reitmair,A., Palazzo,J., Wilson,T., Mak,T. and Fishel,R. (1997) Female embryonic lethality in mice nullizygous for both Msh2 and p53. Nat. Genet., 17, 114–118. [DOI] [PubMed] [Google Scholar]
- Dantzer F., de La Rubia,G., Ménissier-De Murcia,J., Hostomsky,Z., de Murcia,G. and Schreiber,V. (2000) Base excision repair is impaired in mammalian cells lacking poly(ADP- ribose) polymerase-1. Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- Earle E. et al. (2000) Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet., 9, 187–194. [DOI] [PubMed] [Google Scholar]
- Germain M., Affar,E.B., D’Amours,D., Dixit,V.M., Salvesen,G.S. and Poirier,G.G. (1999) Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem., 274, 28379–28384. [DOI] [PubMed] [Google Scholar]
- Healey C.S. et al. (2000) A common variant in BRCA2 is associated with both breast cancer risk and prenatal viability. Nat. Genet., 26, 362–364. [DOI] [PubMed] [Google Scholar]
- Heyer B.S., MacAuley,A., Behrendtsen,O. and Werb,Z. (2000) Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev., 14, 2072–2084. [PMC free article] [PubMed] [Google Scholar]
- Ludwig D.L., MacInnes,M.A., Takiguchi,Y., Purtymun,P.E., Henrie,M., Flannery,M., Meneses,J., Pedersen,R.A. and Chen,D.J. (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res., 409, 17–29. [DOI] [PubMed] [Google Scholar]
- Masutani M., Nozaki,T., Nakamoto,K., Nakagama,H., Suzuki,H., Kusuoka,O., Tsutsumi,M. and Sugimura,T. (2000) The response of Parp knockout mice against DNA damaging agents. Mutat. Res., 462, 159–166. [DOI] [PubMed] [Google Scholar]
- Ménissier de Murcia J. et al. (1997) Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA, 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B.R. (1994) X-chromosome inactivation: molecular mechanisms and genetic consequences. Trends Genet., 10, 230–235. [DOI] [PubMed] [Google Scholar]
- Myslinski E., Amé,J.C., Krol,A. and Carbon,P. (2001) An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res., 29, 2502–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science, 297, 559–565. [DOI] [PubMed] [Google Scholar]
- Sah V.P., Attardi,L.D., Mulligan,G.J., Williams,B.O., Bronson,R.T. and Jacks,T. (1995) A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet., 10, 175–180. [DOI] [PubMed] [Google Scholar]
- Saxena A., Saffery,R., Wong,L.H., Kalitsis,P. and Choo,K.H. (2002a) Centromere proteins Cenpa, Cenpb and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem., 277, 26921–26926. [DOI] [PubMed] [Google Scholar]
- Saxena A., Wong,L.H., Kalitsis,P., Earle,E., Shaffer,L.G. and Choo,K.H. (2002b) Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum. Mol. Genet., 11, 2319–2329. [DOI] [PubMed] [Google Scholar]
- Schreiber V., Amé,J.C., Dollé,P., Schultz,I., Rinaldi,B., Fraulob,V., Ménissier-de Murcia,J. and de Murcia,G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- Shieh W.M., Ame,J.C., Wilson,M.V., Wang,Z.Q., Koh,D.W., Jacobson,M.K. and Jacobson,E.L. (1998) Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem., 273, 30069–30072. [DOI] [PubMed] [Google Scholar]
- Snow M.H.L. (1977) Gastrulation in the mouse: growth and regionalisation of the epiblast. J. Embryol. Exp. Morphol., 42, 293–303. [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surralles J., Hande,M.P., Marcos,R. and Lansdorp,P.M. (1999) Accelerated telomere shortening in the human inactive X chromosome. Am. J. Hum. Genet., 65, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C. and Dawson,V.L. (1998) Role of poly(ADP-ribose) synthetase in inflammation and ischaemia–reperfusion. Trends Pharmacol. Sci., 19, 287–298. [DOI] [PubMed] [Google Scholar]
- Tebbs R.S., Flannery,M.L., Meneses,J.J., Hartmann,A., Tucker,J.D., Thompson,L.H., Cleaver,J.E. and Pedersen,R.A. (1999) Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol., 208, 513–529. [DOI] [PubMed] [Google Scholar]
- Trucco C., Oliver,F.J., de Murcia,G. and Ménissier-de Murcia,J. (1998) DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res., 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J.D., Nath,J. and Hando,J.C. (1996) Activation status of the X chromosome in human micronucleated lymphocytes. Hum. Genet., 97, 471–475. [DOI] [PubMed] [Google Scholar]
- Virag L. and Szabo,C. (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev., 54, 375–429. [DOI] [PubMed] [Google Scholar]
- Wang Z.Q., Stingl,L., Morrison,C., Jantsch,M., Los,M., Schulze-Osthoff,K. and Wagner,E.F. (1997) PARP is important for genomic stability but dispensable in apoptosis. Genes Dev., 11, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S., Smeyne,R.J., Wallace,J.D. and Curran,T. (1996) The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA, 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Scott,F., Fierke,C.A. and Engelke,D.R. (2002) Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem., 71, 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]